Abstract
Regulators of G Protein Signaling (RGS) are key regulators of G protein signaling. RGS proteins of the R4 RGS group are composed of a mere RGS domain and are mainly involved in immune response modulation. In both human and mouse, most genes encoding the R4 RGS proteins are located in the same region of chromosome 1. We show here that the RGS1/RGS16 neighborhood constitutes a synteny group well conserved across tetrapods, and closely linked to the MHC paralogon of chromosome 1. Genes located in the RGS1/RGS16 region have paralogs close to the MHC on chromosome 6 or close to the other MHC paralogons. In amphioxus, a cephalochordate, these genes possess orthologs that are located in the same scaffolds as a number of markers defining the proto-MHC in this species (Abi-Rached et al. 2002). We therefore propose that the RGS1/RGS16 region provides useful markers to investigate the origins and the evolution of the MHC. In addition, we show that some genes of the region appear to have immune functions not only in human, but also in Xenopus.
Keywords: Regulators of G protein signaling, tetrapod evolution, Branchiostoma floridae, proto-MHC, Xenopus tropicalis
Introduction
Regulators of G Protein Signaling (RGS) are key factors in the regulation of a multitude of processes that are initiated by ligand binding to a G protein-coupled receptor (GPCR). They regulate the GDP/GTP exchange on the Gα subunit of the G proteins bound to GPCRs (reviewed by Bansal et al. 2007), thus controlling a vast number of processes requiring cell-to-cell communication including development, neurotransmission, chemotaxis, endocrine regulation, visual and olfactory sensing, and, in fungi, even mating type exclusion.
It has been suggested that the components of GPCR signaling pathways have a prokaryotic origin, the signaling systems of unicellular eukaryotes representing a transitional stage between those found in prokaryotes and higher eukaryotes (Pertseva and Shpakov 2009). Moreover, the presence of RGS domain has been reported in most, if not all, major eukaryotic groups (Anantharaman et al. 2011), and RGS domains therefore constitute very old structural units. The evolution of both RGS and Gα proteins has been dominated by multiple independent lineage-specific expansions (LSE) (Anantharaman et al. 2011), leading to an extensive diversification of their domain architectures. It is generally accepted that an expansion of RGS domains occurred in metazoans with a core list of 12 proteins present in the metazoan common ancestor (Sierra et al. 2002). The RGS protein family, while defined by the presence of a functional RGS domain, is divided into several subfamilies based on sequence homology and the presence or absence of additional protein domains (Bansal et al. 2007; Siderovski and Willard 2005; Sierra et al. 2002). The smallest RGS proteins do not contain any other domains besides the core RGS domain itself, and form the R4 RGS subfamily (Bansal et al. 2007).
Many members of the R4 RGS subfamily are involved in chemotaxis and immunity to infections (reviewed by Bansal et al. 2007). Bacterial endotoxin has been shown to induce the expression of RGS1, 4 and 16 and to downregulate RGS2 and 18 (Lee et al. 2010; Patten et al. 2002; Riekenberg et al. 2009; Shi et al. 2004; Timmusk et al. 2009). RGS2 is upregulated during the intracellular infection of phagocytes with the gram-negative bacteria Brucella abortus, and is in fact required for the efficient replication and survival of the pathogen (Kim et al. 2012). Several R4 RGS can be either up- or downregulated by type I interferons (Giorelli et al. 2002; Tran et al. 2010) and by viral infection (Cahir-McFarland et al. 2004). For example, we have previously shown that RGS16 is upregulated by the interferon inducers poly I:C and CpG as well as by LPS and PHA in porcine PBMCs (Timmusk et al. 2009). Additionally, we demonstrated a direct interaction between RGS16 and a viral protein from the porcine circovirus type 2, which causes immunodeficiency in pigs (Timmusk et al. 2009).
In human, genes encoding the ten R4 RGS proteins are located in groups of two or more genes on chromosome 1, except for RGS3 which is located on chromosome 9: RGS4 and RGS5 are located on 1q23.3, RGS8 and RGS16 on 1q25.3, RGS1, RGS2, RGS13, RGS18 and RGS21 on 1q31.2. This configuration likely results from multiple tandem duplications followed by fragmentation of the region (Sierra et al. 2002). The location of most RGS genes is close to an MHC paralogon on human chromosome 1, which suggests that they may be located in a paralogous region of the extended MHC. The human genome contains three paralogous regions (or ≫ paralogons ≫) of the MHC region; while the MHC itself is located on chromosome 6, its paralogons are for the most part found on chromosomes 1, 9 and 19 (Flajnik and Kasahara. 2001; Kasahara et al. 1996; Kasahara 1999; Katsanis et al. 1996). It is generally accepted that these four regions were produced by the two rounds of whole genome duplication that occurred between the emergence of urochordates and the rise of jawed vertebrates. The MHC is a very large genomic region where many genes are involved in immunity or inflammation (Beck et al. 1999; Trowsdale 2001). It is remarkable that this region and its three paralogons were kept during the evolution of vertebrates as synteny blocks, suggesting that a conserved linkage of genes involved in immunity could be favorable. Alternatively, this configuration could be a mere historical legacy of the vertebrate whole genome duplications. Tracking the history and the limits of these regions is of pivotal importance to understand the origins of the immune system through the evolution of vertebrate genomes.
In this study, we showed that R4 RGS genes identify a region which is conserved across vertebrate genomes and represents an ancient synteny group. This region is linked to one of the paralogons of the MHC, and might therefore provide useful markers to trace the evolution of this genetic complex. In the cephalochordate Amphioxus (Branchiostoma floridae), the counterpart of the R4 RGS region was indeed found closely linked to the proto-MHC.
Materials and methods
Phylogenetic analysis for identification of paralogs/ohnologs
Protein sequences for the 76 genes of the RGS1/RGS16 region were obtained from the human assembly GRCh37.9 in Genbank Refseq database. If more than one protein sequence was available for a given gene, the longest isoform was used. To find paralogs, sequences were then blasted using the first iteration of Domain Enhanced Lookup Time Accelerated BLAST (DELTA-BLAST, Boratyn et al. 2012) against the reference sequences of the human genome to build a list of unique sequences that would display both sequence similarity and similar domain organization to each sequence of the initial set. In most cases, sequences were selected from DELTA Blast hits using a cutoff determined by a sharp drop in either the E values or protein identity. When such a cutoff did not appear, 20 hits with the lowest E values were chosen for phylogenetic analysis. In rare cases like the one of zinc finger proteins (ZNF648 in the human RGS1/RGS16 region), there were too many results all very similar to each other and a relevant phylogenetic analysis could not be performed.
MEGA5 software package was used to compute phylogenetic trees using the retrieved sequences (Tamura et al. 2011; Online Resource 2). The evolutionary history was inferred using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al. 1992). The bootstrap consensus tree was inferred from 1000 replicates (Felsenstein 1985). The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position.
Ensembl BioMart was used to determine the genomic location of each gene used to build the phylogenies. Ohnologs were determined by visual analysis of the trees, and confrontation of the trees and genomic locations (Online Resource 2). Genes that (1) appeared as close paralogs of the RGS1/RGS16 genes in the phylogeny and (2) that were also located in MHC paralogons on chromosomes 1, 6, 9, or 19 as well as 5 and 15 (reviewed by Flajnik and Kasahara 2010) were considered to be potential ohnologs.
Determining the synteny groups in vertebrates
In order to check the conservation of the region in vertebrates other than human, the synteny groups between human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), anole lizard (Anolis carolinensis), western clawed frog (Xenopus tropicalis) stickleback (Gasterosteus acculeatu) and japanese medaka (Oryzias latipes) were determined from the Ensembl genome assemblies version 68.01 of the Genomicus browser described in detail in (Muffato et al. 2010), and updated with some additional genes present in Genbank but not Ensembl itself. To confirm the identity of RGS1/RGS16 region genes shown by the Genomicus browser, we referred to the online database PhylomeDB (Huerta-Cepas et al. 2011; Online Material 1). To confirm the identity of NOTCH2 genes and RGS genes themselves, Maximum Likelihood phylogenies were constructed with bootstrap value 1000 using Mega5 (Online Material 1).
Determining the synteny groups in amphioxus
Cosmids were previously used to identify the amphioxus proto-MHC (Abi-Rached et al. 2002). To determine the equivalent regions in the current genome assembly, these cosmid sequences were blasted against the genomic RefSeq sequences at NCBI. Amphioxus scaffolds with the best coverage and lowest E values were considered. Most similar sequences to the MHC marker genes from these scaffolds were retrieved using protein blast as described above.
Protein sequences corresponding to the 76 genes of the human (GRCh37.9) RGS1/RGS16 region were retrieved from Genbank as described above and blasted against Branchiostoma floridae assembly Version 2 using protein blast (blastp) with the default settings. The best hits were then blasted back against the human assembly with the same settings in order to confirm whether the result had been true or false positive.
Sequences for the previously cloned amphioxus NOTCH-like protein (Holland et al. 2001) and other markers of the proto-MHC (Abi-Rached et al. 2002) located on scaffolds with sequences homologous to RGS1/RGS16 genes were treated similarly. These scaffolds were identified by blasting the mRNA and cosmid sequences of the proto-MHC markers from previous works (Abi-Rached et al. 2002; Holland et al. 2001) against the amphioxus reference genomic sequences using NCBI megablast.
We refer to the online database PhylomeDB (Huerta-Cepas et al. 2011, Supplementary Material 3) for many of our genes of interest. Additional phylogenies were initially created using the online tool Figenix (Gouret et al. 2005; Paganini and Gouret 2012) to get a first idea of the homologs in other organisms. To build the final version of these trees, sequences for relevant genes were retrieved from Genbank and Ensembl and a Maximum Likelihood tree was built using the MEGA5 software package as described above for ohnolog identification (Online Resource 3). Amphioxus orthologs of the human genes were individually confirmed by visual analysis of phylogenies and examination of the domain structure of encoded proteins using NCBI or by EMBL/EBI InterProScan (Quevillon et al. 2005).
Animal handling
Frog virus 3 (FV3, Iridoviridae) was grown in and purified from Baby Hamster Kidney (BHK-21) cell lines incubated at 30°C during the infection, and virus titer was determined on Fathead minnow cells as previously described (Chen et al. 2011). Three years old X. tropicalis outbred animals were obtained from the Xenopus Research Resource for Immunology at the University of Rochester (http://www.urmc.rochester.edu/smd/mbi/xenopus/index.htm) and injected intraperitoneally with 1 million plaque-forming units of FV3 in a volume of 0.1 ml of amphibian PBS. Tissues were collected 6 and 9 days post infection. All animals were handled under strict laboratory and UCAR regulations (Approval number 100577/2003-151), minimizing discomfort at all times.
Sample preparation for qPCR
RNA was extracted from the spleens and kidneys collected from FV3-infected and control frogs using Trizol reagent following the manufacturer's protocol (Invitrogen). 0.5 to 1.0 µg of total RNA in 20 µl was used to synthesize cDNA with the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA).
Quantitative PCR
qPCR reactions were carried out in a total volume of 10 µl containing 5 µl Maxima™ SYBR Green/ROX qPCR Master Mix (2X) (Fermentas), 0.5 µM (0.25 µl) forward primer, 0.5 µM (0.25 µl) reverse primer (Table 1), 1 µl 10x diluted cDNA and 3.5 µl nuclease free water. Thermocycler conditions were as follows: incubation for 2 minutes at 50°C, incubation for 10 minutes at 95°C, 40 cycles of denaturation for 15 seconds at 95°C and annealing/extension for 1 minute at 60°C, followed bt dissociation curve analysis. gapdh was used as the housekeeping gene for determining relative gene expression with the 2ΔΔCt method.
Table 1.
Xenopus tropicalis primers used for qPCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| gapdh | xtGAPDH_ex6F: TGTTGGGGTGAACCATGACAA |
xtGAPDH_ex7-ex8R: AAGGCATGGACGGTAGTCATCA |
| dhx9 | xtDHX9_ex20F: GACCTGGTTTCTGCTTCCATTTATGT |
xtDHX9_ex21R: TCGTGCAGTGGAGTACGGAATATC |
| ier5 | xtIER5_F: CTCTTGCAGCCCAGACGACA |
xtIER5_R: TTTATGCTGGGACGCGGAGT |
| ncf2 | xtNCF2_ex10F: CCGGATGCTACTCCTCCTCAACTA |
xtNCF2_ex11-ex12R: CAGCCACTGCTTCTGTTTCCTGTA |
| ptgs2 | xtPTGS2_ex1F: TGATCGTACTACCCGCCGCT |
xtPTGS2_ex2R: TGGCAGGGATTTGAACAGCA |
| rgs16 | xtRGS16_ex4: GATGGGTAAAAAGGCTCCCCA |
xtRGS16_ex5: TCTCTGGCCCGATGGTCAATA |
| stx6 | xtSTX6_ex3F: GCTGAACTGAGACAAAGGAAAGCC |
xtSTX6_ex4R: TTGAACAGAAGGGCTTGTCATCC |
Statistical analysis
All experiments were repeated 3 times. qPCR was run in triplicates, results from which were averaged (after outlier removal) to obtain the CT value for each sample. After correcting these values in reference to the housekeeping gene gapdh, the resulting deltaCT values were used for p-value calculation using the Student’s unpaired two-tailed t-test.
Results and discussion
The RGS1/RGS16 region is conserved across vertebrate species
Most of the human R4 RGS genes (RGS1, 2, 8, 13, 16, 18 and 21) can be found in the proximity of type I interferon-inducible genes RGS1 and RGS16. For the purpose of the current study we defined the RGS1/RGS16 region in human as the region located between the genes FAM163A and NEK7, i.e. at 1q25.2-q31.3. This region comprises all genes located between RGS16 and RGS2 – including other R4 RGS genes – and 20 additional genes on both sides (Online Resource 1). This region belongs to a single ancestral genomic block (Nakatani et al. 2007; Putman et al. 2008). Following Putman et al., this block corresponds to the human genome segment 1.14, which covers the region 1q23.3–32.1.
To determine whether this region was conserved in other vertebrates, the orthologs of all its genes were searched in mouse, in chicken, in the lizard Anolis carolinensis, in Xenopus tropicalis as well as in two fish species, the stickleback and the medaka. Gene locations were determined in the respective genomes using the Genomicus browser, which supports the viewing on the synteny groups and ancestral blocks in different orthologous and paralogous regions (Muffato et al. 2010). We found that the gene set encoded in the human RGS1/RGS16 region constitutes a synteny group remarkably well conserved across tetrapod species (Figure 1, Online Resource 1). In the chicken, this region has been broken into three subregions later rearranged through intra-chromosomal recombinations and inversions. However, most R4 RGS genes are grouped in a unique cluster on chromosome 8, including RGS4 and RGS5 that are not located in the region in human and mice. Regarding Anolis carolinensis and Xenopus tropicalis, although their chromosomes have not been fully assembled, many of the genes of the RGS1/RGS16 region are located on 3–4 large scaffolds, suggesting that the synteny group is at least partly conserved in these species (Figure 1).
Figure 1. The RGS1/RGS16 region defined on human chromosome 1 is conserved as a synteny group in vertebrate genomes.
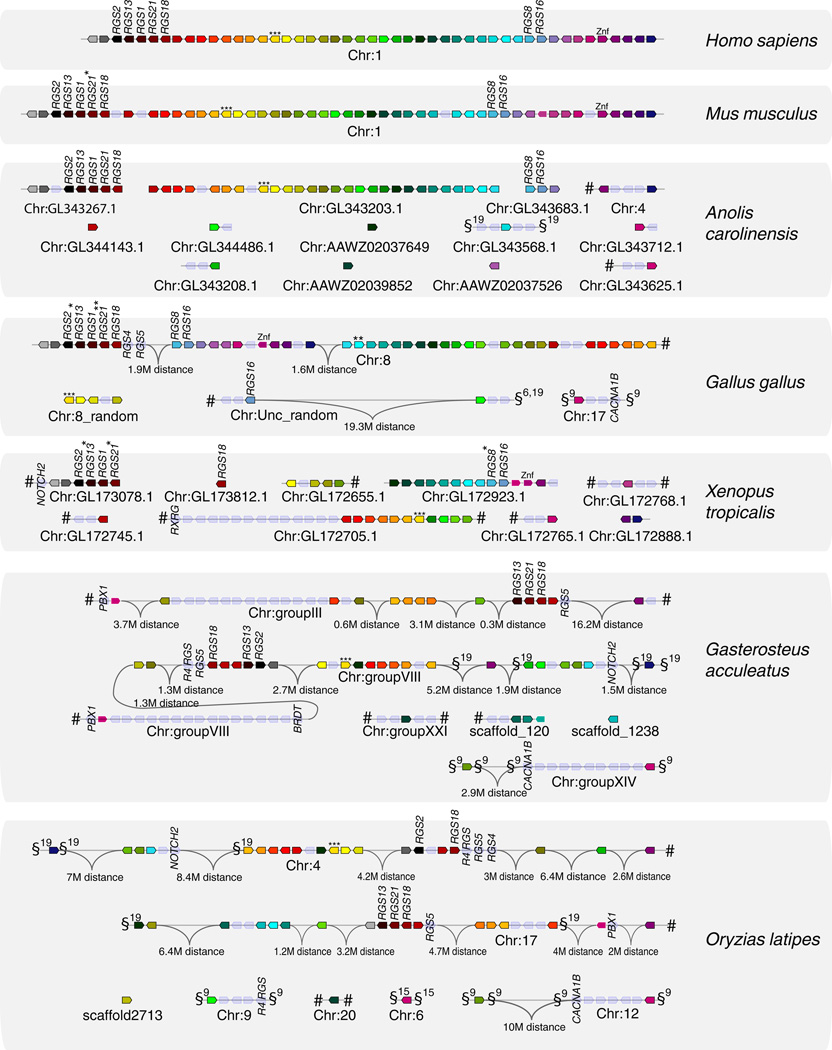
Synteny groups were determined from Ensembl genome assemblies using the Genomicus database and browser version 68.01 (Muffato et al. 2010). Orthologous genes are indicated by the same color in different species. Empty nodes on the figure represent genes not present in the human version of the RGS1/RGS16 region. Gene names on the figure refer to the closest human ortholog. Broken lines for a given species indicate different scaffolds. # - indicates that the next genes on the scaffold are not located in the neigborhood in human. §9 / §19 - genomic region containing orthologs of genes on the human MHC paralogons on chromosomes 9 or19, respectively. * - gene not present in Genomicus/Ensembl, but present in the NCBI assembly. ** - Gene present in NCBI but not in Genomicus/Ensembl. *** - In the current version of Ensembl and Genomicus, human HMCN1 is not recognized as an ortholog of the gene in other species. Due to high similarity between these proteins, it is shown as such on the figure. Znf – Zinc finger protein - representation is based on a Znf gene being located at the correct genetical context, not on phylogenetic analysis. MR1 orthologs in chicken, lizard and xenopus are shown in an additional figure (Online Resource 1). The orthology of the NOTCH and RGS genes across different species was confirmed by phylogenetic analysis (Online Resource 1).
We also retrieved the linkage group in two different teleost fish species. Duplicated copies of the RGS1/RGS16 region could be retrieved in several linkage groups (Figure 1). For example, R4 RGS were found in paralogous regions containing MHC markers and located in medaka chromosomes 4 and 17, which originated from the same ancestral chromosome through the fish-specific whole genome duplication (Kasahara et al. 2007). Importantly, these observations indicate that the RGS1/RGS16 region was present in the common ancestor of fish and tetrapods, which fits with the evolution of genome blocks conserved through vertebrate evolution described in (Nakatani et al. 2007).
The RGS1/RGS16 region belongs to the extended MHC paralogon on human Chromosome 1, and has paralogs on chromosomes 6, 9 and 19
MHC had been first defined functionally as “a group of genes coding for molecules that provide the context of recognition of foreign antigen by T lymphocytes” (Klein 1986), but this definition did not take the genomic dimension into account. The MHC may be also described as a large genetic region of more than 100 genes with a large proportion involved in immunity (Danchin et al. 2004), integrating the presence of many genes in addition to the MHC Class I and Class II genes. Such a genomic definition leads to the notion of “MHC paralogons” describing the four regions on human chromosomes 1, 6, 9 and 19, which were produced from the original proto-MHC through two rounds of global genome duplication that occurred in early vertebrate evolution (Ohno 1970, Kasahara et al. 1996). The main paralogons were initially found on 4 chromosomes, but additional smaller fragments were later identified. In fact, the MHC paralogons in human are therefore located in a total of at least 8 genomic locations on 6 different chromosomes (1p11-p32 and 1q21-25; 6p21-6p22 and 15q13-15q26; 9p13-p24, 9q32-q34 and 5q11-q23; 19p13.1-13.3) (reviewed in Flajnik and Kasahara 2010; Flajnik et al. 2012). The main MHC paralogons are commonly tracked by the presence of sets of markers that have paralog copies on some or all MHC paralogons. These markers include genes such as brd, cacna1, notch, pbx and rxr.
In the human genome, the RGS1/RGS16 region is located on the edge of an MHC paralogon at 1q21-25 and includes CACNA1E, a well-known marker gene that has paralogs on two of the other main MHC paralogons. In addition, RGS1/RGS16 region, which contains multiple RGS genes encoding proteins expressed in the central nervous system, is flanked on the other side by one of the main neurotrophin paralogons at 1q32-q44 (Hallböök et al. 2006). In addition to CACNA1E, other markers for the chromosome 1 MHC paralogon are located in the RGS1/RGS16 neighborhood (Figure 2, Online Resource 2) including (1) the gene MR1 that is a paralog of the HLA genes found in the MHC on chromosome 6; and (2) the gene IER5 which has paralogs on chromosomes 9 and 19. Our phylogenetic analyses (Supplementary Figure S1) show that many genes in the RGS1/RGS16 region have in fact paralogs on chromosomes 5, 6, 9, 15 and/or 19 (Figure 2). When these paralogs were located in the extended MHC area or its counterparts, we considered them as ohnologs. Figure 2 shows that ohnologs of the RGS1/RGS16 genes are indeed distributed within the MHC paralogons. Notably, a gene encoding an R4 RGS protein (RGS3) is found in the MHC paralogon on human chromosome 9, which is also the chromosome that was found to contain the highest amount of ohnologs for the RGS1/RGS16 region genes (Figure 2, Online Resource 2). Interestingly, most of these paralogs mapped to chromosome 9, which is consistent with the idea that among MHC paralogons 9q34 retained much more markers than the other duplicated regions (Vienne et al. 2003).
Figure 2. Ohnologs of genes located in the RGS1/RGS16 region.
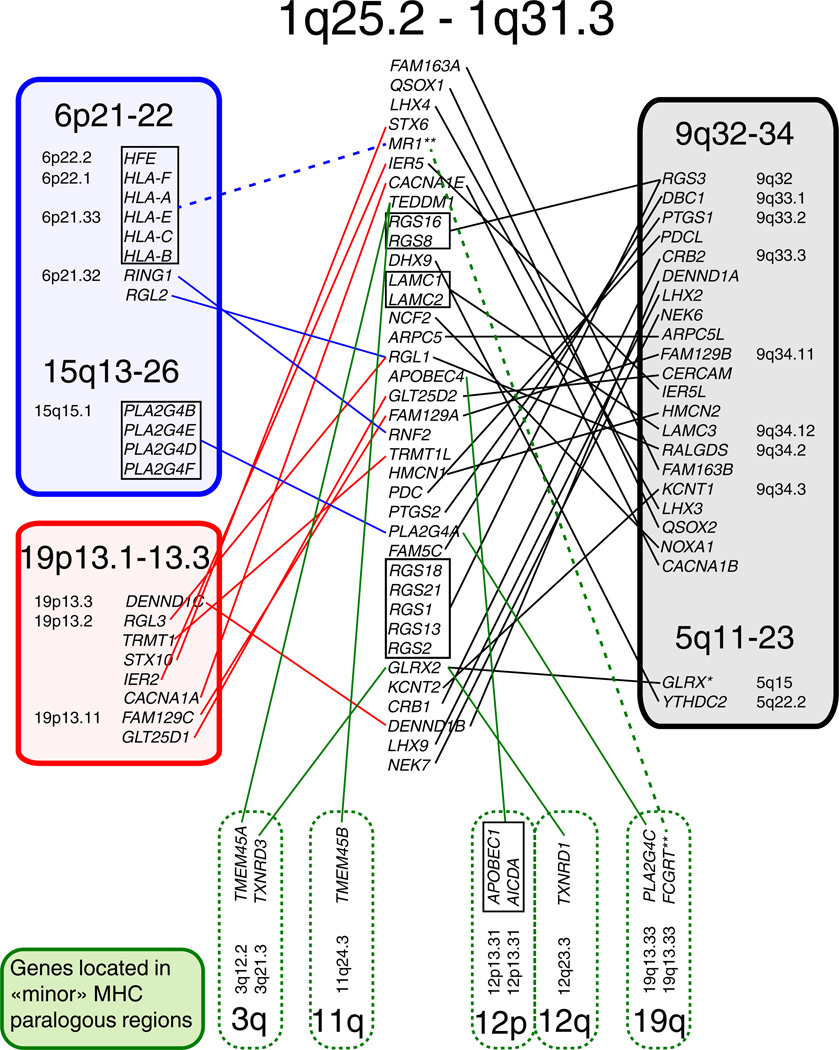
According to phylogenetic analysis, 35 out of 76 genes located around RGS1 and RGS16 in human have copies on the different MHC paralogons, confirming that this region constitutes an extension of the paralogon on the human chromosome 1. The highest number of ohnologs could be retrieved from the MHC paralogon split between genomic locations 9q32-34 and 5q11-23. These results represent a phylogenetic analysis with the maximum likelihood method and bootstrap value 1000. * - The gene GLRX was not amongst the closest paralogs of GLRX2 according to our phylogenetic analysis, yet it was shown as the only paralog found in the database. ** Since phylogenetic analysis indicates that MR1 is rather a recent duplicate of the classical MHC molecules than an ancient paralog, it is linked to those genes by a dashed line on the figure.
The relationship between the RGS1/RGS16 region and MHC paralogons is further supported by observations from other vertebrate genomes: sequences similar to genes from the RGS1/RGS16 region are indeed often found at close proximity of classical MHC marker homologs (Figure 1, Figure 3).
Figure 3. The RGS1/RGS16 synteny group can be traced to the cephalochordate Branchiostoma floridae proto-MHC.
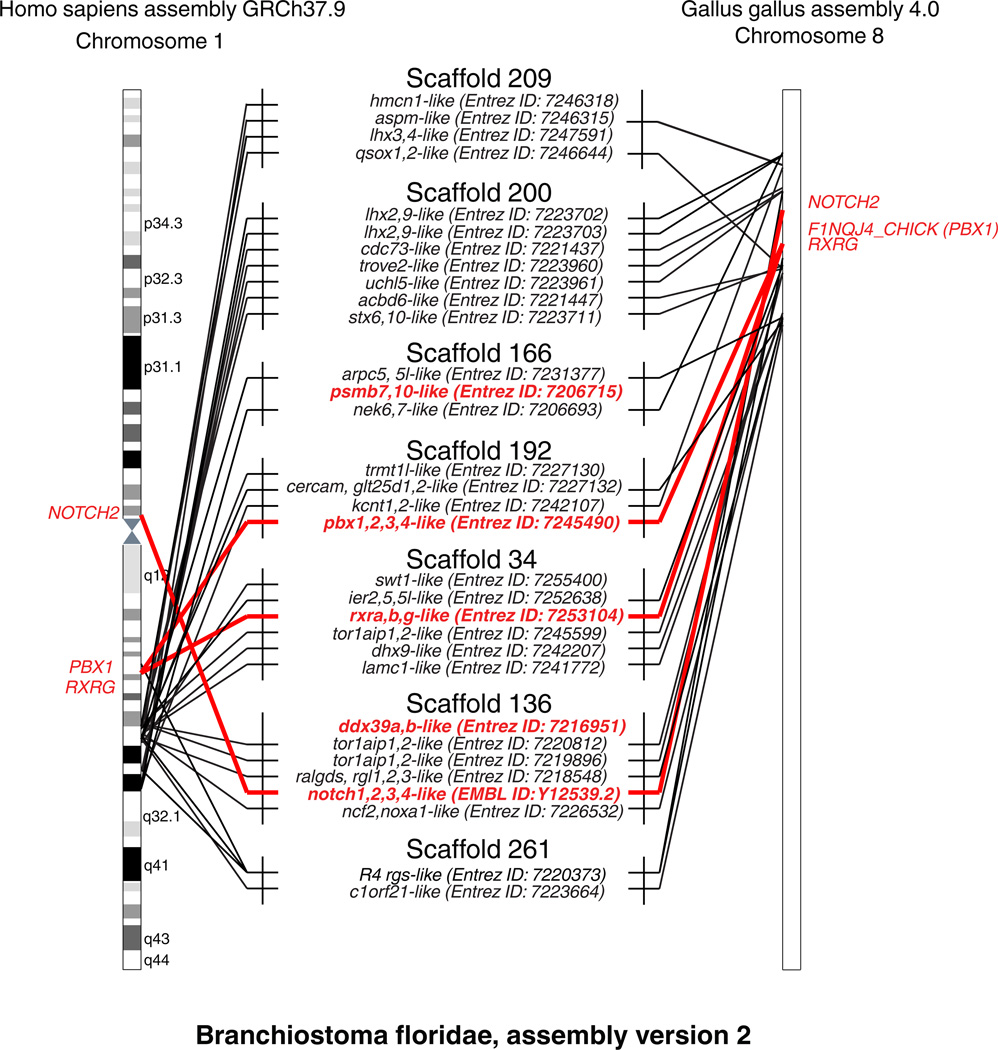
Phylogenetic analysis identifies putative amphioxus orthologs for 24 (30 if taking paralogous genes into account) out of 76 genes of the human RGS1/RGS16 region, located on merely 7 genomic scaffolds. The figure is not in scale, and amphioxus genes are shown only when (a) phylogenetically confirmed orthologs of either human genes from the RGS1/RGS16 region or of MHC marker genes and (b) located on one of these 7 scaffolds. ** - R4 RGS present in NCBI assembly but not Ensembl. For a better overview, lines connect amphioxus gene names with the approximate genomic locations of their orthologs in human chromosome 1 and chicken chromosome 8. Red lines are used to indicate MHC marker genes. As PSMB7,10 and DDX39A,B are not located on the human Chr1 or chicken Chr8, their amphioxus orthologs are mentioned but not linked to their vertebrate counterpart. Four of the seven scaffolds on the figure match those identified as a part of the amphioxus proto-MHC in (Abi-Rached et al. 2002; Castro et al. 2004; Danchin and Pontarotti 2004) and contain gene(s) with the highest similarity to the MHC marker genes DDX39A,B, PBX1,2,3,4, NOTCH1,2,3,4, PSMB7,10 or RXRA,B,G. In addition, one of the scaffolds contains a protein phylogenetically similar to the vertebrate R4 RGS proteins (see also Online Resource 1).
In chicken, a copy of gluthamine synthase GLUL, a gene of the RGS1/RGS16 region, is located near the MHC marker CACNA1B on the small 11Mb chicken chromosome 17 supporting the notion that avian microchromosomes provide a good representation of the ancestral configuration (Burt 2002; Nakatani et al. 2007). Additionally, a copy of GLUL and CACNA1B are also found close to each other in both stickleback and medaka, and an additional MHC marker, a NOTCH2-like sequence maps together with the RGS1/RGS16 region genes in both Xenopus and fish. The MHC paralogon found on human chromosome 1 appears to have been split into two parts, resulting in several of the genes including the human NOTCH2 located in the region 1p11-1p32, while other markers are located closer to the RGS1/RGS16 region at the location 1q21-25. However, this is likely a result of recent intrachromosomal rearrangements (Kasahara 1999) and thus does not represent the ancestral configuration. Notably, a NOTCH2-like sequence is not only located next to the R4 RGS genes in Xenopus tropicalis, but is also located next to some other genes of the RGS1/RGS16 region in both medaka and stickleback (Figure 1). Phylogenetic analysis indicates that these NOTCH2-like sequences are indeed orthologs of the human NOTCH2 (Online Resource 1).
A copy of the MHC marker gene PBX is next to a GLUL homolog in both medaka and stickleback and in stickleback, a homolog of the marker BRDT is located at close proximity to a second pair of GLUL and PBX (Figure 1). More generally, fish RGS1/RGS16 region contains a number of homologues to genes found in the human MHC paralogon on Chromosome 9 (Figure 1).
Several genes of the RGS1/RGS16 region also have paralogs on „minor“ MHC paralogous regions as defined in (Flajnik et al. 2012). These regions include for example human 19q and 12p (Figure 2, Online Resource 2), which contain the leukocyte receptor complex (LRC) and NK cell receptor genes (NKC), respectively. Our findings are well consistent with previous studies, which have provided clear evidence that such regions were indeed linked to the MHC before the divergence of tetrapods and teleosts (Flajnik and Du Pasquier 2008; Ohashi et al. 2010).
The RGS1/RGS16 region origin can be tracked to cephalochordates
The origin of MHC as an antigen-presenting unit is generally seen as the result of the co-option of pre-existing genes combined with the recruitment of new genes. The conserved clustering of many genes inside MHC paralogons across vertebrate evolution strongly suggests that (some of) the pre-existing genes were already located in the same region before the emergence of the MHC. Selection pressures keeping them in a conserved synteny group are still a matter of debate; it may be either related to immunity-related functions, which could be co-regulated, or to the presence of genomic elements in the vicinity, that would determine critical developmental patterns and then freeze the regional architecture (Engström et al. 2007).
Since the RGS1/RGS16 region belongs to the same conserved block as the ancestral MHC, its genes could represent useful markers to track the origin of the proto-MHC before the emergence of vertebrates, as shown for example with a gene orthologous to LHX4 (and LHX3) which maped near the MHC marker PBX in Ciona (Kasahara et al. 2004). To further test this idea, we searched for homologues of these genes in genomic scaffolds of the amphioxus, in which a proto-MHC has been previously identified (Abi-Rached et al. 2002, Castro et al. 2004; Danchin and Pontarotti 2004). A full genome assembly is not available for this species, and our analysis was therefore limited to scaffolds. To identify amphioxus orthologs of the human genes present in the RGS1/RGS16 region, we used NCBI blastp to find similar sequences. The correspondence of protein domain structure was then checked for each hit, and phylogenies present in the PhylomeDB or built using Figenix and Mega5 (Online Resource 3) examined to select true orthologs. In addition to scaffolds containing only one gene of interest, seven scaffolds were found with two or more orthologs of genes from the RGS1/RGS16 region. We retrieved 24 markers (30 if taking the paralogs of RGS and LAMC into account) out of the 76 genes located in the human RGS1/RGS16 region (Figure 3, Supplementary Table S2).
In addition, scaffolds containing amphioxus proto-MHC markers previously identified by (Abi-Rached et al. 2002; Holland et al. 2001) were retrieved from Genbank (Supplementary Table S2A and S2B), and four scaffolds out of seven with genes from the RGS1/RGS16 region matched those with MHC markers: the scaffold 166 (Genbank ID: NW_003101439.1) containing a PSMB7,10-like gene, the scaffold 192 (Genbank ID: NW_003101413.1) containing a PBX1,2,3,4-like gene, the scaffold 34 (Genbank ID: NW:003101413.1) containing a RXRA,B,G-like gene and the scaffold 136 (Genbank ID:NW_003101539.1) containing both a DDX39A,B-like gene and a NOTCH1,2,3,4-like gene (Figure 3). Importantly, previous studies have shown that the scaffolds containing amphioxus markers of the proto-MHC are all linked together on a single chromosome (Castro et al. 2004; Danchin and Pontarotti 2004), thus indicating that the amphioxus orthologs of RGS1/RGS16 region genes are also found on this linkage group. However, the current annotation of the amphioxus genome does not always match the protein models published by Abi-Rached and colleagues in 2002. For example, the current annotation of the amphioxus NOTCH-like gene predicts only a gene consisting of two EGF domains, while the mRNA of a complete amphioxus NOTCH with a NOTCH domain and other additional domains has been already previously characterized (Holland et al. 2001). This sequence’s similarity to the human NOTCH genes was confirmed phylogenetically (Online Resource 3) and it was also used here in the current study (Figure 3). In fact, although the copy of NOTCH on human chromosome 1 is quite far from RGS genes in mammals, it is indeed close to RGS homologs in other vertebrates such as Xenopus and chicken, and has been recognized as part of conserved vertebrate linkage group (CVL) 2, which formed a single ancestral block together with CVL block 5 in the gnathostome ancestor as proposed by (Nakatani et al. 2007).
Taken together, these observations strongly suggest that genes from the RGS1/RGS16 region have ancestors located in the proto-MHC, and could therefore be used as markers for tracking MHC evolution.
The RGS1/RGS16 region contains other IFN-induced/virus responsive genes and genes involved in immune responses to pathogens in human and in mice
To investigate whether the genes located in the RGS1/RGS16 region may have functions related to immunity as many genes found in the MHC we performed a systematic in silico functional analysis, mining human micro-array data available about expression changes imposed by viral infections in the EMBL/EBI Gene Expression Atlas 2.0.151. Many genes in the region were found to be involved in immune responses, especially to virus/host interactions (Online Resource 4). These observations suggest that the RGS1/RGS16 region may be enriched in genes involved in immunity, which is consistent with its MHC-linked origin. A complete demonstration of such an enrichment would require a comparison with global transcriptome data in a number of infectious contexts.
As a first step to determine if the immunity related function of selected markers of the RGS1/RGS16 region is a conserved feature, we studied their expression profile in a frog (Xenopus tropicalis) infected by the Ranavirus FV3 (Frog Virus 3). The genes ier5, mr1, ptgs2, ncf2 and stx6 were chosen, for which human micro-array data were available about expression changes imposed by viral infections in the EMBL/EBI Gene Expression Atlas 2.0.152 (Online Resource 4). The induction of rnaseL, an important gene for antiviral immunity located close to RGS, could not be tested since it was absent from the frog sequence. The Xenopus host immune response against FV3 peaks at day 6 (e.g., height of T cell proliferation in the spleen and T cell infiltration in the kidney), whereas viral clearance is notable at day 9 (Robert, et al., 2005). Since FV3 targets mainly the kidney in both X. laevis (Robert et al. 2005) and X. tropicalis (Chen and Robert 2011), the expression was measured in kidney and spleen on day 6 and 9 post infection, in comparison with non-infected controls. As expected, the greatest changes in expression of RGS1/RGS16 related genes were seen in the kidneys, the main site of infection. The upregulation of ncf2 (neutrophil cytosolic factor 2) and ptgs2 (cyclooxygenase 2) in the kidneys of infected frogs at the peak of the anti-FV3 immune response (Figure 4). was consistent with our previous study reporting the infiltration of FV3-infected X. laevis kidneys by CD8 T cells and activated MHC class II positive leukocytes between 3 and 6 days post-infection (Morales and Robert 2007). In naïve frog, most kidney leukocytes are either blood-derived lymphocytes from the blood vessels or resident macrophages. Thus, the increased expression of ncf2 and ptgs2 likely results from both an increased number of infiltrating leukocytes and upregulation of gene expression at the site of infection. Accordingly, ncf2 and ptgs2 were more upregulated in the kidneys than in the spleen. These results provide further evidence of the involvement of monocytic cells in anti-FV3 immune response and suggest that genes of the rgs1/rgs16 region are involved in the response of myeloid cells.
Figure 4. Expression of rgs1/rgs16 region genes in FV3-infected frogs.
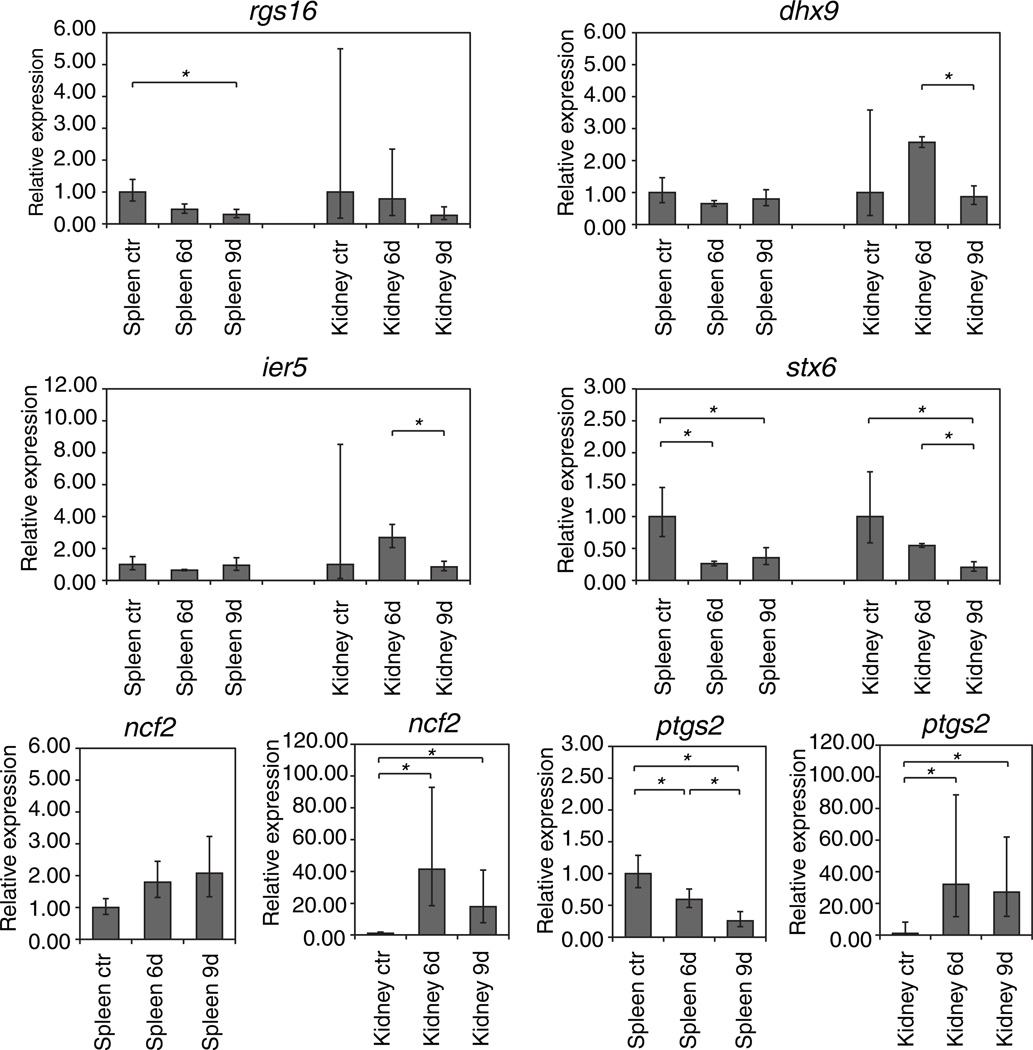
6d - 6 days post infection. 9d - 9 days post infection. ctr - control (non-infected) frogs. The expression of markers of interest was normalized on gapdh expression. Error bars represent standard deviation of results obtained from 3 similarly treated frogs. p values were calculated using the unpaired two-tailed Student’s t-test. Asterisks indicate significant changes in gene expression (p<0.05).
The increased amounts of dhx9 and ier5 transcripts at 6 dpi may suggest a local response of the kidney epithelium. Similarly, the decrease of rgs16 expression in the kidneys is likely due to the effects of type I interferon: in mammals, RGS16 has been shown to be upregulated by interferon-inducing agents (Timmusk et al. 2009) or downregulated by type I interferon itself (Giorelli et al. 2002), suggesting a complex mechanism of regulation. As a matter of fact, the increased expression of type I IFN-inducible Myxovirus-resistance1 (mx1) gene at 6 days post-infection in the Xenopus kidney (De Jesús Andino et al. 2011; Grayfer et al. 2012) is indicative of a strong type IFN response at the site of FV3 infection. In addition, STX6 was found significantly downregulated in both spleens and kidneys of the infected frogs as seen in infected human.
Taken together, our results suggest that at least some genes of the X. tropicalis RGS1/RGS16 region are modulated during a viral infection similar to human genes, providing hints of putative functional basis of the evolutionary conservation of the extended MHC and MHC paralogons. However, there is still no direct evidence that the genomic configuration of the MHC tetrad of paralogs has been maintained by selection pressures related to functional issues.
Conclusion
The RGS1/RGS16 region appears as a conserved synteny group with paralogs in the neighborhood of MHC and other paralogons. This study supports the conclusions of Nakatani et al. (Nakatani et al. 2007), who have previously shown that CVL block 5, together with all the blocks that correspond to MHC paralogons on human chromosomes 1, 5, 6, 9, 15 and 19, have originated from a single ancestral chromosome that existed in the common vertebrate ancestor. The RGS1/RGS16 region contains genes involved in immunity at least in human and frog, which suggests a possibility that they may participate to a conserved functional unit connected to the MHC. Finally, markers from the RGS1/RGS16 region can be useful to track the proto-MHC in non-vertebrates.
Supplementary Material
Acknowledgements
We thank Pierre Pontarotti and Louis du Pasquier for helpful discussions, and Muriel Perron for providing critical reagents. This work was supported by Tallinn University of Technology and the Institut National Recherche Agronomique, by Estonian Research Council Targeted Financing Project SF0140066s09, and by grants ETF8914 from Estonian Science Foundation, IOB-0923772 from the National Science Foundation and R24-AI-059830 from the National Institutes of Health.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat Genet. 2002;31:100–105. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Abhiman S, de Souza RF, Aravind L. Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene. 2011;475:63–78. doi: 10.1016/j.gene.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal G, Druey KM, Xie Z. R4 RGS proteins: regulation of G-protein signaling and beyond. Pharmacol Ther. 2007;116:473–495. doi: 10.1016/j.pharmthera.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Geraghty D, Inoko H, Rowen L. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt DW. Origin and evolution of avian microchromosomes. Cytogenet Genome Res. 2002;96:97–112. doi: 10.1159/000063018. [DOI] [PubMed] [Google Scholar]
- Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol. 2004;78:4108–4119. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LF, Furlong RF, Holland PW. An antecedent of the MHC-linked genomic region in amphioxus. Immunogenetics. 2004;55:782–784. doi: 10.1007/s00251-004-0642-9. [DOI] [PubMed] [Google Scholar]
- Chen G, Robert J. Antiviral immunity in amphibians. Viruses. 2011;3:2065–2086. doi: 10.3390/v3112065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Ward BM, Yu KH, Chinchar VG, Robert J. Improved knockout methodology reveals that frog virus 3 mutants lacking either the 18K immediate-early gene or the truncated vIF-2alpha gene are defective for replication and growth in vivo. J Virol. 2011;85:11131–11138. doi: 10.1128/JVI.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin E, Vitiello V, Vienne A, Richard O, Gouret P, McDermott MF, Pontarotti P. The major histocompatibility complex origin. Immunol Rev. 2004;198:216–232. doi: 10.1111/j.0105-2896.2004.00132.x. [DOI] [PubMed] [Google Scholar]
- Danchin EG, Pontarotti P. Towards the reconstruction of the bilaterian ancestral pre-MHC region. Trends Genet. 2004;20:587–591. doi: 10.1016/j.tig.2004.09.009. [DOI] [PubMed] [Google Scholar]
- De Jesús Andino F, Chen G, Li Z, Grayfer L, Robert J. Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus Frog-Virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology. 2012;432:435–443. doi: 10.1016/j.virol.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity. 2001;15:351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Du Pasquier L. Evolution of the immune system. In: Paul WE, editor. Fundamental Immunology. 6th edn. Philadelphia: Wolters Kluwer / Lippincott Williams & Wilkins; 2008. pp. 56–124. [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Tlapakova T, Criscitiello MF, Krylov V, Ohta Y. Evolution of the B7 family: co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7's historical relationship with the MHC. Immunogenetics. 2012;64:571–590. doi: 10.1007/s00251-012-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorelli M, Livrea P, Defazio G, Iacovelli L, Capobianco L, Picascia A, Sallese M, Martino D, Aniello MS, Trojano M, De Blasi A. Interferon beta-1a counteracts effects of activation on the expression of G-protein- coupled receptor kinases 2 and 3, beta-arrestin-1, and regulators of G-protein signalling 2 and 16 in human mononuclear leukocytes. Cell Signal. 2002;14:673–678. doi: 10.1016/s0898-6568(02)00011-6. [DOI] [PubMed] [Google Scholar]
- Gouret P, Vitiello V, Balandraud N, Gilles A, Pontarotti P, Danchin EG. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayfer L, Andino Fde J, Chen G, Chinchar GV, Robert J. Immune evasion strategies of ranaviruses and innate immune responses to these emerging pathogens. Viruses. 2012;4:1075–1092. doi: 10.3390/v4071075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallböök F, Wilson K, Thorndyke M, Olinski RP. Formation and evolution of the chordate neurotrophin and Trk receptor genes. Brain Behav Evol. 2006;68:133–144. doi: 10.1159/000094083. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Abi-Rached L, Tamme R, Holland ND, Kortschak D, Inoko H, Shiina T, Burgtorf C, Lardelli M. Characterization and developmental expression of the amphioxus homolog of Notch (AmphiNotch): evolutionary conservation of multiple expression domains in amphioxus and vertebrates. Dev Biol. 2001;232:293–507. doi: 10.1006/dbio.2001.0160. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Capella-Gutierrez S, Pryszcz LP, Denisov I, Kormes D, Marcet-Houben M, Gabaldón T. PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acid Res. 2011;39:D556–D560. doi: 10.1093/nar/gkq1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T. Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc Natl Acad Sci U S A. 1996;93:9096–9101. doi: 10.1073/pnas.93.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M. The chromosomal duplication model of the major histocompatibility complex. Immunol Rev. 1999;167:17–32. doi: 10.1111/j.1600-065x.1999.tb01379.x. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Suzuki T, Du Pasquier L. On the origins of the adaptive immune system: novel insights from invertebrates and cold-blooded vertebrates. Trends Immunol. 2004;25:105–111. doi: 10.1016/j.it.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, Jindo T, Kobayashi D, Shimada A, Toyoda A, Kuroki Y, Fujiyama A, Sasaki T, Shimizu A, Asakawa S, Shimizu N, Hashimoto S, Yang J, Lee Y, Matsushima K, Sugano S, Sakaizumi M, Narita T, Ohishi K, Haga S, Ohta F, Nomoto H, Nogata K, Morishita T, Endo T, Shin-I T, Takeda H, Morishita S, Kohara Y. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Fitzgibbon J, Fisher EM. Paralogy mapping: identification of a region in the human MHC triplicated onto human chromosomes 1 and 9 allows the prediction and isolation of novel PBX and NOTCH loci. Genomics. 1996;35:101–108. doi: 10.1006/geno.1996.0328. [DOI] [PubMed] [Google Scholar]
- Kim DH, Lim JJ, Lee JJ, Kim DG, Lee HJ, Min W, Kim KD, Chang HH, Endale M, Rhee MH, Watarai M, Kim S. RGS2-mediated intracellular Ca2+ level plays a key role in the intracellular replication of Brucella abortus within phagocytes. J Infect Dis. 2012;205:445–452. doi: 10.1093/infdis/jir765. [DOI] [PubMed] [Google Scholar]
- Klein J. Natural history of the major histocompatibility complex. New York: Wiley; 1986. [Google Scholar]
- Lee HK, Yeo S, Kim JS, Lee JG, Bae YS, Lee C, Baek SH. Protein kinase C-eta and phospholipase D2 pathway regulates foam cell formation via regulator of G protein signaling 2. Mol Pharmacol. 2010;78:478–485. doi: 10.1124/mol.110.064394. [DOI] [PubMed] [Google Scholar]
- Morales HD, Robert J. Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. J Virol. 2007;81:2240–2248. doi: 10.1128/JVI.01104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffato M, Louis A, Poisnel CE, Roest Crollius H. Genomicus: a database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics. 2010;26:1119–1121. doi: 10.1093/bioinformatics/btq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Takizawa F, Tokumaru N, Nakayasu C, Toda H, Fischer U, Moritomo T, Hashimoto K, Nakanishi T, Dijkstra JM. A molecule in teleost fish, related with human MHC-encoded G6F, has a cytoplasmic tail with ITAM and marks the surface of thrombocytes and in some fishes also of erythrocytes. Immunogenetics. 2010;62:543–559. doi: 10.1007/s00251-010-0460-1. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Paganini J, Gouret P. Reliable Phylogenetic Trees Building: A New Web Interface for FIGENIX. Evol Bioinform Online. 2012;8:417–421. doi: 10.4137/EBO.S9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten M, Bunemann J, Thoma B, Kramer E, Thoenes M, Stube S, Mittmann C, Wieland T. Endotoxin induces desensitization of cardiac endothelin-1 receptor signaling by increased expression of RGS4 and RGS16. Cardiovasc Res. 2002;53:156–164. doi: 10.1016/s0008-6363(01)00443-6. [DOI] [PubMed] [Google Scholar]
- Pertseva MN, Shpakov AO. The prokaryotic origin and evolution of eukaryotic chemosignaling systems. Neurosci Behav Physiol. 2009;39:793–804. doi: 10.1007/s11055-009-9190-y. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekenberg S, Farhat K, Debarry J, Heine H, Jung G, Wiesmuller KH, Ulmer AJ. Regulators of G-protein signalling are modulated by bacterial lipopeptides and lipopolysaccharide. FEBS J. 2009;276:649–659. doi: 10.1111/j.1742-4658.2008.06813.x. [DOI] [PubMed] [Google Scholar]
- Robert J, Morales H, Buck W, Cohen N, Marr S, Gantress J. Adaptive immunity and histopathology in frog virus 3-infected Xenopus. Virology. 2005;332:667–675. doi: 10.1016/j.virol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi GX, Harrison K, Han SB, Moratz C, Kehrl JH. Toll-like receptor signaling alters the expression of regulator of G protein signaling proteins in dendritic cells: implications for G protein-coupled receptor signaling. J Immunol. 2004;172:5175–5184. doi: 10.4049/jimmunol.172.9.5175. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra DA, Gilbert DJ, Householder D, Grishin NV, Yu K, Ukidwe P, Barker SA, He W, Wensel TG, Otero G, Brown G, Copeland NG, Jenkins NA, Wilkie TM. Evolution of the regulators of G-protein signaling multigene family in mouse and human. Genomics. 2002;79:177–185. doi: 10.1006/geno.2002.6693. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S, Merlot E, Lovgren T, Jarvekulg L, Berg M, Fossum C. Regulator of G protein signalling 16 is a target for a porcine circovirus type 2 protein. J Gen Virol. 2009;90:2425–2436. doi: 10.1099/vir.0.008896-0. [DOI] [PubMed] [Google Scholar]
- Tran T, Paz P, Velichko S, Cifrese J, Belur P, Yamaguchi KD, Ku K, Mirshahpanah P, Reder AT, Croze E. Interferon beta-1b Induces the Expression of RGS1 a Negative Regulator of G-Protein Signaling. Int J Cell Biol. 2010;2010:529376. doi: 10.1155/2010/529376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- Vienne A, Shiina T, Abi-Rached L, Danchin E, Vitiello V, Cartault F, Inoko H, Pontarotti P. Evolution of the proto-MHC ancestral region: more evidence for the plesiomorphic organisation of human chromosome 9q34 region. Immunogenetics. 2003;55:429–436. doi: 10.1007/s00251-003-0601-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


