SUMMARY
The 3′ ends of most eukaryotic mRNAs are produced by an endonucleolytic cleavage followed by synthesis of a poly(A) tail. Poly(A) polymerase (PAP), the enzyme that catalyzes the formation of the tail, is subject to tight regulation involving several post-translational modifications. Here we show that the enzyme poly(ADP-ribose) polymerase1 (PARP1) modifies PAP, and regulates its activity both in vitro and in vivo. PARP1 binds to and modifies PAP by poly(ADP-ribosyl)ation (PARylation) in vitro, which inhibits PAP activity. In vivo we show that PAP is PARylated during heat shock, leading to inhibition of polyadenylation in a PARP1-dependent manner. The observed inhibition reflects reduced RNA binding affinity of PARylated PAP in vitro, and decreased PAP association with non-heat shock protein-encoding genes in vivo. Our results both provide direct evidence that PARylation can control processing of mRNA precursors and also identify PARP1 as a regulator of polyadenylation during thermal stress.
INTRODUCTION
Eukaryotic mRNA precursors undergo several processing events before the mature mRNA is transported out of the nucleus and translated into protein. Transcription, capping, splicing and polyadenylation are all complex reactions that require numerous protein factors, and which we now know are all interconnected (reviewed in Hirose and Manley, 2000; Moore and Proudfoot, 2009). Consistent with this, polyadenylation contributes to many aspects of mRNA metabolism, including transcription termination by RNA polymerase II (RNAP II), mRNA stability, mRNA export to the cytoplasm and the efficiency of translation (reviewed in Millevoi and Vagner, 2010; Richard and Manley, 2009). The importance of polyadenylation is emphasized by the growing appreciation of its role in gene control and by the association of a number of human diseases with aberrant polyadenylation (reviewed in Danckwardt et al., 2008; Di Giammartino et al., 2011).
Polyadenylation consists of two reactions: an endonucleolytic cleavage followed by synthesis of the poly(A) tail onto the 5′ cleaved product. While these two reactions are catalyzed by the action of only two enzymes (CPSF73 and PAP, respectively), they are supported by a large number of protein factors, reflecting the necessity of finely regulating the process and coordinating it with other nuclear events. A proteomic analysis revealed the complexity of the molecular apparatus responsible for the generation of mature mRNA 3′ ends, identifying ~80 proteins that are associated with the 3′ processing complex (Shi et al., 2009). These comprise several new core 3′ processing factors as well as other proteins that may mediate crosstalk between pre-mRNA maturation and other cellular events.
Among the factors detected by Shi et al. (2009) that had not been previously identified in the mammalian 3′ processing complex was PARP1. PARP1 is an abundant nuclear enzyme that has been implicated in the DNA damage detection and repair pathway and in regulation of gene expression, especially through chromatin modification and transcription regulation (Ji and Tulin, 2010; Krishnakumar and Kraus, 2010; Rouleau et al., 2010). PARP1 is responsible for initiation, elongation and branching of ADP-ribose units from donor NAD+ molecules onto target proteins, leading to the post-translational modification known as PARylation. Although PARP1 is the major target of its own activity, through an automodification reaction, a number of other covalently PARylated proteins have been described including histones, chromatin remodeling proteins and transcription factors. PARylation influences the activity of target proteins by modulating their protein-nucleic acid interactions, enzymatic activity, protein-protein interactions and/or subcellular localization.
PARP1 is known to be activated by a variety of stresses (reviewed by Luo and Kraus, 2012). These include exposure to reactive oxygen, alkylating agent, ionizing radiation and heat shock. In Drosophila, PARP1 has been shown to be potently activated upon heat shock and is crucial for the formation of heat shock-induced puffs on heat shock protein (hsp) genes (Tulin and Spradling, 2003). In humans, PARP1 is involved in regulation of the highly inducible hsp70 gene (Ouararhni et al., 2006). Many of the proteins implicated in 3′ processing are subject to post-translational modifications (reviewed in Ryan and Bauer, 2008). PAP, in particular, is subject to phosphorylation, which has been shown to inhibit PAP activity in vitro and during M phase (Colgan et al., 1996), acetylation, which disrupts its nuclear localization and association with the 3′ processing complex (Shimazu et al., 2007), and sumoylation, which is important for PAP’s nuclear localization and stability, and can also downregulate its activity (Vethantham et al., 2008).
Here we present evidence that PAP is a direct, physiologically significant PARP1 target. We show that PARP1 can bind and PARylate PAP in vitro, and that this inhibits PAP polyadenylation activity. Furthermore, we show that PAP is PARylated by PARP1 in vivo in response to heat shock, and that this is responsible for an observed inhibition of polyadenylation that occurs during heat shock. Finally, our data indicates that the mechanism of inhibition relies on decreased RNA binding by PARylated PAP. In vivo this is reflected by dissociation of PAP from transcribed genes upon heat shock, although interestingly, hsp genes are resistant to this inhibition. Our data thus provide evidence that PARP1-mediated PARylation functions directly in control of pre-mRNA processing, and also define PARylation as a regulator of PAP and mRNA 3′ formation during heat shock.
RESULTS
Activation of PARP1 inhibits polyadenylation in vitro
Our discovery that PARP1 can associate with the polyadenylation machinery raised the question of whether PARP1 might be able to regulate 3′ processing. We first wished to determine if activation of PARP1 affects 3′ processing in vitro. To this end, we added increasing amounts of NAD+ to HeLa nuclear extracts (NE) to activate endogenous PARP1 (Ogata et al., 1981). Extracts were then incubated with a 32P-labeled polyadenylation substrate (SV40 late RNA, SVL) under conditions that allow either cleavage only or coupled cleavage and polyadenylation. Strikingly, NAD+ potently inhibited polyadenylation, both the extent and length of the poly(A) tail, but not cleavage of the substrate (Figure 1A, left panel). Quantitation (right panel) indicates that addition of increasing concentrations of NAD+ resulted in a dose-dependent decrease in polyadenylation such that 0.5 mM NAD+ inhibited polyadenylation by ~40%. To assess whether the effect of NAD+ was specific and indeed dependent on activation of PARP1, we repeated the coupled cleavage/polyadenylation assay as above adding NAD+ either alone, which again resulted in polyadenylation inhibition, or in the presence of increasing amounts (0.1–0.5 uM) of the PARP inhibitor XI, which led to restoration of polyadenylation activity (Figure 1B). Another PARP inhibitor (3-ABA) gave the same results (Figure S1). Moreover, to confirm that the NAD+ effect was dependent on the presence of PARP1, NEs were prepared from HeLa cells treated for 72 hrs with siRNA against PARP1 or a non-targeting siRNA (Figure 1C). Figure 1D shows that upon PARP1 knockdown, NAD+ had almost no effect on polyadenylation, confirming that PARP1 was responsible for the observed inhibition. (The slight inhibitory effect of NAD+ after knockdown of PARP1 can be attributed to the presence of low levels of PARP1 remaining following siRNA treatment; see Figure 1C). Polyadenylation in the absence of NAD+ was not affected by PARP1 knockdown (compare lines 1 and 4 in Figure 1D), indicating that PARP1 does not have a constitutive role in 3′ processing.
Figure 1. Activation of PARP1 by NAD+ inhibits polyadenylation in vitro and induces PARylation of PAP in NE.
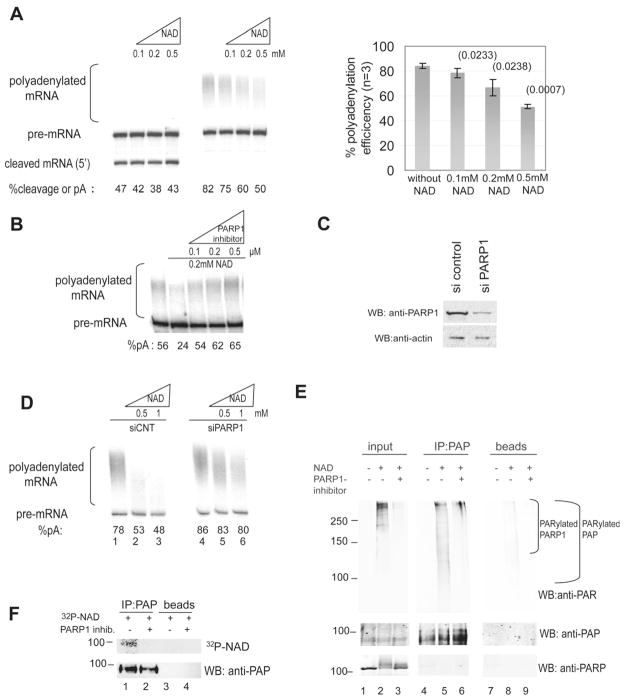
(A) 3′ cleavage and polyadenylation assays were carried out using internally 32P labeled RNA substrate and HeLa NE in the presence of the indicated concentrations of NAD+. RNAs were purified, resolved by denaturing PAGE and visualized by autoradiography. Positions of precursor and products are indicated. Percentage of cleavage/polyadenylation efficiency is indicated at the bottom and was calculated by dividing pA signal by total signal (which equals pA plus pre-mRNA). The graph (right) represents the mean percentage of 3 experiments, error bars represent standard deviation and p-values are indicated in parenthesis (B) Polyadenylation assay in the presence of the indicated amounts of NAD+ and PARP1 inhibitor XI. Polyadenylation efficiency is indicated at the bottom (C) HeLa cells were transfected with a non-targeting siRNA or siRNA against PARP1. Western blots (WB) of NE were carried out with the indicated antibodies. (D) NEs were made from the cells transfected with a non-targeting siRNA (lanes 1 to 3) or siRNA against PARP1 (lanes 4 to 6) and subsequently used in polyadenylation assays with increasing amounts of NAD+. Polyadenylation efficiency is indicated at the bottom (E) PAP was IPed from NE, either without incubation with NAD+ (lane 4), with incubation with 0.5 mM NAD+ (lane 5) or after incubation with both NAD and 0.5 uM of PARP inhibitor XI (lane 6). Western blots were carried out using the indicated antibodies. Lanes 1–3, input samples corresponding to lanes 4–6. Lanes 7–9, samples incubated with sepharose A beads without antibody. MWs (in KDa) are indicated on the left. (F) PAP was IPed from NE with (lanes 2 and 4) or without (lanes 1 and 3) PARP inhibitor XI, in the presence of 0.4 uM 32P-NAD+. The membrane was exposed to a phosphorscreen (upper panel) followed by western blot with an anti-PAP antibody (lower panel).
Since the inhibitory effect of activated PARP1 was evident only on the second step of 3′ processing, we reasoned that PAP might be a PARP1 substrate. To test this, we immunoprecipitated (IPed) PAP from NE or NE that had been incubated with 0.5 mM NAD+ for 30 mins at 30°C. Western blot of the NEs with an anti-PAR antibody detected a high molecular weight smear, reflecting PARylated proteins (mostly auto-PARylation) in NE that contained NAD+ (Figure 1E, lanes 1–3 in upper panel), indicating that PARP1 was successfully activated by this treatment. Because of the heterogeneity in the length of the ADP-ribose chain, PARylation is typically detected as a smear starting extending upwards from the modified protein (e.g., Hossain et al., 2009). Consistent with this, and suggesting that PAP was indeed PARylated in NE containing NAD+, a smear extending upward from the position of PAP was detected in the PAP IP from NAD+-containing NE blotted with the anti-PAR antibody (Figure 1E, lanes 4–6, upper panel). This smear did not come from PARylated PARP1 that might have co-IPed with PAP, as PARP1 was not detected by western blotting with an anti-PARP1 antibody (lanes 4–6, lower panel). Moreover, addition of 0.5 uM PARP inhibitor XI abolished PAP modification (compare lanes 5 and 6 in Figure 1E, upper panel). These results indicate that under conditions in which polyadenylation was impaired by activation of PARP1 with NAD+, PAP was indeed efficiently PARylated.
To provide additional evidence that PAP was PARylated in NE, we used 32P-NAD+ in the assay to enhance sensitivity and allow better visualization of modified PAP. Specifically, we used a low concentration of total NAD+ (0.4 μM) to limit extension of PAR chains and therefore allow detection of the target protein as a discrete band rather than a smear (e.g. Lonn et al., 2010). Following IP with anti-PAP antibodies, SDS-PAGE and transfer, the membrane was exposed first to a phosphor screen (Figure 1F, upper panel) and then subjected to western blot with anti-PAP (Figure 1F, lower panel). Significantly, a radioactive band was indeed detected at the position of PAP (lane 1, upper panel), which was not observed in the presence of inhibitor XI (lane 2) or when using only proteinA sepharose in the IP (lanes 3 and 4). These results provide strong confirmatory evidence that PAP is indeed PARylated in HeLa NE.
Purified PARP1 PARylates PAP in vitro and inhibits its intrinsic activity
We next wished to examine more directly the effect of PARP1-catalyzed PARylation on PAP activity. We first asked whether PAP can be PARylated using recombinant proteins purified from E. coli. Figure S2A shows a coomassie-stained gel of the two purified proteins, MBP-PARP1 and His-PAP. We employed an in vitro PARylation assay in which His-PAP was incubated with MBP-tagged PARP1 in the presence of MgCl2, NAD+ and sheared salmon sperm DNA. Under these conditions PARP1 uses NAD+ as a substrate to attach ADP-ribose units onto itself and target proteins, generating ADP-ribose chains that can be as long as 200 ADP-ribose units (de Murcia et al., 1983; Gagne et al., 2001). As expected (Ogata et al., 1981), PARP1 was a good acceptor of ADP-ribose units, resulting in auto-modification, as detected by anti-PAR western, when incubated under activating conditions (Figure 2A, compare lane 2 to lane 1). When PAP was included in the reaction, an additional smear starting from the molecular weight of PAP and extending upward, representing PARylated PAP, was detected upon PARP1 activation (Figure 2A, compare lane 4 to lane 3).
Figure 2. PARP1 interacts with and PARylates PAP in vitro.
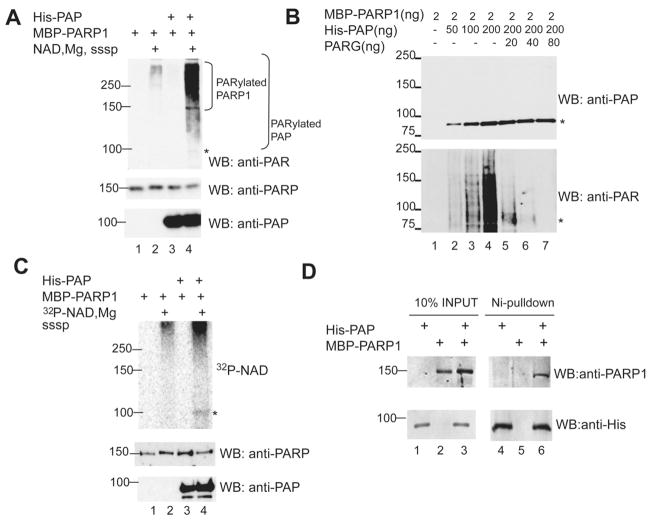
(A) In vitro PARylation reactions were carried out with the indicated purified recombinant proteins. Reaction mixtures in lanes 1 and 2 contained PARP1 alone; those in lanes 3 and 4 also contained PAP. In lanes 2 and 4 PARP1 was activated. Proteins were resolved by SDS-PAGE and western blot were carried out with the indicated antibodies.(B) In vitro PARylation as in (A) using increasing amounts of PAP (lanes 1 to 5) followed by incubation with increasing amounts of PARG (lanes 5 to 7) (C) In vitro PARylation reactions as in (A) with the exception that 4 uM of 32P-NAD+ was used along with 0.05 mM NAD+ (D) Purified recombinant MBP-PARP1 and His-PAP were used in “pull-down” assays using nickel beads. Following SDS-PAGE proteins were detected by western blot with the antibodies indicated. MWs (in KDa) are indicated on the left. In panels A, B and C, an asterisk indicates PAP.
To confirm and extend these findings, we performed two additional experiments. First, we repeated the above assay using increasing amounts of PAP. Importantly, this increased the signal detected by the anti-PAR antibody (Figure 2B lanes 1 to 4), indicating that the PAR detected was from PARylated PAP (since the PARP concentration was kept constant). We then added increasing amounts of purified PARG, which hydrolyzes ADP-ribose units, to reaction mixtures, which caused the signal of PAR incorporation to collapse to the molecular weight (MW) of PAP (Figure 2B, lanes 5 to 7), confirming that the PAR signal came from PARylated PAP. Second, we again used 32P-NAD+ as a source of NAD+, which resulted in detection of a radioactive band at the MW of PAP (Figure 2C upper panel, lane 4). Together, these results confirm that PAP is a PARP1 target in vitro.
We next tested whether PAP and PARP1 stably associate with each other in vitro. To this end, His-PAP was bound to nickel beads and its ability to bind MBP-PARP1 determined by incubating the two proteins at 4°C, followed by washes with high salt buffer (500 mM NaCl). Proteins were resuspended in denaturing loading buffer and resolved by SDS-PAGE. Western blot with anti-PARP1 antibodies revealed that MBP-PARP1 indeed bound to His-PAP on nickel beads (Figure 2D, lane 6) but not to nickel beads alone (Figure 2D, lane 5).
We next wished to determine the effect of PARylation on PAP intrinsic activity. To this end, we re-purified PARylated PAP after in vitro PARylation and tested its activity in a non-specific polyadenylation assay (e.g., Takagaki et al., 1988). In this assay, the presence of Mn2+ renders PAP independent from other 3′ processing factors, and purified PAP in the presence of ATP can therefore polyadenylate by itself essentially any RNA substrate. Instead of purifying total PAP after PARylation, which would include both modified and unmodified protein, we isolated specifically the PARylated fraction of PAP after in vitro PARylation (NAD+ was not added in a control sample). We first used amylose beads to remove MBP-PARP1 (Figure S2B). The supernatant containing PAP was then incubated with an ADP-ribose affinity resin. The resin was washed and modified PAP eluted with free ADP-ribose (for the control, the free ADP-ribose was added directly to the supernatant). Following dialysis, the unmodified control and PARylated PAP proteins were quantified by dot blotting (Figure S2C; see Experimental Procedures), and their concentration equalized. A dot blot also demonstrated that the recovered PAP was indeed PARylated (Figure 3A). Consistent with the results obtained with NAD+-supplemented NE (Figure 1), the non-specific polyadenylation assay revealed that PARylated PAP was essentially inactive (Figure 3B; compare lanes 2 and 3 to lanes 4 and 5). Together, our findings indicate that PARP1 directly PARylates PAP, thereby inhibiting its activity.
Figure 3. Heat shock inhibits polyadenylation in a PAP-dependent manner.
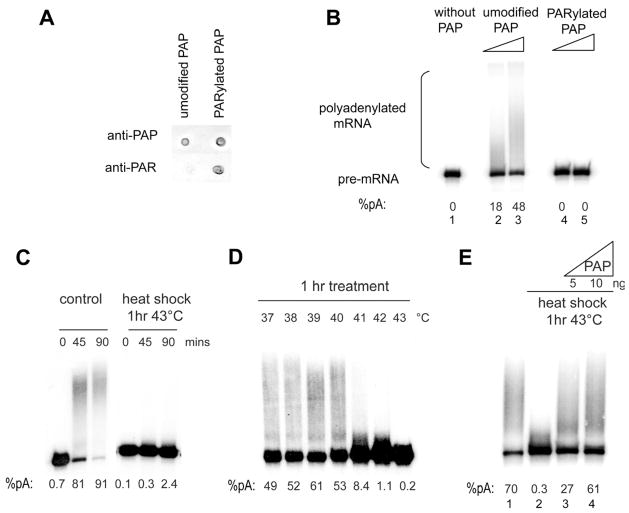
(A) Dot blot of 2 ul of PAP protein that was re-purified after in vitro PARylation or mock PARylation as described in Experimental Procedures. Anti-PAP or anti-PAR antibodies were used to visualize the extent of recovery and PARylation of the re-purified proteins. (B) Non-specific polyadenylation assays with 32P-labeled SVL RNA were performed in the absence of PAP (lane 1) or with increasing amounts (2 and 4 ng) of purified mock-PARylated (lanes 2 and 3) or PARylated PAP (lanes 4 and 5). (C) In vitro polyadenylation was carried out for the indicated times using SVL RNA and HeLa NEs made from untreated cells or cells that were heat shocked for 1 hr at 43 °C. (D) In vitro polyadenylation as in (A) using NEs made from cells that were treated at the indicated temperatures for 1 hr. (E) Polyadenylation assays as in (A) using NE from untreated cells (lane 1), or from cells that were heat shocked 1 hr at 43 °C (lanes 2–4). NEs in lanes 3 and 4 were supplemented with the indicated amounts of recombinant His-PAP In panels B-E, polyadenylation efficiencies are indicated at the bottom.
Heat shock inhibits polyadenylation in a PAP- and PARP1-dependent way
We next investigated whether the link between PARP1 and polyadenylation we characterized in vitro also exists in vivo. Given that PARP1 is activated in vivo by a variety of stimuli, such as DNA damage, oxidative stress and heat shock (Luo and Kraus, 2012), we exposed cells to several conditions known to activate endogenous PARP1, and as an initial approach tested whether any of these treatments affected the 3′ processing activity of NEs prepared from these cells. NEs were prepared from HeLa cells exposed to hydrogen peroxide, γ-IR or heat shock, and used in polyadenylation assays with a pre-cleaved RNA substrate to examine polyadenylation uncoupled from cleavage. PARP1 was indeed activated by all of these conditions (Figure S3A), although it is possible that other PARPs may have contributed to the observed increase in PAR. While hydrogen peroxide or γ-IR treatments had no detectable effect on polyadenylation activity (Figures S3B and C), a drastic inhibition in activity was observed with NEs prepared from cells that had been exposed to 43°C for one hour (Figure 3C). Polyadenylation was strongly inhibited even with a milder heat shock, carried out at 41°C (Figure 3D). If this inhibition reflected specifically repression of PAP, then addition of purified PAP should restore activity. Figure 3E shows that addition of 5 ng of purified His-PAP restored polyadenylation completely, demonstrating that PAP activity was indeed impaired by heat shock.
We next investigated the effect of heat shock on polyadenylation in vivo. For this, we used a previously described HEK 293 cell line that stably expresses a tetracycline-(tet) inducible β-globin transgene integrated in the genome through site-specific recombination (de Almeida et al., 2010). This system was particularly suitable for our purposes because it enabled us to measure the effect of heat shock on 3′ processing in a way that was largely independent of effects that heat shock might have on transcription. Moreover, since transcription is inducible, any effects that a short heat shock might have on polyadenylation would not be masked by polyadenylated transcripts that accumulated before heat treatment. After 3 hrs of induction β-globin mRNA expression was induced ~8 fold (Figure 4A). Cells were then incubated for 30 mins at 37 or 43°C followed by nuclear RNA extraction and reverse transcription either with random hexamer primers (to measure total mRNA) or with an oligo(dT) primer (to measure polyadenylated mRNAs). Figure 4B shows that the ratio of polyadenylated to total β-globin mRNA was indeed reduced after heat shock treatment (as measured by real-time PCR). To determine if the inhibition of polyadenylation was dependent on PARP activity, we treated cells with a cell-permeable PARP1 inhibitor (3-ABA) at the time of β-globin induction and during heat shock, and analyzed transcripts as above. Polyadenylation activity was fully restored by the PARP inhibitor (Figure 4B). Similar results were obtained with another PARP inhibitor, PJ34 (Figure S3D). Together, these results provide strong evidence that heat shock-induced inhibition of polyadenylation requires PARP activity.
Figure 4. Heat shock inhibits polyadenylation in vivo in a PARP-dependent manner.
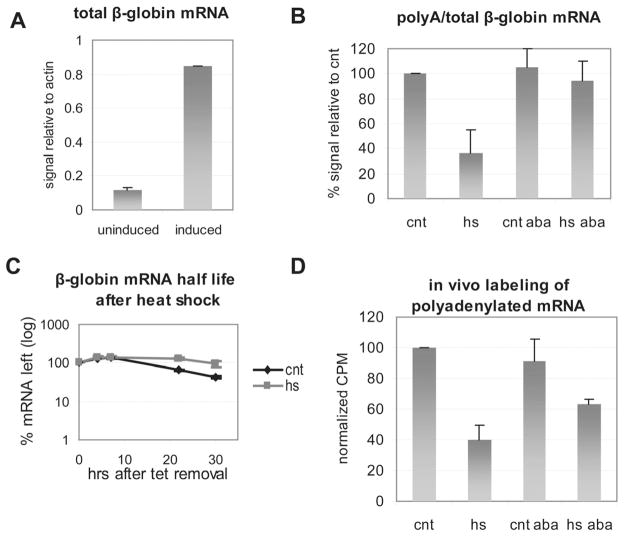
(A) RNA was extracted from 293 cells stably transfected with an inducible β-globin transgene following 3 hr induction with tet or without induction. Following qPCR the amount of induced β-globin mRNA relative to endogenous actin mRNA was calculated before and after induction. (B) Nuclear RNA was extracted from cells induced for 3 hrs as in (A) that were either kept at 37°C (cnt) or heat shocked at 43°C (hs) for 30 mins. Where noted the PARP inhibitor 3-ABA was added at the time of induction. qPCR was used to calculate the relative amount of β-globin polyadenylated mRNA compared total β-globin RNA as described in the Extended Experimental Procedures. (C) The β-globin transgene was induced for 3 hrs as in (A). Cells were either treated 30 mins at 43°C (hs) or left untreated (cnt). To measure β-globin half life RNA samples were extracted at the indicated times following tet removal. (D) Following 30 mins labeling with 3H uridine, nuclear RNA was extracted from HeLa cells that were either kept at 37°C (cnt) or heat shocked at 43°C (hs) for 30 mins. Where noted the PARP inhibitor 3-ABA was added together with 3H uridine. Poly (A)+ fraction was isolated and quantitated by scintillation counting. Count per minute (CPM) relative to cnt are shown. Results from three independent experiments are shown represented as mean and standard error.
To rule out the possibility that the observed decrease in polyadenylated mRNA during heat shock reflects degradation of β-globin mRNA that accumulated prior to heat shock, rather than inhibition of polyadenylation, we measured the β-globin mRNA half life following the heat treatment. For this purpose the β-globin gene was induced for 3 hrs and cells were either incubated at 37 °C or 43 °C for 30 mins (as in Figure 4B), followed by extensive washes to remove tet from the medium and stop transcription. mRNA was extracted at the indicated time points following tet removal and the percentage of remaining mRNA was plotted against time. As shown in Figure 4C, there was no detectable change in half life after heat shock.
We next wished to provide additional evidence that polyadenylation is inhibited during heat shock, and that this inhibition is a general phenomenon, not specific to the β-globin gene reporter used above. To this end, we employed 3H uridine labelling and oligo(dT) selection to measure newly synthesized polyadenylated mRNA during heat shock. HeLa cells were heat shocked for 30 mins (3H uridine added at the beginning of treatment), nuclear RNA was extracted, polyadenylated RNA selected by oligo(dT) and quantitated by scintillation counting. The results (Figure 4D) reveal a 60% decrease in accumulation of nuclear polyadenylated RNA during heat shock. As with the β-globin gene, addition of 3-ABA restored, albeit partially polyadenylation (Figure 4D). The 3-ABA resistant fraction may reflect some inhibition of transcription (see Discussion), but together our results provide strong evidence that polyadenylation is inhibited in a PARP-dependent manner during heat shock.
PARP1 PARylates PAP in vivo during heat shock
We next asked whether PAP is in fact PARylated during heat shock. For this analysis, we used two different cell lines, HeLa (Figure S4A) and MCF-7 (Figure 5A). Cells were subject to heat shock, and PAP IPed from cell extracts. Using an anti-PAR antibody for Western, we detected PARylated PAP in the samples from heat-shocked cells but not from control cells or from cells heat shocked in the presence of 3-ABA (compare lane 3 with 9, and lane 3 with 4 in Figure 5A). The signal did not derive from PARylated PARP1 because PARP1 did not co-IP with PAP under the conditions used (Figure 5A; see also Figure 1E). As the PAP precipitate revealed a faint signal at the PARP1 MW, which could be auto-modified PARP1, we treated the PAP precipitate with PARG (Figure S4B), but still did not detect PARP1 co-IPing with PAP. Also, in agreement with the fact that polyadenylation was not inhibited by hydrogen peroxide-mediated activation of PARP1 (see above), PAP was not PARylated under this condition (Figure S4C).
Figure 5. PARP1 PARylates PAP during heat shock in vivo.
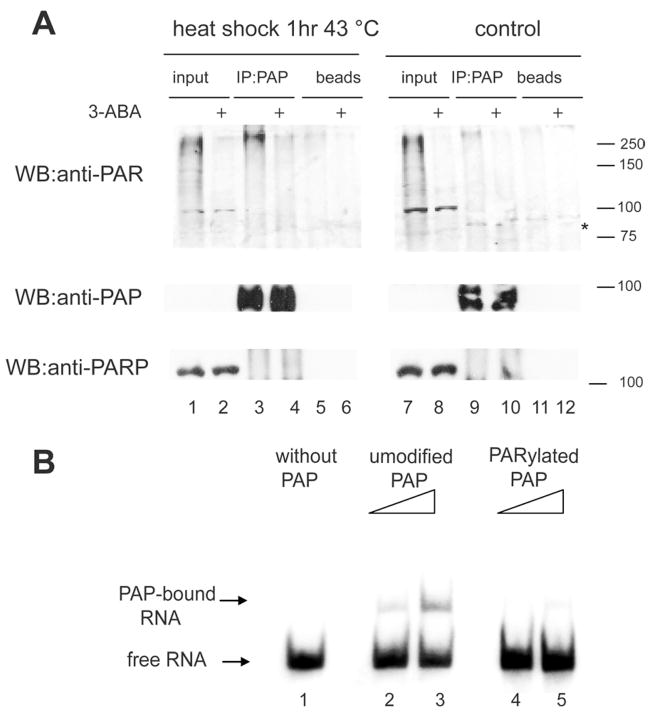
(A) PAP was IPed from untreated MCF-7 cells (lanes 7 to 12) or from cells that were heat shocked for 1 hr at 43°C (lanes 1 to 6). Where indicated, 3-ABA was added just before the heat shock (lanes 4, 6, 10 and 12). Following IP, samples were subjected to SDS-PAGE and western blots (WB) were carried out with the indicated antibodies. The asterisk indicates the position of PAP (B) Gel shift assay without PAP (lane 1) or with increasing amounts of mock-PARylated (lanes 2 and 3) or PARylated (lanes 4 and 5) PAP samples that were re-purified after in vitro PARylation as in Figure 2C and incubated with a 32P-labeled RNA (SVL). Samples were resolved in a 5% non-denaturing polyacrylamide gel. The gel was dried and exposed to a PhosphorImager screen
PAP dissociates from mRNA transcripts during heat shock
We next wished to investigate the mechanism by which PARylation inhibits PAP activity. Given that PARylation can affect protein-protein interactions of modified targets (e.g., Huang et al., 2006; Kim et al., 2004), we first asked whether PARylation of PAP affects assembly of the 3′ processing complex. NEs were prepared from control cells or cells incubated 1 hr at 43°C, incubated with a 32P-labelled RNA under conditions that allow formation of the 3′ processing complex, and loaded on a non-denaturing agarose gel. The results (Figure S5A) indicate that heat shock did not detectably affect the assembly or stability of the 3′ processing complex.
PARylation is also known to affect modified proteins by causing their dissociation from DNA (reviewed by Kraus, 2008). We therefore asked whether PARylation of PAP alters the enzyme’s ability to bind its substrate RNA. To address this, we first used an in vitro assay. We incubated unmodified or PARylated PAP (after re-purification as in Figure 3A) with SVL RNA under conditions used for polyadenylation but omitting ATP and MgCl2/Mn2+ to prevent poly(A) synthesis, and then loaded the samples onto a non-denaturing polyacrylamide gel (Figure 5B). Unmodified PAP bound the RNA in a concentration-dependent manner (lanes 2 and 3), while binding by PARylated PAP was greatly diminished (lanes 4 and 5), indicating that PARylation interferes with PAP binding to the RNA substrate.
We next asked whether PAP substrate binding is compromised by PARylation in vivo. For this analysis, we used the inducible β-globin cell line described above. We examined first whether PAP association with the 3′ end of the activated β-globin gene could be detected and, if so, whether it was reduced following heat shock. Chromatin immunoprecipitation (ChIP) was performed using anti-PAP antibodies and PAP association with the 3′ end of the gene was quantified by real-time PCR (Figure 6A). In the absence of induction, a signal slightly above background was detected, likely reflecting a low level of transcription in the absence of tet (see Figure 4A and Figure 6D). When tet was added for 6 hrs, PAP association with the 3′ end of the β-globin gene significantly increased, providing evidence that PAP was present at the 3′ end of the actively transcribed gene. Strikingly, PAP chromatin association was reduced rapidly, after only 5 mins heat shock at 43°C. This effect was dependent on PARP1, as addition of 3-ABA during the last 4 hrs of tet induction prevented the decrease in PAP crosslinking. ChIP with anti-PAP antibodies before and after heat shock was also performed on the 3′ ends of two endogenous genes, C-MYC and GAPDH, and a similar decrease in PAP crosslinking to these genes was observed (Figure S5B).
Figure 6. PARylated PAP is unable to bind RNA in vitro and dissociates from the 3′ end of non-hsp genes during heat shock in vivo.
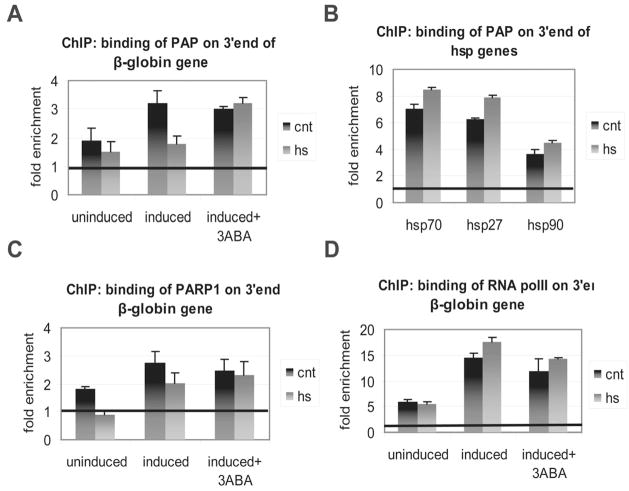
(A) ChIP was carried out using an anti-PAP antibody and amplifying a 3′ end region of the β-globin gene from the inducible 293 cell line. Cells were either uninduced or induced with tet for 6 hrs, with or without 3-ABA for the last 4 hrs of induction, as indicated. Cells were heat shocked for 5 mins at 43°C. Results were analyzed as described in the Extended Experimental Procedures and quantified as fold change over background. (B) ChIP in uninduced 293 cells using anti-PAP antibody as above. Cells were either untreated or heat shocked for 5 mins at 43°C. Primers specific for the 3′ end of the indicated hsp genes were used for qPCR. The results were analyzed as in Figure 6A. (C) ChIP using anti-PARP1 antibody was carried out and analyzed as in (A). (D) ChIP using antibody against RNA polymerase II was carried out and analyzed as in (A). Results from three independent experiments are shown represented as mean and standard error.
In order for cells to cope with stress, hsp genes must be transcribed and their transcripts processed by polyadenylation. Given the reduced association of PAP with non-hsp genes described above, we next asked how PAP association with the 3′ ends of three hsp genes: hsp70, hsp90 and hsp27 (i.e., HSPA1A, HSP90AA1 and HSPB1) is affected by heat shock. The same cells, conditions for ChIP and qPCR analysis were used as in Figure 6A. Figure 6B shows that, in contrast with the results obtained with the non-hsp genes, association of PAP with each of the hsp genes did not decrease, and in fact slightly increased, after a heat shock of 5 mins.
To gain a better understanding of the mechanism by which PARP1 inhibits PAP, we performed ChIP with an anti-PARP1 antibody to examine PARP1 association with the β-globin gene. The results (Figure 6C) indicate that PARP1 was also present at the 3′ end of the gene. Following heat shock, a 30% decrease in PARP1 chromatin association was observed, indicating that, similarly to PAP, although not as sharply, PARP1 was released from the 3′ end of β-globin upon activation by heat shock. To exclude the possibility that PAP and PARP1 chromatin association was reduced following heat shock because of a possible reduction in transcription, we performed a ChIP experiment using an antibody against RNAP II. Heat shock did not reduce RNAP II density at the 3′ end of the gene (Figure 6D) nor at its promoter (Figure S5C), and a slight increase (~20%) in RNAP II occupancy was indeed observed.
While we discuss below how hsp genes might evade PARP1-mediated inhibition of PAP recruitment and polyadenylation following heat shock, our findings together implicate PARylation of PAP as a significant aspect of the cellular response to stress.
DISCUSSION
Our results have provided several insights into mechanisms of gene control. First, our findings have expanded the function of PARP1 and PARylation, into the area of posttranscriptional regulation and specifically polyadenylation of mRNA precursors. Second, we have provided yet another mechanism by which PAP can be regulated, highlighting the importance of controlling PAP activity. Finally, we provide evidence that polyadenylation is inhibited following heat shock. Similar to splicing inhibition, inhibition of polyadenylation provides an additional layer of protection against the deleterious effects of heat shock, by ensuring that production of mature mRNAs is repressed so that new proteins will not be produced in a stressed environment prone to misfolding and other detrimental effects. Below we discuss the implications of our findings with respect to the role of both polyadenylation and PARylation in the regulation of gene expression.
As mentioned in the introduction, PAP is a well-known target for several post-translational modifications. Consistent with the need to regulate PAP activity tightly under different conditions, PAP was previously reported to be phosphorylated, sumoylated and acetylated, and now we have identified PARylation as an additional PAP modification. The reversible nature of these modifications is particularly suitable to regulating gene expression by modulating PAP, allowing the cell to respond efficiently to a changing cellular context such as during an environmental stress (e.g., PARylation during heat shock) or during the cell cycle (e.g., phosphorylation during M phase). All of the previously characterized post-translational modifications occur in the C-terminal domain of PAP; they are in fact situated very closely and sometimes overlap (reviewed by Ryan and Bauer, 2008). PARylation, in contrast, does not seem to occur in this region, as a truncated version of PAP that lacks this domain can still be PARylated in vitro (unpublished data). Since PARylated PAP looses its ability to bind RNA, we speculate that the most reasonable site for PARylation is the RNA-binding region itself.
An interesting question is how and when does PARP1 associate with PAP to block polyadenylation. Here we show that PARP1 colocalizes with PAP on the 3′ end of the β-globin gene. PARP1 has been shown previously to associate both with the 3′ processing machinery (Shi et al., 2009) and, similar to several components of the 3′ processing complex (reviewed in Calvo and Manley, 2003), with the promoters of numerous genes. Indeed, ChIP-chip experiments coupled to gene expression microarrays indicated that PARP1 binding is enriched at 90% of promoters of actively transcribed genes in human MCF-7 cells (Krishnakumar et al., 2008). In addition, and also analogous to components of the 3′ processing complex (reviewed in Hsin and Manley, 2012), PARP1 was found to interact with RNAP II (Carty and Greenleaf, 2002). This is consistent with the idea that PARP1 on promoters associates with RNAP II, which then facilitates its recruitment to the 3′ processing complex co-transcriptionally. However, it is also possible that PARP1 associates independently with promoters and then with PAP near the 3′ end of genes.
Several of the multiple factors required for mRNA 3′ end formation have previously been shown to associate with transcribed genes. These include in mammals CPSF, CstF and CFI (Glover-Cutter et al., 2008; Rozenblatt-Rosen et al., 2009; Venkataraman et al., 2005). PAP has long been known to associate only loosely with the other core polyadenylation factors (Takagaki et al. 1988), but recently it was reported to crosslink to both 5′ and 3′ end of genes in yeast, where it is necessary for gene looping (Medler et al., 2011). This data is consistent with our results showing that PAP can be recruited to the 3′ ends of transcribed genes in human cells, implying that, even if polyadenylation occurs after release of the mRNA from RNAP II, PAP joins the 3′ processing complex co-transcriptionally. An attractive model is that PAP and PARP1 are recruited to transcribed genes together, and then upon PARP1 activation, for example by heat shock, PAP and PARP1 PARylation occurs rapidly, leading to dissociation of both from non-heat shock protein-encoding genes (see Figure 7).
Figure 7. Model for PARP-mediated regulation of polyadenylation during heat shock.
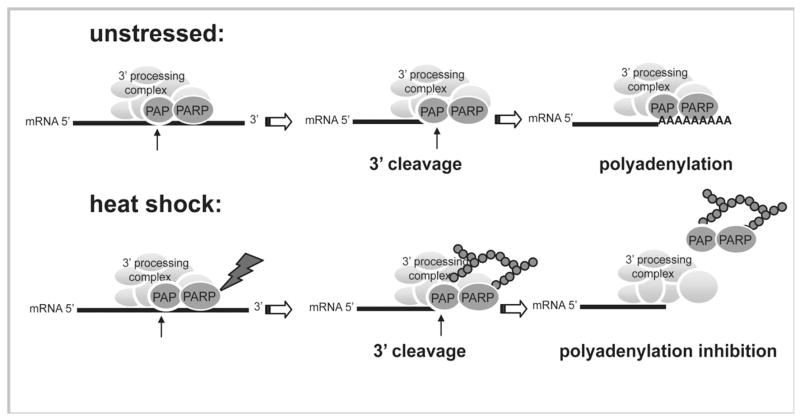
In unstressed cells 3′ cleavage and polyadenylation occur normally. During thermal stress PARP1 becomes activated and PARylates PAP (and itself). The modified PAP and PARP1 dissociate from the 3′ end of genes, leading to polyadenylation inhibition and therefore arresting the production of new proteins under thermal stress.
Gene expression in mammalian cells is regulated at multiple levels during heat shock. From a post-transcriptional perspective, extensive studies have shown that pre-mRNA splicing (Shin et al., 2004 and refs therein) and protein translation (Cuesta et al., 2000 and refs therein) are inhibited following heat shock, and our data add polyadenylation to this list. Transcriptional regulation, however, appears to be more complex, and the effect that heat shock has on ongoing transcription in mammalian cells seems to be gene-specific. Early studies showed that following heat treatment of HeLa cells, rRNA but not mRNA transcription was inhibited (Sadis et al., 1988; Warocquier and Scherrer, 1969). Interestingly, mRNA export into the cytoplasm was found to be repressed (Sadis et al., 1988), which is consistent with a defect in 3′ processing. In addition, transcription of specific genes, e.g., c-fos (Andrews et al., 1987), was shown not to be affected by heat shock. Our own experiments showing a decrease in accumulation of newly synthesized polyadenylated nuclear RNA following heat shock may reflect inhibition of transcription as well as polyadenylation. The fact that 3-ABA only partially rescued the inhibition indicates that PARylation, likely of PAP, might not be the only determinant of repression, and we suggest that the 3-ABA resistant fraction reflects inhibition of transcription of a subset of genes.
In contrast to most genes, heat shock protein-encoding genes must be expressed robustly during heat shock. A mechanism must therefore exist to ensure that PARP1 activation does not negatively affect polyadenylation of hsp transcripts. Consistent with this, our results showed that PAP association with hsp genes was not reduced, and actually increased, following heat shock. We propose two possible mechanisms that explain how PAP association with hsp and non-hsp genes is differentially regulated following heat shock. In the first, HSF1 (heat shock factor 1) plays the key role in determining specificity. HSF1 is constitutively expressed but becomes rapidly activated during heat shock by entering the nucleus and binding as a trimer to HSE (heat shock element) sequences present in heat-inducible promoters (reviewed in Shamovsky and Nudler, 2008). Notably, HSF1 has been shown to associate with 3′ processing components during heat shock (Xing et al., 2004). Therefore, in addition to its function in stimulating transcription of hsp genes, HSF1 may also act to enhance polyadenylation of the resulting hsp transcripts. Intriguingly, HSF1 has been shown to contain a PAR-binding motif (Fossati et al., 2006), and it is therefore tempting to speculate that this motif allows HSF1 to bind PARylated PAP. This might then explain our observation that PAP is not only retained on activated hsp genes but its association enhanced after stress, ensuring that even if PAP is PARylated, hsp mRNAs will still be polyadenylated. Another possibility is that PAP PARylation does not occur on hsp genes. By this model, clearance of PARP1 from hsp promoters during heat shock (as has been shown for hsp70.1; Ouararhni et al., 2006) prevents the enzyme from associating with the 3′ processing complex on hsp transcripts, thereby preventing PAP PARylation on hsp genes and allowing polyadenylation of hsp transcripts to occur unabated.
PARylation has also been implicated in control of alternative splicing (Ji and Tulin, 2009). However, the effects on splicing appear to be indirect and not mediated by PARylation of the splicing factors involved. While these previous studies, along with ours, support a role for PARP1 in regulating gene expression through modulation of mRNA processing, our experiments have provided evidence that direct PARylation of a core processing factor, PAP, can modulate its activity and thereby influence processing of mRNA precursors.
Our results show that only heat shock, and not γ-IR or oxidative stress, was able to redirect PARP1 activity toward PAP and inhibit polyadenylation. Our current understanding of how PARP1 is activated by different stimuli is partial, and it is therefore difficult to explain this specificity. Three different modes of PARP1 activation have been described: DNA damage, post-translational modification and binding to protein partners (reviewed in Luo and Kraus, 2012). The only mechanistic insight available is with respect to DNA damage, where binding of PARP1 to damaged DNA induces a structural distortion that destabilizes its catalytic domain, leading to activation (Langelier et al., 2012). While any of the above three modes may apply, the molecular mechanism of PARP1 activation during heat shock is currently unknown.
In conclusion, our data supports a model in which PARP1 activation following heat shock leads to PARylation of PAP, which in turn prevents PAP from associating with most mRNA transcripts, inhibiting their polyadenylation (Figure 7). Moreover, our ChIP results suggest that PAP is released from mRNAs concomitantly with PARP1. Ultimately, this mechanism, together with heat-induced repression of splicing, inhibits the maturation of newly synthesized transcripts, thereby preventing protein production in an environment otherwise prone to protein misfolding and aggregation. Our discovery that PAP is PARylated during heat shock provides an additional mechanism by which gene expression can be modulated at the post-transcriptional level, and also adds another layer of complexity to the functions of PARP1.
EXPERIMENTAL PROCEDURES
Cell culture and cell treatments
HeLa and MCF-7 were cultured in DMEM with 10% fetal bovine serum. HEK 293 cells stably expressing a tetracycline (tet)-inducible β-globin transgene were grown as previously reported. 200 pmol of siRNA against PARP1 (AAGAUAGAGCGUGAAGGCGAA) or non-targeting control (Dharmacon) were transfected into ~3×106 HeLa cells using Oligofectamine (Invitrogen). Cells were harvested 72 hrs post-transfection directly into loading buffer or used to prepare NE. To analyze polyadenylation during heat shock in vivo, the HEK 293 cells stably expressing the inducible β-globin transgene were treated for 3 hrs with 1 ug/ml tet. Where noted, cells were incubated during the time of induction with 5 mM 3-ABA (Calbiochem). HEK 293 cells were heat shocked for 30 mins at 43°C in an incubator, and nuclear RNA was then extracted with 10 mM Tris pH7.4, 100 mM NaCl, 2.5 mM MgCl2 and 50 ug digitonin followed by purification with TRIZOL reagent (Invitrogen) and DNAseI treatment (Fermentas). For the in vivo labeling experiment 3X106 HeLa cells were incubated with 200 uCi of 3H uridine for 30 mins either at 37°C or 43°C. Where noted 5 mM 3-ABA was added together with the tritiated-uridine. HeLa nuclear RNA was extracted in NP-40 buffer (10 mM Tris pH 7.4, 0.15% NP-40, 150 mM NaCl) followed by purification with TRIZOL. Poly(A) selection was performed using magnetic oligo(dt) beads (Novagen) and the eluted RNA was collected for scintillation counting.
To measure the half-life of β-globin transcripts following heat shock, 293 cells stably expressing the inducible β-globin transgene were treated for 3 hrs with 1 ug/ml tet, cells were then heat shocked for 30 mins at 43°C in an incubator, after which they were washed several times and incubated with tet-free DMEM. RNA was extracted at the indicated time points using TRIZOL. Samples were subjected to DNAseI treatment, phenol/chloroform, ethanol precipitation and reverse transcription.
Immunoprecipitation of PARylated PAP and western blotting
HeLa or MCF-7 cells (~8×106) were heat shocked for 1 hr at 43°C. Where noted 10 mM 3-ABA was added just before heat shock. After two washes with cold PBS containing 0.1 mM tannic acid (a PARG inhibitor), cells were resuspended in two volumes of IP lysis buffer (see Extended Experimental Procedures) Cells were lysed for 45 mins on ice and then centrifuged at 13000g for 15 mins at 4°C. After measuring protein concentrations using the Bradford assay and equalizing protein amounts, a combination of two anti-PAP antibodies were added to the supernatants and tubes were rotated for 1 hr at 4°C. 20 ul of protein A sepharose beads (GE healthcare) was then added and samples incubated overnight with rotation at 4°C. Samples were then washed 4 times in lysis buffer and resuspended in 2x loading buffer. Where noted, the PAP precipitate was then incubated with 40 ng of PARG for 10 mins at 37°C in a buffer containing 10 mM NaH2PO4 pH7.5, 10 mM KCl and 1 mM DTT. See Extended Experimental Procedures for western blot and IP of PARylated PAP from NE protocols.
In vitro 3′ processing assays
32P-labeled simian virus 40 late (SVL) full-length or pre-cleaved RNA substrates were prepared as described previously (Ryner et al., 1989). For 3′ cleavage assays, reaction mixtures consisted of 40% NE (prepared as described in Kleiman and Manley, 2001), 0.2 to 0.5 ng labeled RNA, 0.25U RNasin (Promega), 2 mM EDTA, 250 ng tRNA, 2.5% polyvinyl alcohol (PVA), 20 mM creatine phosphate, 8 mM Tris pH 7.9, 10% glycerol, 25 mM NH4(SO4)2, 0.2 mM DTT, 0.2 mM PMSF. Polyadenylation assays contained the same reagents with the omission of EDTA and addition of 1 mM MgCl2 and 1 mM ATP. See Extended Experimental Procedures for non-specific polyadenylation protocol.
In vitro PARylation
The indicated amounts of His-PAP and MBP-PARP1 were incubated in Buffer D with 1 mM NAD+ (or 0.05 mM NAD+ and 0.4 uM 32P-NAD+), 400 ng sssp (sheared salmon sperm DNA) and 10 mM MgCl2. PARylation reactions were carried out at 37°C for 10 mins and stopped by adding 2X loading buffer. Where noted, the indicated amount of PARG was then added to reaction mixtures for an additional 10 mins at 37°C. After 5 mins boiling, proteins were resolved by SDS-PAGE and subjected to ECL with the relevant antibodies (anti-PAR from Biomol, anti-PAP and anti-PARP as above) or subjected to autoradiography.
Supplementary Material
PARP1 binds and PARylates PAP in vitro, which inhibits its polyadenylation activity
Polyadenylation is inhibited during heat shock in a PARP1-dependent manner
PAP is PARylated in vivo during heat shock
Inhibition of polyadenylation reflects decreased RNA binding by PARylated PAP
Acknowledgments
We thank Dr. Maria Carmo-Fonseca for providing us with the β-globin inducible stable 293 cell line and Dr. W. Lee Kraus for providing the His-PARG and His-PARP1 constructs. We are also grateful to members of our lab for helpful discussion and to Dr. Emanuel Rosonina for critical reading of the manuscript. This work was supported by NIH grant R01 GM28983.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews GK, Harding MA, Calvet JP, Adamson ED. The heat shock response in HeLa cells is accompanied by elevated expression of the c-fos proto-oncogene. Mol Cell Biol. 1987;7:3452–3458. doi: 10.1128/mcb.7.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty SM, Greenleaf AL. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol Cell Proteomics. 2002;1:598–610. doi: 10.1074/mcp.m200029-mcp200. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Garcia-Sacristan A, Custodio N, Carmo-Fonseca M. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res. 2010;38:8015–8026. doi: 10.1093/nar/gkq703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia G, Jongstra-Bilen J, Ittel ME, Mandel P, Delain E. Poly(ADP-ribose) polymerase auto-modification and interaction with DNA: electron microscopic visualization. EMBO J. 1983;2:543–548. doi: 10.1002/j.1460-2075.1983.tb01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati S, Formentini L, Wang ZQ, Moroni F, Chiarugi A. Poly(ADP-ribosyl)ation regulates heat shock factor-1 activity and the heat shock response in murine fibroblasts. Biochem Cell Biol. 2006;84:703–712. doi: 10.1139/o06-083. [DOI] [PubMed] [Google Scholar]
- Gagne JP, Shah RG, Poirier GG. Analysis of ADP-ribose polymer sizes in intact cells. Mol Cell Biochem. 2001;224:183–185. doi: 10.1023/a:1011910329010. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hossain MB, Ji P, Anish R, Jacobson RH, Takada S. Poly(ADP-ribose) Polymerase 1 Interacts with Nuclear Respiratory Factor 1 (NRF-1) and Plays a Role in NRF-1 Transcriptional Regulation. J Biol Chem. 2009;284:8621–8632. doi: 10.1074/jbc.M807198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Chen WH, Chang YL, Wang HT, Chuang WT, Lee SC. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 2006;34:2398–2407. doi: 10.1093/nar/gkl241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Curr Opin Genet Dev. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonn P, van der Heide LP, Dahl M, Hellman U, Heldin CH, Moustakas A. PARP-1 attenuates Smad-mediated transcription. Mol Cell. 2010;40:521–532. doi: 10.1016/j.molcel.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medler S, Al Husini N, Raghunayakula S, Mukundan B, Aldea A, Ansari A. Evidence for a complex of transcription factor IIB with poly(A) polymerase and cleavage factor 1 subunits required for gene looping. J Biol Chem. 2011;286:33709–33718. doi: 10.1074/jbc.M110.193870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981;256:4135–4137. [PubMed] [Google Scholar]
- Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci U S A. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Bauer DL. Finishing touches: post-translational modification of protein factors involved in mammalian pre-mRNA 3′ end formation. Int J Biochem Cell Biol. 2008;40:2384–2396. doi: 10.1016/j.biocel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner LC, Takagaki Y, Manley JL. Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol Cell Biol. 1989;9:4229–4238. doi: 10.1128/mcb.9.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadis S, Hickey E, Weber LA. Effect of heat shock on RNA metabolism in HeLa cells. J Cell Physiol. 1988;135:377–386. doi: 10.1002/jcp.1041350304. [DOI] [PubMed] [Google Scholar]
- Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cell Mol Life Sci. 2008;65:855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Horinouchi S, Yoshida M. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J Biol Chem. 2007;282:4470–4478. doi: 10.1074/jbc.M609745200. [DOI] [PubMed] [Google Scholar]
- Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 2008;22:499–511. doi: 10.1101/gad.1628208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warocquier R, Scherrer K. RNA metabolism in mammalian cells at elevated temperature. Eur J Biochem. 1969;10:362–370. doi: 10.1111/j.1432-1033.1969.tb00699.x. [DOI] [PubMed] [Google Scholar]
- Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J Biol Chem. 2004;279:10551–10555. doi: 10.1074/jbc.M311719200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


