Abstract
The metabolome refers to the entire set of small molecules, or metabolites, within a biological sample. These molecules are involved in many fundamental intracellular functions and reflect the cell’s physiological condition. The ability to detect and identify metabolites and determine and monitor their amounts at the single cell level enables an exciting range of studies of biological variation and functional heterogeneity between cells, even within a presumably homogenous cell population. Significant progress has been made in the development and application of bioanalytical tools for single cell metabolomics based on mass spectrometry, microfluidics, and capillary separations. Remarkable improvements in the sensitivity, specificity, and throughput of these approaches enable investigation of multiple metabolites simultaneously in a range of individual cell samples.
Introduction
The cell metabolome is oftentimes defined as the complete set of intracellular and cell membrane metabolites—molecules with masses ranging in size from a few Daltons to several thousand Daltons. These include lipids and carbohydrates, but exclude elements, protein-derived peptides, proteins, RNA, and DNA. The total number of existing metabolites is estimated to reach one million (in plants) [1], but only a small percentage has been identified. And in fact, the metabolome of individual cells, which contain a smaller number of compounds, remains unknown.
A goal of single cell analysis is to achieve a detailed understanding of cell function. Doing so requires obtaining information about the chemical components of the cell, as well as their metabolic flux. Single cell investigations often endeavor to reveal cell-to-cell differences within cellular populations. These variances are typically classified as either being due to cellular heterogeneity or biological variation among similar cells. Although these terms have been used interchangeably, they are not the same. We define cellular heterogeneity as the functional differences between similar cells, as well as different cell types. For example, in a specific brain region, astrocytes and neurons are obviously distinct cells, but two biochemically similar neurons can have different nociceptive fields and therefore, be functionally different. Thus, the chemical heterogeneity of cells reflects functional differences in their elementomes, metabolomes, transcriptomes, proteomes, and genomes. In contrast, biological variation oftentimes refers to differences among so-called identical cells that arise from their physical and chemical environment, as well as stochastic processes.
The detection and identification of defined analytes or their classes in single cells has been performed for many years using a range of histochemical and cytochemical methods (e.g., immunostaining, 4’6-diamidino-2-phenylindol staining of DNA), some of them developed more than a century ago and still in use today [2]. Single cell metabolomics expands the knowledge that can be acquired using these methods while introducing new capabilities in multianalyte qualitative and quantitative detection. Metabolomics, along with elementomics, proteomics, transcriptomics, and genomics, cover most of the chemical constituents within a single cell. Each of these ‘omics measurement fields evolved from the introduction of new technologies that enabled broad analyte coverage in a single experiment. In addition, advances in statistical analysis and mathematical modeling have made the processing of very large, multiparametric data sets and the mining of meaningful results possible. Systems biology, in particular, relies heavily on the qualitative and quantitative results of metabolomic experiments [3].
In contrast with proteomics, transcriptomics, and genomics, metabolomics and elementomics investigate chemical species, including some xenobiotics that have multiple cellular and extracellular origins and relocalize between different cells, extracellular spaces, and intracellular compartments. Metabolites produced in one cell can be found in many other cells. While this is also true for some peptides, proteins, and RNAs, it is more common for metabolites. Metabolite mobility makes it more difficult to predict (and model) the composition and dynamics of the cellular metabolome using transcriptomics and proteomics data [4].
Despite significant advances in metabolome coverage, currently no single analytical method can successfully investigate a large percentage of the cellular metabolome in a single measurement. Key challenges include the large number of unidentified metabolites in the majority of biological samples, their diversity and relatively small size, significant difficulties in determining their structure, and the wide dynamic range of analyte amounts per cell (from single molecules to billions of molecules). Another issue, compared with RNA and DNA, is the inability to increase analyte copies using amplification; however, experimental increases in metabolic precursor concentration may allow for investigations of previously undetectable metabolites [5,6].
Metabolomics also deals with a much greater variety of diverse molecular classes compared with genomics and transcriptomics. The two most common approaches for measuring the untargeted metabolome of tissues and larger samples are those based on separations coupled to mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy [7,8]. While the information content of NMR is unmatched, its poor detection limit has mostly precluded its application to individual cells, although a few examples of small-volume NMR have been reported [9-11].
An additional challenge to single cell metabolomics is the incomplete state of current databases, even though they list tens of thousands of compounds. For example, the NIST electron ionization mass spectral library includes 243,893 mass spectra for 212,961 unique compounds, but not all of them are endogenous metabolites. However, the number of metabolomic databases available for matching the physicochemical parameters of known metabolites to experimentally observed mass spectra is increasing, along with improved quality and coverage (reviewed in [12]; e.g., NIST Chemistry WebBook). Importantly, projects such as the Metabolomics Standards Initiative (http://msi-workgroups.sourceforge.net/; [13]) are leading a strong effort towards standardization, compatibility, and integration of metabolomic data on all levels, including its compilation into databases. However, issues such as inconsistent inter-database compound nomenclature still remain, and as a result, facile database comparisons can be difficult.
One goal of single cell metabolomics is the quantitative and qualitative determination of the metabolite content of individual cells. The temporal and spatial dynamics of cellular metabolite levels, as well as metabolite functional involvement, are key factors in metabolomics, but logically relate to wider classical and interdisciplinary fields such as biochemistry and cellular physiology. There are a number of analytical technologies used to elucidate the metabolome of a single cell, including microfluidics, capillary LC, and capillary electrophoresis (CE), combined with a variety of detection modalities such as laser-induced fluorescence and MS. However, the high diversity in abundances and physicochemical parameters of the different metabolite classes present in a single cell often require the use of multistage technologies that combine a variety of analyte separation and detection modalities. Additionally, sample preparation, especially metabolome stabilization, is an important part of a metabolomic experiment. Isolation of single cells from tissues and the alternative, direct sampling from individual cells (without perturbing the cellular metabolome) can be technically difficult. For example, metabolites such as nitric oxide and carbon monoxide are short-lived and need to be preserved during sample preparation if they are to be studied. There are many preservation and stabilization approaches, often specific for particular experiments, as described below and elsewhere in the cited literature (e.g., [11,14]).
Here we focus on selected methods for investigating metabolites in single cells as well as their applications, with an emphasis on reports over the past two years, 2011 and 2012. There are a number of other reviews detailing the progress in single cell metabolomics prior to this time period [2,11,15,16]. We highlight approaches based on small-volume separations coupled with fluorescence or mass spectrometric detection. We also explore direct MS approaches because of their applicability to individual cell studies. The discussion is organized by the specific type of cell to be analyzed—bacterial, fungal, protist and plant, animal—rather than the technology. Specific single cell approaches applied to biotechnology can also be found in a recent review [17].
Bacteria
Bacteria are an abundant, widespread, dynamic, and genetically and biochemically diverse group of organisms. Considering also the functional heterogeneity inherent to bacterial populations, single cell analysis offers important advantages for following the dynamics of their cellular metabolome. With few exceptions, individual bacteria are quite small, 0.5–5.0 μm in length, and thus contain only trace amounts of most analytes. This makes them difficult targets for single cell bioanalyses; therefore, the need for highly sensitive detection methods is paramount. Raman microspectroscopy (RMS), secondary ion mass spectrometry (SIMS), and Fourier transform infrared spectroscopy (FTIRS) are among the bioanalytical technologies that are capable of detecting metabolites in individual bacterial cells (reviewed in [18], [19], and [20]).
RMS is nondestructive and offers fast data acquisition, as short as 1 millisecond. It was used for differentiation and identification of microbial cells [21,22], and investigation of their different functional parameters [23]. A laser is used to probe the cellular chemical composition in spot profiling and imaging modes. When complex biological samples are investigated, RMS generates chemical information on the abundance of classes of chemicals (e.g., lipids and proteins), a restricted number of individual analytes, and the incorporation of specific labels such as stable isotopes. For example, a red shift occurring after the incorporation of 13C (as 13CO2) into intracellular carotenoids helped to elucidate CO2 fixation by photosynthetic microorganisms such as Synechocystis sp. and Synechococcus elongates [23]; a 532 nm laser was used for Raman excitation of carotenoids, which absorb light in the 400–550 nm range. Three characteristic, dominant Raman bands were observed in these experiments. Incorporation of 13C into the carotenoids resulted in a microbial species-specific and concentration-dependent red shift of all three bands. These findings have furthered the investigation of heterogeneous microorganism populations and determination of fixed CO2 levels for different species. That said, RMS detection can only help to identify a small number of individual metabolites in a single cell, and thus offers low metabolome coverage.
SIMS is another bioanalytical tool used in single cell bacterial studies, providing good detection limits, high chemical specificity and high spatial resolution. SIMS imaging, an extension of this approach, probes the spatial distribution of analytes within intact or semi-intact cells with a lateral spatial resolution than can surpass 50 nm [24]. Recent and ongoing development of “cluster” ion sources such as C60+, Bi3+ and Au4004+ has also enhanced SIMS detection limits for intact biomolecules in the metabolite mass range, but generally low secondary ion yields for biomolecules still impose a significant limit on label-free detection at single cell and subcellular length scales. Since SIMS detection of elemental and diatomic secondary ions gives much higher yields and therefore offers lower (trace level) detection limits, one viable solution is to incorporate stable isotope tags such as 13C and 15N into a cell via nutrients, then visualize the distribution of the isotopes by the characteristic atoms and labeled small fragments. (See [25] for review and representative images.) For example, the determination of diatomic ion abundance ratios (e.g., 12C14N− and 12C15N−) has allowed the analysis of nitrogen gas fixation by the cellulolytic bacterium Teredinibacter turnerae [26]. Bacteria grown in an environment containing an experimentally added 15N2 tracer exhibited a time-dependent increase in the 15N/14N ratio. In contrast, this ratio remained stable over time for Enterococcus faecalis cells exposed to the same condition.
Protists, fungi and plants
Protists, fungi, and plants are important targets of study for a variety of applied and fundamental reasons, including their essential role in the development of biotechnologies. Specifically, the availability of relatively large, mechanically stable, and well investigated protist and plant cellular models has been instrumental in recent advances in single cell metabolomics.
As one example, cells of the algae Closterium acerosum are large—they vary between 250–790 μm in length and can be 25–84 μm wide—making them good models for the development and optimization of single cell metabolomics approaches. Not only does their size make them especially convenient for the visual observation of morphology, analyte extraction is relatively straightforward when using sample volumes that are easy to handle. Metabolites in single cells of this organism were investigated using negative-mode matrix-assisted laser desorption/ionization (MALDI) MS by the Zenobi group [27]. Cells were deposited on modified MALDI sample plates and the protocols used allowed quenching of metabolic activity, analyte extraction, instrument mass calibration, and assessment of relative quantities of measured analytes. Four metabolites, ADP, ATP, GTP and UDP-glucose, were subjected to targeted MS investigation in two populations of cells, both activated and inactivated by cold, at several time points. Successful semiquantitative detection of the compounds, and analyses of the resulting data using principal component analysis (PCA) and support vector machines analysis, revealed significant differences in metabolite levels between cells of the two groups.
MALDI-MS was also instrumental in the investigation of 12C and 13C stable isotope endogenous incorporation into uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) and its localization in the single, ~10 μm in diameter, hyphae of the red bread mold Neurospora crassa mycelium [28]. Twenty-four metabolites were observed in another fungi—the yeast Saccharomyces cerevisiae —using laser-based nanophotonic ionization, which utilizes microfabricated nanopost arrays [29].
It is worth mentioning that selection of the appropriate biological model is integral to the success of metabolomics investigations, as evidenced in a study of differential metabolite distribution between cytoplasm and vacuoles. To overcome typical organelle size limitations, Oikawa et al. [30] used a giant internodal cell of Chara australis (also known as Chara coralline), reaching up to 20 cm in length, more than 300 um in diameter, and 50 μL in volume, in their subcellular investigation of the vacuole metabolome. After both ends of the studied cell were cut, samples were collected by gravity-assisted flow. Interestingly, the purity of the isolated vacuolar samples was successfully determined by testing for vacuole- and cytoplasm-specific enzyme activity. Using CE-MS, 125 known metabolites were detected. Hierarchical cluster analysis of the data revealed a set of 11 vacuole-specific metabolites, including hydrophobic amino acids. These substances can be considered as vacuolar biomarkers in the C. australis. The large single cell size permitted the examination of several functional correlations in the organelle metabolome. It was found that the signal abundances of many vacuolar and cytoplasmic metabolites fluctuated in opposite directions under various environmental conditions, including the light/dark cycle (Figure 1), heat stress, and CO2 deficiency.
Figure 1.
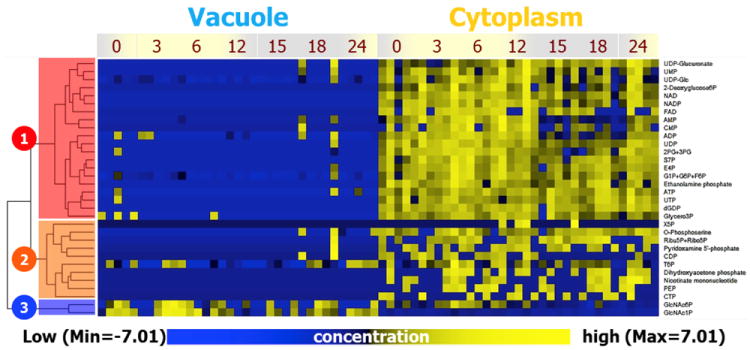
Analyses of vacuolar and cytoplasmic phosphate metabolites of Chara australis based on hierarchical cluster analysis shows that they are distinct. Some dependence on the 24 h light/dark cycle is detected. Three major data clusters represent cytoplasm-type metabolites (clusters 1 and 2) and vacuole-type metabolites (cluster 3). Reprinted from an open access source [30].
Masujima and colleagues [31] demonstrated subcellular metabolite analysis of smaller live plant cells using glass micropipette sampling of intracellular content, followed by MS analysis (Figure 2). The same sampling pipette containing the analytes was also utilized in the MS step as an electrospray ion nanosource. The sample was diluted inside the micropipette with 1–5 nL of a formic acid solution in acetonitrile, then electrosprayed into a high resolution LTQ Orbitrap XL (ThermoScientific). Despite the small sample volume, around a thousand signals were detected in the metabolite molecular mass range in different types of individual cells of the garden geranium (Pelargonium zonale). Visual control of the sampling process enabled the selection of cells based on their morphological features and localization. Metabolites in single leaf, stem, and petal cells were investigated and the resulting data analyzed with t-test and PCA. Statistical tests revealed differences in the metabolomic composition of cells in accordance with their tissue origin in the plant. Factors contributing to the identification of some metabolites were the high sensitivity of detection, high accuracy of mass measurement (0.3–0.7 ppm), use of analyte standards, and low dilution of the samples using single cell high-energy collision-induced dissociation (CID), with the mass spectrometer operating in tandem MS (MS/MS) mode. Additional structural information for isomers that were difficult to distinguish in the MS/MS mode was obtained with CID and the MS3 mode. However, single cell samples did not contain enough material for successful investigation in the MS3 mode; therefore, larger samples were sequentially accumulated from 2–3 cells of the same origin and analyzed. Using this sampling approach, analyte standards, and CID, one of the observed metabolites was identified as blumenol B.
Figure 2.
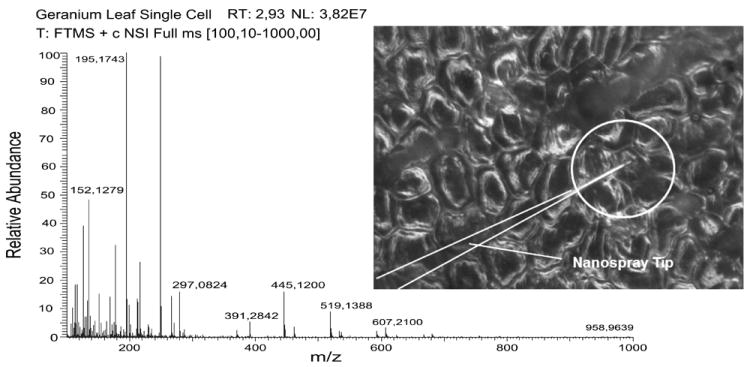
Live single-cell mass spectrometry was used to examine metabolite content in a variety of cells, including leaf cells from Pelargonium zonale. A representative mass spectrum of sample collected from an individual cell using a nanoelectrospray tip (tip is encircled). Adapted with permission from [31]. Copyright 2012 American Chemical Society.
Improved ionization of analytes, and therefore metabolome coverage, can be attained by combining laser ablation with electrospray ionization (or LAESI) with MS, used successfully to investigate metabolites in single onion (Allium cepa) epidermal cells [32]. Among the variety of analytes detected was cyanin, the purple pigment molecule of the tissue. The cell-by-cell molecular image of the cyanin distribution correlated well with the color of cells in the tissue, thus validating the semi-quantitative MS measurements. However, only a fraction of cellular content is collected with this approach.
Although there are obvious advantages to the ability to use different analytical approaches to analyze the same individual cell, these methods require low sample consumption to ensure minimal analyte loss during the initial stages of analysis, as well as compatible single cell sample preparation approaches. Clearly, the development of analytical technologies for multiplatform, multidimensional microanalysis will greatly benefit single cell metabolomics. For example, in a collaborative study by the Sweedler and Bohn groups [33], sequential investigation of the same cells by nondestructive confocal Raman microscopy (CRM) with high spatial resolution, partially destructive SIMS imaging, and relatively highly-destructive laser desorption ionization (LDI) MS imaging, enabled the determination of cellulose and lignin localization in the perennial grass Miscanthus x giganteus with cellular and subcellular spatial resolution (Figure 3). The lateral spatial resolution of the SIMS imaging and CRM was about 1 μm or less; the LDI imaging utilized a larger probe (~50 μm diameter) and thus provided lower spatial resolution, but ionized and detected larger molecules. The results importantly demonstrated that the intracellular globular structures observed in the study were composed of hemicellulose-rich lignin complexes. Furthermore, this study combined two analytical platforms—CRM and MS—to unambiguously identify metabolites of interest, whereas each platform on its own would not have done so. The focus was on the analysis of lignocellulosic materials as potential biofuel feedstock, but a wide variety of additional metabolite-related chemical information was collected in this work as well, and has yet to be presented. A similar combination of LDI-MS and RMS imaging was also implemented to investigate metabolites in individual unicellular organisms (Euglena gracilis) [34].
Figure 3.
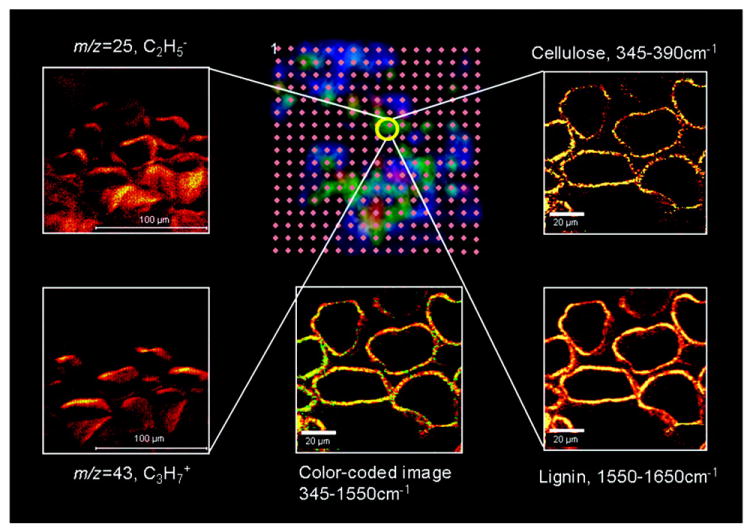
Sequential investigation of samples with several analytical technologies yields increased chemical information content and metabolome coverage. The LDI-MS/SIMS/CRM spatial correlation strategy is illustrated using tall perennial grass Miscanthus x giganteus cross sections. The LDI-MS grid (center top) is color-coded, corresponding to the intensity of m/z = 45 ions, obtained by laser desorption/ionization excitation spots on 100 μm centers. The yellow circle highlights the spot where high-resolution imaging was performed by both negative (m/z = 25, C2H-, top left) and positive (m/z = 43, C3H7+, bottom left) ion SIMS, as well as CRM, characterized by the cellulose band, 345–390 cm-1 (top right), and the lignin band, 1550–1650 cm-1 (bottom right). (Bottom center) Composite CRM image combining information from both cellulose (green) and lignin (yellow) bands. Reprinted with permission from [33]. Copyright 2010 American Chemical Society.
Animals
Individual animal cells are an important target for single cell fundamental and applied metabolomics research, but also present distinct and significant challenges. Their relative fragility, location in cohesive (i.e., difficult to dissociate) tissues, and the rapid changes they undergo in the cellular environment in response to different influences, including sample preparation, are among these challenges.
The increasing potential of single cell metabolomics in animals was shown in experiments where more than 300 distinct signals in the metabolite molecular mass range were observed in individual neurons using CE-ESI-MS [35]; recently, this system was used to document differences in the metabolome between freshly isolated and cultured neurons [36]. In both studies, identified Aplysia californica neurons were isolated and their metabolites immediately extracted in a 0.5% (v/v) acetic acid methanol solution, which also served to quench intracellular enzymatic activity. A small portion, in some cases just 0.1%, of the content of an individual neuron was loaded onto the CE capillary and separated. The high mass resolution and ~5 ppm mass accuracy obtained allowed comparisons of data sets from different cells, as well as metabolite identification, which was also assisted by matching the fragmentation patterns and migration times of corresponding standards and endogenous molecules. Of the more than 300 ion signals, >30 metabolites in single cell samples were unambiguously identified. Additionally, PCA analysis of the data sets representing 144 signals revealed significant differences in the metabolomes of different identified Aplysia neurons. As an example, data points of presumably homologous but differently located neurons grouped together and did not overlap with data representing other cell types. Absolute concentrations of several metabolites in the isolated neurons were determined using calibration curves produced by corresponding standards; intracellular glutamic acid concentration was 11 mM in one type of neuron and 4 mM (± 20%) in others.
Presently, CE-ESI-MS is a relatively low-throughput approach in that each cell analysis takes more than 15 min. However, we expect this timeframe will be reduced with further development and application of faster measurement schemes, which may include parallel separation and MALDI MS detection [37,38] in spot probing and imaging modes. For example, remarkable improvement in metabolite identification in individual HeLa cells was achieved by direct MALDI MS in experiments utilizing high mass resolution (the experimental mass resolving power ranged from 20,000 to 100,000) and a highly sensitive mass spectrometer [39]. The desorption and ionization of analytes from a 7-μm wide spot on a single cell were sufficient to identify 20 molecular features, including adenine, guanine, cholesterol, and two phospholipids, PC(16:0/16:1) and PC(16:0/18:1). Moreover, the spatial distributions of these and other detected compounds were determined by MS operating in imaging mode.
Vertebrate studies in biotechnology-related fields range from agriculture research to examining the fundamental mechanisms of the human body’s response to pharmaceuticals (pharmacodynamics). There are a number of important examples of new bioanalytical technologies that have been developed and applied to the study of metabolites in single cells, such as metabolomic cytometry, developed by the Dovichi group [5]. This unique CE system, equipped with a two-spectral channel laser-induced fluorescence detector having a six orders of magnitude dynamic range, allowed the simultaneous investigation of two glycosphingolipid metabolic pathways in individual primary rat cerebellar cells, including neurons. The sensitivity of the analyte detection reached 1 zmol for BODIPY-FL and 500 ymol for tetramethylrhodamine standard solutions. The micellar electrokinetic capillary chromatography employed in these experiments exhibited high separation efficiency in the 400,000–600,000 range. In this work, GM1-TMR and LacCer-BODIPY-FL fluorescently labeled lipids were used as exogenous substrates to examine the glycosphingolipid metabolic pathways. Incubation of cells in a mixture of these compounds led to their accumulation and intracellular metabolism by a variety of enzymes. For individual cell analysis, cultured cells were loaded into a CE capillary, lysed with Triton X-100, followed by analyte separation and detection. GM1-TMR catabolites, such as GD1a, GM2, GM3, glucosylceramide (GlcCer) and ceramide (Cer), were detected, as where catabolites and anabolites of LacCer-BODIPY, including GlcCer, Cer, GM1, GD1a, and GM3, after the analysis of multicellular homogenates. Analyte identification was accomplished by matching analytes and corresponding standards by electrophoretic mobility. Significant variation in the signal intensities of incorporated substrates and intracellularly-produced metabolites was detected among the cells studied (Figure 4), indicating heterogeneity in the cell population.
Figure 4.
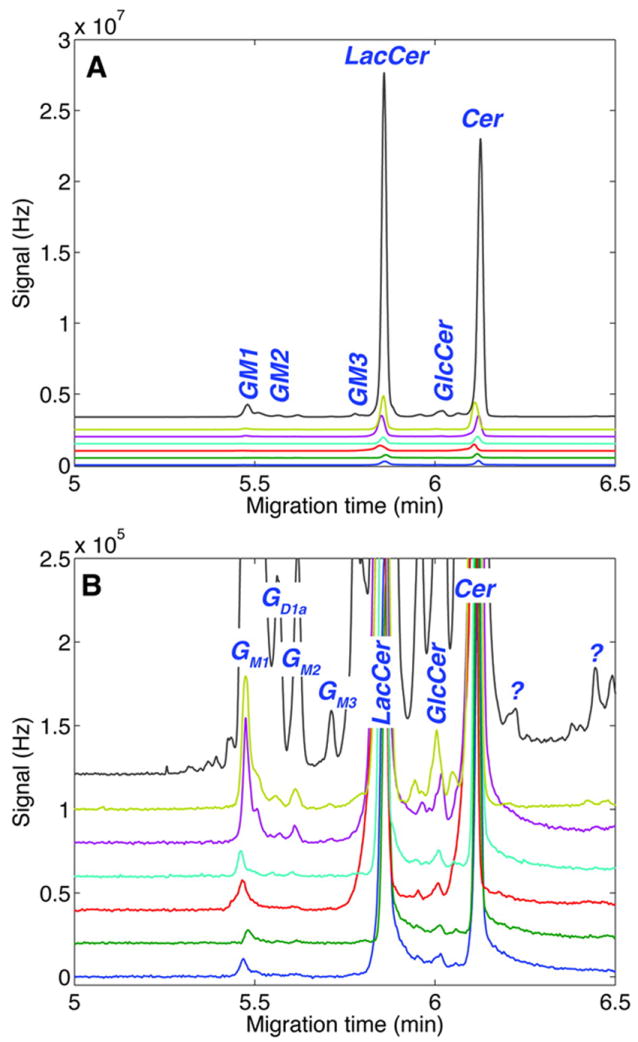
LacCer-BODIPY-FL metabolism in seven individual granule neurons is revealed by capillary electrophoresis. Representative electropherograms are shown at (A) full scale and (B) expanded scale. Several unidentified compounds marked with a question mark are also observed. (Reprinted (adapted) with permission from [5]. Copyright 2012 American Chemical Society)
High resolution chemical imaging techniques such as SIMS and atom probe tomography (APT) [40] present an exciting opportunity to study metabolites and/or their components in single cells, with the added value of spatial information. Single cell SIMS imaging experiments cannot easily be hyphenated to chromatographic or electrophoresis separations (e.g., LC and GC) without some loss of spatial information. As a result, mass spectra generated by single cell SIMS imaging are complex and are complicated by analyte fragmentation and ion suppression; nonetheless, the observed spectral complexity encodes rich and unique spatiochemical information that can be mined for metabolomic data. For relative quantitation of specific metabolites, as in the case of bacteria, incorporation of stable isotope label nutrients into the mammalian cell and subcellular structures can be tracked using SIMS. The Ewing group [41] recently employed this approach to measure the accumulation of extracellular membrane phospholipids into pheochromocytoma PC12 cell membranes as a function of the environmental concentrations of phospholipids. PC12 cells were cultured in medium enriched with deuterated membrane phospholipids—either phosphatidylcholine (PC) or phosphatidylethanolamine (PE)— and then the cells were freeze-fractured; those with exposed outer membranes were imaged by SIMS. A deuterated fatty acid fragment ion, C3D7+ (m/z 50.1), was found to be the most reliable signal for measurement of enriched lipid content, and relative quantitation was performed using the ratio of the C3D7+ and non-deuterated phosphocholine headgroup (m/z 184.1) signals. Results showed that uptake and membrane incorporation of the labeled lipids ranged from 0.5%–2% for PC and 1–9% for PE, depending on environmental concentrations. This study demonstrated that SIMS imaging is a potent tool for the detection of very small changes in the metabolite composition of cell membranes. The results also suggest that the observed changes can have significant effects on cell function (e.g., exocytosis), as demonstrated previously by the same group [42]. In another recent study, dietary 13C-palmitate was administered to Drosophila and then visualized with SIMS imaging in intestinal tissue (enterocytes) and body fat (adipocytes) [43]. Remarkable high-resolution chemical images representing the 13C/12C ratio showed preferential incorporation of the fatty acid into enterocytes. Following the labeled palmitate with an unlabeled chase helped to determine the turnover rate of the analyte.
Single cell atom probe tomography (APT) is a promising imaging technology, providing an opportunity to determine the localization of atomic and small molecular ions in cells [40]. Three-dimensional imaging with almost 1 nm spatial resolution is possible on conical focused ion beam (FIB)-milled single cell samples. The approach has the potential to elucidate the subcellular localization of stable isotope-labeled metabolites. APT has a limited molecular mass range for analyte detection (<100 Da), so metabolome coverage is restricted to small analytes.
Challenges and future breakthroughs in single cell metabolomics
Thus far, the majority of metabolomic single cell investigations have yielded information on a small fraction of the estimated full cell metabolome. However, more recently, several hundred metabolites were detected in a single cell [31,35]. Is determination of the complete single cell metabolome even possible? Theoretically it is, assuming we achieve adequate analytical capabilities, specifically with regard to mass resolution, sensitivity, and specificity. Key elements of the optimal microanalytical toolset should be sequential separation, extraction, and/or derivatization of analytes, including lipophilic, hydrophilic, small, and large compounds. Excellent detection limits are paramount for success, with the ultimate level of sensitivity being single molecule detection. Currently, single molecule detection is a fast developing field [44,45], and can be performed on platforms capable of on-line analyte separation and modification such as microfluidic devices [44]. That said, most studies using these detection methods have focused on larger molecules such as DNA and proteins. Metabolites typically are smaller, may contain fewer fluorophores (if any), and are therefore harder to characterize with these approaches, and they require analyte preselection. Thus, although single molecule approaches provide one exciting avenue, they currently do not yield the information content of many other platforms.
Perhaps the highest throughput single cell measurement approach is flow cytometry (FC) (for review see [46]). This technology can be used to analyze thousands of cells in a short time period. Usually FC uses fluorescence detection; to increase the number of independent measurement channels, MS-based detection is possible. One new embodiment is “mass cytometry” [47], which utilizes FC with ICP-MS ionization and molecular probe labels containing rare earth elements. The isotopes incorporated into the molecular probes are not present at high concentrations in the cells of interest in vivo. Therefore, recognition of cellular proteins with specific antibodies containing these atoms (e.g., ytterbium 171, neodynium, or samarium 152 [37]), and consecutive analysis using ICP-MS, allow reliable detection of these marker atoms and therefore, antigens. As a result, hyphenated FC-ICP-MS can be used to simultaneously perform quantitative analysis of more than 34 parameters, including the binding of 31 antibodies [48], cell viability, DNA content, and relative cell size at up to 1000 cells/s. Metabolites and pharmaceuticals also can be investigated with mass cytometry, including cases where a marker atom is incorporated into the analyte molecule itself, rather than in an affinity probe [49]. As the major effort of mass cytometry work has been to investigate proteins in single cells, there are no examples of it being used to probe metabolites, despite the abundant availability of antibodies for small molecules.
A wide variety of metabolites play important roles in most major cellular functions. The ability to detect, identify, and quantify a broad range of metabolites in a single, multistage experiment is one of the technological goals under current development in single cell bioanalytical research. A variety of bioanalytical technologies have been recently advanced for individual cell metabolome studies, some of which have been described here. Despite progress in the capability to detect a number of analytes, single cell metabolome coverage remains relatively low. New bioanalytical technologies that hyphenate distinct analyte extraction, separation, and detection approaches are needed in order to improve single cell metabolome coverage. Single cell metabolome data is becoming an essential component of systems biology modeling, and therefore, additional efforts are necessary in the development and implementation of analyte identification and quantification capabilities into each single cell metabolomics experiment.
Highlights.
-
►
Single cell metabolomics provides information on the heterogeneity of populations of cells
-
►
Current technologies allow hundreds of metabolites to be measured from a single cell
-
►
Quantitative metabolome information at the single cell level aids systems biology modeling
-
►
Sample preparation remains a significant challenge in single cell metabolomics
Acknowledgments
This work was supported by Award No. P30 DA018310 from the National Institute on Drug Abuse, and Award No. DE-SC0006642 from the Department of Energy. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. We also thank Stephanie Baker for assistance with manuscript.
Footnotes
The authors declare they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schwab W. Metabolome diversity: Too few genes, too many metabolites? Phytochemistry. 2003;62:837–849. doi: 10.1016/s0031-9422(02)00723-9. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Bodovitz S. Single cell analysis: The new frontier in ‘omics’. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kell DB. Metabolomics and systems biology: Making sense of the soup. Curr Opin Microbiol. 2004;7:296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Paul Lee WN, Wahjudi PN, Xu J, Go VL. Tracer-based metabolomics: Concepts and practices. Clin Biochem. 2010;43:1269–1277. doi: 10.1016/j.clinbiochem.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Essaka DC, Prendergast J, Keithley RB, Palcic MM, Hindsgaul O, Schnaar RL, Dovichi NJ. Metabolic cytometry: Capillary electrophoresis with two-color fluorescence detection for the simultaneous study of two glycosphingolipid metabolic pathways in single primary neurons. Anal Chem. 2012;84:2799–2804. doi: 10.1021/ac2031892. A exciting example of wide dynamic range capillary electrophoresis applied to investigate labeled compound metabolism in individual mammalian cells. The work exemplifies the use of CE in a functional metabolomics study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essaka DC, Prendergast J, Keithley RB, Hindsgaul O, Palcic MM, Schnaar RL, Dovichi NJ. Single cell ganglioside catabolism in primary cerebellar neurons and glia. Neurochem Res. 2012:1–7. doi: 10.1007/s11064-012-0733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barding GA, Jr, Salditos R, Larive CK. Quantitative NMR for bioanalysis and metabolomics. Anal Bioanal Chem. 2012;404:1165–1179. doi: 10.1007/s00216-012-6188-z. [DOI] [PubMed] [Google Scholar]
- 8.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Olson DL, Peck TL, Webb AG, Magin RL, Sweedler JV. High-resolution microcoil H-1-NMR for mass-limited, nanoliter-volume samples. Science. 1995;270:1967–1970. [Google Scholar]
- 10.Grant SC, Aiken NR, Plant HD, Gibbs S, Mareci TH, Webb AG, Blackband SJ. NMR spectroscopy of single neurons. Magn Reson Med. 2000;44:19–22. doi: 10.1002/1522-2594(200007)44:1<19::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Profiling metabolites and peptides in single cells. Nat Methods. 2011;8:S20–S29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Go EP. Database resources in metabolomics: An overview. J Neuroimmune Pharmacol. 2010;5:18–30. doi: 10.1007/s11481-009-9157-3. [DOI] [PubMed] [Google Scholar]
- 13.Fiehn O, Robertson D, Griffin J, van der Werf M, Nikolau B, Morrison N, Sumner LW, Goodacre R, Hardy NW, Taylor C, et al. The metabolomics standards initiative (MSI) Metabolomics. 2007;3:175–178. [Google Scholar]
- 14.Villas-Boas SG, Hojer-Pedersen J, Akesson M, Smedsgaard J, Nielsen J. Global metabolite analysis of yeast: Evaluation of sample preparation methods. Yeast. 2005;22:1155–1169. doi: 10.1002/yea.1308. [DOI] [PubMed] [Google Scholar]
- 15.Heinemann M, Zenobi R. Single cell metabolomics. Curr Opin Biotechnol. 2011;22:26–31. doi: 10.1016/j.copbio.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Amantonico A, Urban PL, Zenobi R. Analytical techniques for single-cell metabolomics: State of the art and trends. Anal Bioanal Chem. 2010;398:2493–2504. doi: 10.1007/s00216-010-3850-1. [DOI] [PubMed] [Google Scholar]
- 17.Fritzsch FS, Dusny C, Frick O, Schmid A. Single-cell analysis in biotechnology, systems biology, and biocatalysis. Annu Rev Chem Biomol Eng. 2012;3:129–155. doi: 10.1146/annurev-chembioeng-062011-081056. [DOI] [PubMed] [Google Scholar]
- 18.Wagner M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411–429. doi: 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]
- 19.Read DS, Whiteley AS. Identity and function of single microbial cells within a community by Raman microspectroscopy and related single-cell techniques. In: Sen K, Ashbolt NJ, editors. Environmental microbiology: Current technology and water applications. Caister Academic Press; 2011. pp. 163–178. [Google Scholar]
- 20.Wagner H, Dunker S, Liu Z, Wilhelm C. Subcommunity FTIR-spectroscopy to determine physiological cell states. Curr Opin Biotechnol. 2012 doi: 10.1016/j.copbio.2012.1009.1008. [DOI] [PubMed] [Google Scholar]
- 21.Guicheteau J, Christesen S, Emge D, Tripathi A. Bacterial mixture identification using Raman and surface-enhanced Raman chemical imaging. J Raman Spectrosc. 2010;41:1632–1637. [Google Scholar]
- 22.Xie C, Mace J, Dinno MA, Li YQ, Tang W, Newton RJ, Gemperline PJ. Identification of single bacterial cells in aqueous solution using conflocal laser tweezers Raman spectroscopy. Anal Chem. 2005;77:4390–4397. doi: 10.1021/ac0504971. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Canniffe DP, Jackson PJ, Davison PA, FitzGerald S, Dickman MJ, Burgess JG, Hunter CN, Huang WE. Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J. 2012;6:875–885. doi: 10.1038/ismej.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kollmer F, Paul W, Krehl M, Niehuis E. Ultra high spatial resolution SIMS with cluster ions — approaching the physical limits. Surf Interface Anal. 2012 doi: 10.1002/sia.5093. [DOI] [Google Scholar]
- 25.Musat N, Foster R, Vagner T, Adam B, Kuypers MMM. Detecting metabolic activities in single cells, with emphasis on nanosims. FEMS Microbiol Rev. 2012;36:486–511. doi: 10.1111/j.1574-6976.2011.00303.x. [DOI] [PubMed] [Google Scholar]
- 26.Lechene CP, Luyten Y, McMahon G, Distel DL. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science. 2007;317:1563–1566. doi: 10.1126/science.1145557. [DOI] [PubMed] [Google Scholar]
- 27.Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R. Single-cell MALDI-MS as an analytical tool for studying intrapopulation metabolic heterogeneity of unicellular organisms. Anal Chem. 2010;82:7394–7400. doi: 10.1021/ac1015326. [DOI] [PubMed] [Google Scholar]
- 28.Hu JB, Chen YC, Urban PL. On-target labeling of intracellular metabolites combined with chemical mapping of individual hyphae revealing cytoplasmic relocation of isotopologues. Anal Chem. 2012;84:5110–5116. doi: 10.1021/ac300903x. [DOI] [PubMed] [Google Scholar]
- 29.Walker BN, Stolee JA, Vertes A. Nanophotonic ionization for ultratrace and single-cell analysis by mass spectrometry. Anal Chem. 2012;84:7756–7762. doi: 10.1021/ac301238k. [DOI] [PubMed] [Google Scholar]
- 30*.Oikawa A, Matsuda F, Kikuyama M, Mimura T, Saito K. Metabolomics of a single vacuole reveals metabolic dynamism in an alga chara australis. Plant Physiol. 2011;157:544–551. doi: 10.1104/pp.111.183772. A demonstration of the utility of a large single cell model for investigation of the metabolome and determination of functional changes in the metabolite profiles of subcellular regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Lorenzo Tejedor M, Mizuno H, Tsuyama N, Harada T, Masujima T. In situ molecular analysis of plant tissues by live single-cell mass spectrometry. Anal Chem. 2012;84:5221–5228. doi: 10.1021/ac202447t. This study demonstrates both intracellular sampling and ESI-MS detection of metabolites from single plant cells. Observation of hundreds of compounds and identification of several metabolites are presented. The utility of high resolution ESI-MS for single cell analysis is demonstrated. [DOI] [PubMed] [Google Scholar]
- 32.Shrestha B, Patt JM, Vertes A. In situ cell-by-cell imaging and analysis of small cell populations by mass spectrometry. Anal Chem. 2011;83:2947–2955. doi: 10.1021/ac102958x. [DOI] [PubMed] [Google Scholar]
- 33**.Li Z, Chu LQ, Sweedler JV, Bohn PW. Spatial correlation of confocal Raman scattering and secondary ion mass spectrometric molecular images of lignocellulosic materials. Anal Chem. 2010;82:2608–2611. doi: 10.1021/ac100026r. An example of a multiplatform, chemically-information rich investigation of plant tissue with single cell resolution and complementary chemical information output. The work presents a strategy that allows Raman and mass spectrometry imaging on the same samples. [DOI] [PubMed] [Google Scholar]
- 34.Urban PL, Schmid T, Amantonico A, Zenobi R. Multidimensional analysis of single algal cells by integrating microspectroscopy with mass spectrometry. Anal Chem. 2011;83:1843–1849. doi: 10.1021/ac102702m. [DOI] [PubMed] [Google Scholar]
- 35**.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Metabolic differentiation of neuronal phenotypes by single-cell capillary electrophoresis-electrospray ionization-mass spectrometry. Anal Chem. 2011;83:6810–6817. doi: 10.1021/ac2015855. The combination of small volume samples with CE and ESI-MS analysis achieved extended metabolome coverage so that hundreds of peaks were obtained for each cell studied; the identification of 36 intracellular metabolites, along with quantitative analysis, highlight the versatility of this strategy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Single-cell metabolomics: Changes in the metabolome of freshly isolated and cultured neurons. ACS Chem Neurosci. 2012;3:782–792. doi: 10.1021/cn300100u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaidyanathan S, Goodacre R. Quantitative detection of metabolites using matrix-assisted laser desorption/ionization mass spectrometry with 9-aminoacridine as the matrix. Rapid Commun Mass Spectrom. 2007;21:2072–2078. doi: 10.1002/rcm.3063. [DOI] [PubMed] [Google Scholar]
- 38.Rubakhin SS, Sweedler JV. Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal Chem. 2008;80:7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schober Y, Guenther S, Spengler B, Rompp A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal Chem. 2012;84:6293–6297. doi: 10.1021/ac301337h. [DOI] [PubMed] [Google Scholar]
- 40.Narayan K, Prosa TJ, Fu J, Kelly TF, Subramaniam S. Chemical mapping of mammalian cells by atom probe tomography. J Struct Biol. 2012;178:98–107. doi: 10.1016/j.jsb.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanekoff I, Sjövall P, Ewing AG. Relative quantification of phospholipid accumulation in the PC12 cell plasma membrane following phospholipid incubation using TOF-SIMS imaging. Anal Chem. 2011;83:5337–5343. doi: 10.1021/ac200771g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchiyama Y, Maxson MM, Sawada T, Nakano A, Ewing AG. Phospholipid mediated plasticity in exocytosis observed in PC12 cells. Brain Res. 2007;1151:46–54. doi: 10.1016/j.brainres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, Lechene CP. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–519. doi: 10.1038/nature10734. SIMS-based, high-resolution chemical imaging allowed studies on individual cells while retaining stable isotope- containing functional tracers. Subcellular spatial resolution and quantitative analysis are among the methodological achievements of this work. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C, Qu Y, Luo Y, Fang N. Recent advances in single-molecule detection on micro- and nano-fluidic devices. Electrophoresis. 2011;32:3308–3318. doi: 10.1002/elps.201100159. [DOI] [PubMed] [Google Scholar]
- 45.Sako Y, Hiroshima M, Pack CG, Okamoto K, Hibino K, Yamamoto A. Live cell single-molecule detection in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2012;4:183–192. doi: 10.1002/wsbm.161. [DOI] [PubMed] [Google Scholar]
- 46.Diaz M, Herrero M, Garcia LA, Quiros C. Application of flow cytometry to industrial microbial bioprocesses. Biochem Eng J. 2010;48:385–407. [Google Scholar]
- 47.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Bendall SC, Simonds EF, Qiu P, Amir EAD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang CN, Ho KS, Sun HZ, Chan WT. Tracking bismuth antiulcer drug uptake in single Helicobacter pylori cells. J Am Chem Soc. 2011;133:7355–7357. doi: 10.1021/ja2013278. [DOI] [PubMed] [Google Scholar]


