Abstract
RNA polymerase II-like enzymes carry out transcription of genomes in Eukaryota, Archaea, and some viruses. They also exhibit fundamental similarity to RNA polymerases from bacteria, chloroplasts, and mitochondria. In this review we take an inventory of recent studiesilluminating different steps of basic transcription mechanism, likely common for most multi-subunit RNA polymerases. Through the amalgamation of structural and computational chemistry data we attempt to highlight the most feasible reaction pathway for the two-metal nucleotidyl transfer mechanism, and to evaluate the way catalysis can be linked to translocation in the mechano-chemical cycle catalyzed by RNA polymerase II.
Keywords: RNA polymerase, nucleotidyl transfer, two-metal catalysis, transcription, molecular dynamics
Introduction
Transcription of cellular genomes in all domains of life is carried out by essentially orthologous enzymes, multi-subunit DNA-dependent RNA polymerases (RNAPs). The archetypal eubacterial core enzyme, represented by that of Escherichia coli, consists of two large subunits (β and β′)1,2, that carry out the chemical (NTP condensation) and mechanical (translocation) steps3,4, and three smaller ones, α2 dimer, and ω, that play roles in the assembly of the enzyme and regulation of transcription1,2,4,5 (Fig. 1A). Variant forms of the core are described for some Eubacteria, such as Francisella (featuring α1α2 heterodimer6), and Bacillus (adding δ and alternate ω subunits7), whereas in Helicobacter, Wolbachia and Wolinella RNAPs β and β′ subunits are fused together1. RNAP subunit initially described as γ in Nostoc, Anabaena, and other Cyanobacteria turned out to be a product of a split in the ancestral gene, coding for β′ ortholog8,9. A similar split exists in the β′ subunit of plastid-encoded RNAPs in chloroplasts, featuring a reduced bacterial-type core α2ββ′ (ω subunit appears to be encoded by plastid genomes only in Rhodophyta algae)10. Interestingly, this core architecture predominates in early chloroplast development (e.g. in etioplasts), whereas in mature chloroplasts it is augmented by up to 30 additional subunits11. Mitochondrial RNAPs are predominantly related to those of bacteriophages, but the relatively large mitochondrial genomes of some protists (namely Jakobidae and Malawimonadidae) encode orthologs of bacterial α, β, and β′ subunits12,13.
Figure 1. Orthologous bacterial RNA polymerase-like core in Eubacteria (A, 3lu0), Eukarya (B, 1nik), and Archaea (C, 3hkz).
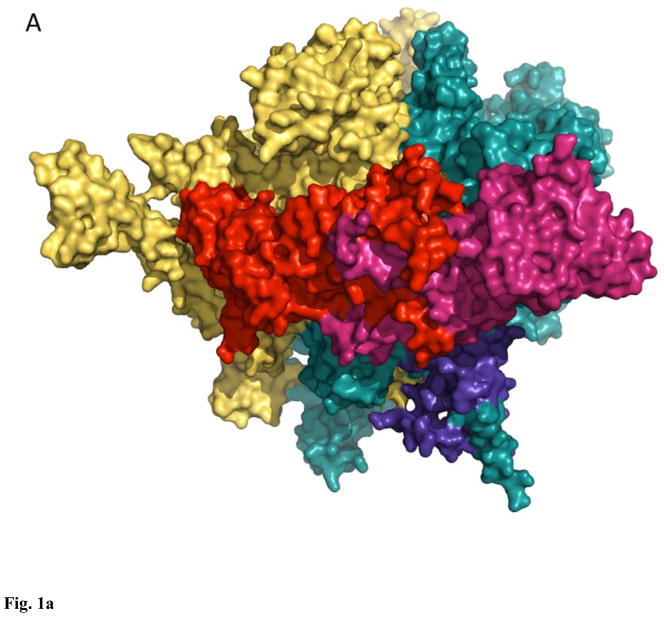
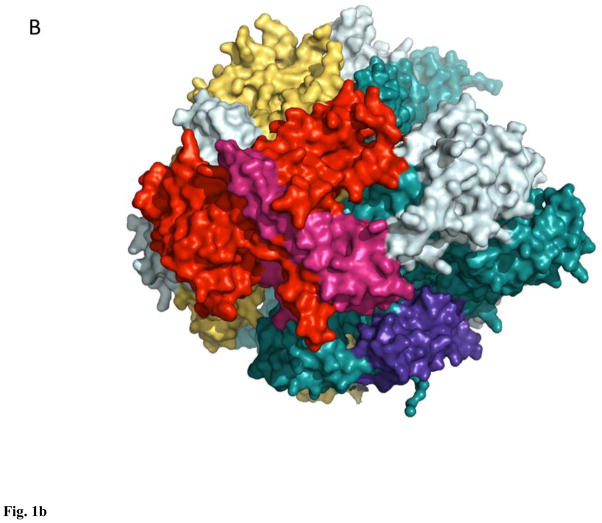
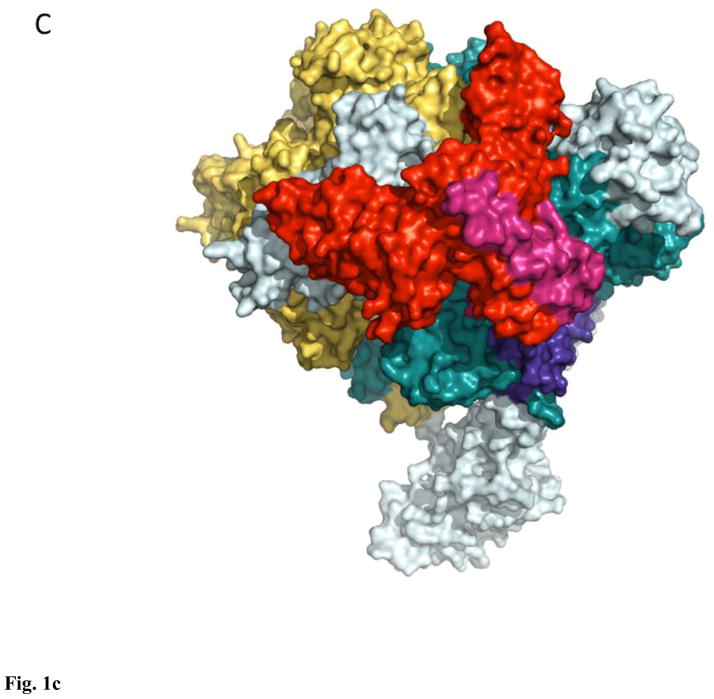
All enzymes are surface, light teal, β′-like subunits are deep teal, β-like subunits are yellow-orange, α-like subunits are red and warm pink, ω-like subunits are purple-blue.
Nuclear RNAPs in Eukaryota are represented by a minimal set of three classes: RNAPI transcribing ribosomal RNA genes, RNAPII carrying out the synthesis of messenger RNA and a subset of small non-coding RNAs, and RNAPIII synthesizing transfer RNAs, 5S RNA, and the bulk of small non-coding RNAs14–17. RNAPs from each class contain 12+ subunits, with a core orthologous to bacterial-type enzyme: yeast RNAPII largest subunits, Rpb1 and Rpb2, correspond to β′ and β, respectively, Rpb3 and Rpb11 are divergent orthologs of α, and Rpb6 is an ω ortholog18–20 (Fig. 1B). In plants the set of nuclear RNAPs is extended to 5, RNAPIV and V conforming to the overall architecture of RNAPI-III21–23, but exhibiting an unexpected divergence in the mechano-chemical core24,25, otherwise universally conserved in both pro- and eukaryotes, which led some to question the competence of these enzymes to act as bona fide RNAPs24 (this competence still eludes in vitro demonstration22). Genomes of large viruses from the order Megavirales encode RNAPs, related to the nuclear RNAPII, but with a reduced complement of subunits, typically numbering 826. Remarkably, whereas some enzymes, such as that of Mimivirus, feature a fully conserved α2ββ′ω-like core, namely the orthologs of Rpb1, 2, 3/11, and 6 (together with Rpb5, 9, and 10 homologs)27, RNAPs from poxviruses lack an apparent α ortholog28,29, indicating a potentially dramatic change in enzyme architecture and assembly. Lane and Darst, based on the results of their large-scale multiple sequence alignment for β,β′-like subunits, argued that poxvirus RNAPs were related to the nuclear RNAPI enzymes1. Finally, Archaea contain one type of RNAPs, similar in size (11–13 subunits) and composition to RNAPII: it comprises orthologs of Rpb1 (split into two subunits), 2–8, 10, and 1230,31 (Fig. 1C). Further in depth analysis of structural and sequence data pertaining to RNAPs from bacteria, archaea, and eukaryotes is provided by Cramer, Darst, Kornberg, Murakami, Yokoyama and colleagues1,2,18–20,30–33. In this review we will focus on the basic mechanism of transcription, as it emerges from studies of yeast RNAPII, complemented when necessary with the relevant data for bacterial enzymes.
Nucleotide addition cycle
Synthesis of RNA by RNAP comprises three distinct stages, initiation, elongation, and termination33–37. The second and longest stage, elongation, is a reiterative mechano-chemical process that consist of a single NTP condensation reaction, that extends nascent RNA by one NMP with concomitant release of pyrophosphate (PPi), followed by enzyme translocation along the template by one nucleotide (nt)36,38,39.
RNAP is different from the classic Cleland enzymes in that it does not dissociate one of the reaction products, RNA, each cycle, it remains bound to the enzyme, and becomes a substrate as the new cycle begins. Nascent RNA is annealed to the template strand of DNA for 8–9 nts, forming so-called RNA-DNA hybrid, which is a major determinant of elongation complex stability20,40,41. The length of the hybrid is maintained constant throughout the cycle: as the 3′ end extends by one nt, another from the 5′ end of the hybrid peels off the template and is threaded through the RNA exit channel. Read-out of the template, governed by the Watson-Crick complementarity of the incoming NTP and the templating nt, requires melting of the double-stranded DNA to allow formation of the RNA-DNA hybrid. Locally melted DNA segment is known as transcription bubble, with single-stranded DNA chains reaching 11–12 nt in length being engaged by RNAP and the hybrid. Melting of DNA at the front edge of RNAP and its annealing at the back edge are thought to occur simultaneously with translocation and to be neutral in terms of free energy, if averaged over the length of the transcript.
Translocation returns 3′ end of RNA into the catalytically active position (so-called nucleophilic, or i site) and resets the enzyme for accepting the next NTP, so it is imperative that it spans only one nt of template DNA. Hypertranslocation occurs in vitro (stimulated by addition of a cognate NMP), as does retrograde translocation (backtracking), both in vitro and in vivo, indicating that there is no significant activation barrier to either of them42–46. The mechanism of limiting translocation to one nt step is not known, and so far translocation has not been replicated in molecular dynamics (MD) simulations. Crystallographic models exist, that may have captured translocation intermediates43,47, but the insights they provided are not sufficient to build an explicit MD or molecular mechanics (MM) trajectory of the translocating RNAP. It has been proposed that the “gating tyrosine” (Rpb2 Tyr769) physically delimits backtracking by sterically hindering base stacking beyond the first backtracked RNA nt42. It is also tempting to speculate that deformability of the so-called bridge helix (Rpb1 815–845), long implicated in various aspects of the transcription cycle, determines the single nt step of the forward translocation47–50. Both models await further experimental and computational interrogations. The size of the transcription bubble also appears to be maintained constant, even during backtracking or hypertranslocation, although it may expand by extending the size of the RNA-DNA hybrid leading to the formation of so called R-loops, detrimental to genome stability in vivo51.
NTP condensation is believed to occur via a general two-metal-ion mechanism, common for all nucleotidyl transferases, first put forth by Thomas Steitz52. The central feature of this mechanism is the employment of two magnesium cations, coordinated by at least two aspartate residues located in the active site. According to this general model the first magnesium (A) promotes deprotonation of the RNA 3′OH, facilitating 3′ O− attack on the substrate NTP α-phosphate, which in turn leads to formation of a new phosphodiester bond and a leaving group, PPi. The second magnesium ion (B) participates in stabilization of the pentacovalent transition state. This mechanism was further elaborated upon by Goldfarb and colleagues, who proposed the universal central role of two Mg cations coordinated in the RNAP active site in other reactions catalyzed by this enzyme: exo- and endo-ribonuclease, and exo- and endo-pyrophosporylase53,54. Detailed reaction geometries at the pseudo-atomic resolution were developed, consistent with the bulk biochemical interrogation of the E. coli RNAP. These reactions, although not situated in the main pathway of NTP condensation, play important roles in transcription fidelity, reactivation of arrested complexes, etc3,55.
Multi-subunit RNAPs feature an invariant triad of aspartate residues54,56 (Rpb1 Asp481, 483, 485), that coordinates both the persistently bound MgA and the NTP-bound MgB, thought to enter and leave the active site during each NTP addition cycle. Structural analysis of various NTP-bound RNAP complexes extended the boundaries of the active site to include a number of other residues, participating in catalysis, metal coordination, or substrate binding.
Composition of the active site and the details of coordination/interaction network remain a subject of some controversy, stemming from the occasional incongruity of structural models featuring mechanistically identical complexes. The atomistic details inferred from such models must be treated with a caveat in mind, namely that crystallographic models by their nature deal with catalytically inactive mimics of the on-pathway intermediates. In order to prevent catalysis in the NTP-bound RNAP crystals NTPs are often represented by their non-hydrolysable analogs (such as α,β-methylene derivatives GMPCPP or AMPCPP), in other cases the reactive 3′ OH group have been removed from the RNA57–59. These changes directly affect coordination of the RNA and/or NTP in the active site of the enzyme, whereas these and other interactions are subject to artifacts of crystallization, quality of the electron density and of the subsequent computational structure solution. In one of his recent papers addressing variations in NTP binding mode Patrick Cramer noted that “the exact positioning of the NTP depends on the experimental design”58. This remark highlights the fact that crystallographic structural models provide not only many crucial insights into the reaction geometry and dynamics, but also an occasional artifact, singular to a given experimental setup, that can dramatically affect the interpretation of the data. Unfortunately there are limitations on the degree to which such artifacts can be ameliorated through the improvements in the design of crystallographic experiments. So far no crystallographic model of any multi-subunit RNAP features intermediates of the NTP addition reaction, with all available structures representing either pre- or post-chemistry states. An intriguing possibility to explore the reaction mechanism by crystallographic means has been opened up by the successful application of time-resolved X-ray crystallography to the dNTP condensation by DNA polymerase η60. Having created crystals poised for catalysis by replacing catalytic Mg cations with inert Ca ones, Nakamura et al. were able to start the dNTP addition by restoring Mg cations to the system and stop it at different times by rapidly cooling the crystals to 77 K. As a result new details of the reaction mechanism emerged, such as previously unobserved and unforeseen third Mg cation in the active site of the enzyme coincident with PPi formation60. Confidence in the assigning electronic density to reactants or catalytic metals is significantly boosted in this case by the high resolution of the crystallographic models, ~ 1.5 Å, whereas the reverse is true for RNAP II, which structures are routinely resolved at 3–4 Å. Hence although NTP condensation by RNAPII should be amenable to similar time-resolved crystallographic analysis, the fine atomistic and mechanistic details of this reaction can only be derived with confidence from the models resolved at < 2 Å. Complementary methods, experimental as well as computational, have to be deployed in order to fully explore the atomic, kinetic and thermodynamic details of the reaction mechanism61. It bears noting that even the considerable power of modern MD and MM computations cannot mitigate the problems present in the starting structural models, such as poor resolution and/or refinement, which give rise to technically competent but materially spurious simulations. The growing appreciation of the fact that the quality of “structures” (crystallographic models) deposited with the Protein Data Bank varies greatly (but imperceptibly for many of the downstream users) led to the introduction of new and improved structure validation protocols62,63, whereas comprehensive structure evaluation and rebuilding/refinement are available from such resources as PDBREPORT (http://swift.cmbi.ru.nl/gv/pdbreport/) and PDB_REDO (http://www.cmbi.ru.nl/pdb_redo/)64,65. These tools can assist with assessment and choice of the crystallographic models, an issue of critical importance as the strength of the structure-based arguments (including computational ones) in many ways depends on the quality of the starting model.
RNA polymerase active site
The minimal composition of the RNAPII active site (Fig. 2) as it emerges from the crystallographic models of the enzyme42,43,57,58,66–73 in addition to the catalytic triad (Rpb1 Asp481, 483, 485) includes Rpb2 residues Asp837, that coordinates MgB, Lys987, and Arg1020, which form H-bonds with the substrate triphosphate, Rpb1 His1085 as a candidate general acid57,74, Arg446, Asn479 and Gln1078 that contact 2′ OH of the substrate and may participate in discrimination of NTPs over dNTPs, and Leu1081 which stacks on the NTP base. Non-peptide elements of the active site consist of 2 catalytic Mg ions, templating nt, that forms Watson-Crick pair with incoming NTP, and 3′ terminal nt in the RNA, which prior to the nucleophilic attack stacks on the base of the substrate NTP57–59,75. Gln1078, Leu1081, and His1085 are embedded into a metamorphic structural module of the RNAP, the so called trigger loop (TL, Rpb1 residues 1070–1100)57,76. TL is widely believed to adopt two obligatory conformations: open, with an unstructured/loop tip in absence of NTP, and closed, essentially an α-helical hairpin, which forms upon binding of substrate NTP38,57–59,77–79 (Fig. 3). The obligatory transition between two forms of the trigger loop, one of the most deformable/mobile elements of RNAP80 is most clearly seen in the bacterial enzyme59, whereas in yeast RNAPII it can adopt different conformations even in the presence of the substrate57,58. Notably, the TL in these complexes tended to adopt the closed conformation in presence of UTP or GTP, and remain in open one when bound to α,β-methylene derivative GMPCPP (Fig. 3), emphasizing the importance of proper phosphate coordination by the trigger loop residues (predominantly His1085)79 in binding and catalysis.
Figure 2. Active site of yeast RNA polymerase II (2e2h).
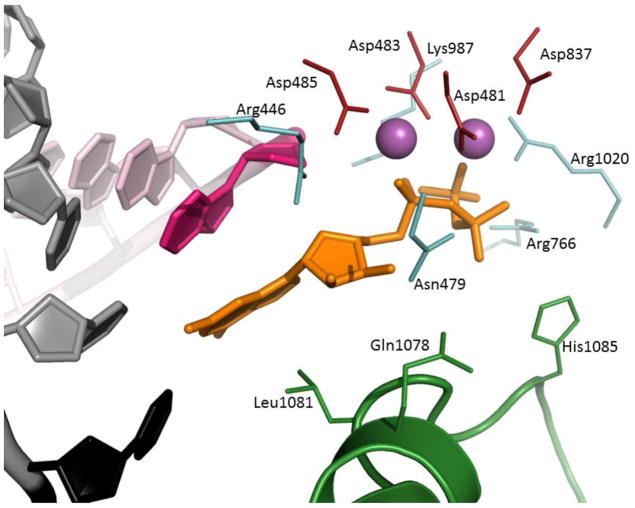
Template DNA is cartoon grey, except for the templating nucleotide (black), RNA is cartoon light pink, except for the 3′ nucleotide (hot pink). Substrate GTP is orange sticks. Catalytic Mg2+ cations are solid magenta spheres, catalytic tetrad (Asp481, 483, 485, and 837) is red sticks. Trigger loop is green cartoon, residues Gln1078, Leu1081, and His1085 are green sticks. The rest of amino acids are light teal sticks.
Figure 3. Structural variants of the trigger loop in yeast RNA polymerase II.
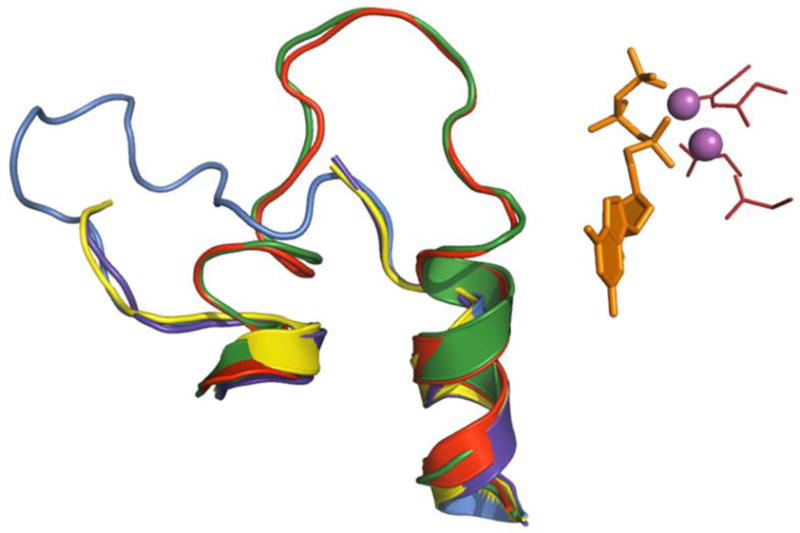
Elements of the active site (from GTP-bound 2e2h) are represented as in Fig. 2. (substrate is orange sticks, catalytic Mg2+ cations are solid magenta spheres, catalytic tetrad is red sticks) Superimposed are trigger loops (cartoon) from NTP-free enzyme (1sfo, purple blue), GMPCPP-bound 2e2j (yellow) and 2nvt (marine), and UTP-bound 2nvz (red).
Role of the invariant Asn479 in ribo-discrimination, based on structural and biochemical studies of bacterial RNAPs59,75, was called into question by Kornberg and colleagues based on structural analysis of yeast RNAPII57. Recently interactions between 2′ OH and Asn479 were observed in NTP-bound initiation complex of RNAPII58 and the apparent discrepancies were explained by the differential NTP coordination due to absence of the 3′ OH in the complexes crystallized by Wang et al.57
The Watson-Crick complementarity of the incoming NTP and the templating DNA nt is of paramount and obvious importance for the overall fidelity of transcription. However, the catalytic role of these interactions was not apparent until Kellinger et al. tested a set of non-polar isosteres of the templating nt in the NTP condensation81. These nucleotide analogs, matching the conventional DNA nts in size (within 1 Å) were completely devoid of the H-bond donors/acceptors, eliminating the need to consider alternate/wobbling bonding patterns. Remarkably, removal of the Watson-Crick complementarity with the incoming NTP reduced the apparent binding constant (KM) only 6-fold with concomitant drop in the kcat of about 5600-fold81. This prominent effect on catalysis clearly indicates that the Watson-Crick complementarity with its planar conformation of the bonding DNA-NTP pair orients the incoming substrate for deprotonation and subsequent nucleophilic attack, arguing for the inclusion of the templating nt among the catalytic elements of the RNAP active site.
Mechanism of NTP condensation
The theory of nucleotidyl transfer via two-metal catalysis does not specify the identity of the proton acceptor for the 3′ OH, nor the same of the proton donor for the PPi, whereas the proton inventory experiments suggested the general two-proton transfer mechanism for DNA and RNA polymerases and indicated that the PPi leaves active site in the protonated form74,82. The available structures of NTP-bound yeast and bacterial RNAPs57–59,69,73,83 allow only the exclusion of certain residues from direct participation in catalysis (e.g. as being too far removed from the active site), but not the unambiguous elucidation of reaction mechanism and intermediates. Some of these structures have been recently used in MD and other computational interrogations of the basic mechanism of NTP condensation.
Using the 4.3 Å resolution crystal structure of RNAPII elongation complex (2nvz57) as the starting model for MD simulations Zhu and Salahub explored some of the possible mechanisms of 3′ OH deprotonation84. To this end they employed a first-principles-based reactive force field, ReaxFF, parameterized for proteins and nucleic acids, and a backbone model, including incoming NTP, 3′-terminal nt of the RNA, catalytic loop (Rpb1 481–485), His1085, and two Arg residues (766 and 1020). This model was employed in simulation of the general base variation on the nucleophilic attack, where the 3′ OH attacks the α-phosphate of the incoming NTP and is simultaneously deprotonated by the action of the catalytic residue, Asp485. The activation barrier for this reaction pathway was estimated at 19 kcal/mol.
In an alternate simulation (involving only the catalytic loop, and the 3′ terminal nt of RNA) 3′ OH was deprotonated by the bulk solvent (water) generating nucleophile prior to the attack. Activation barrier to this step was only 7 kcal/mol. Further emphasizing the feasibility of this step was the observation of water molecules clustering around the 3′ OH in the equilibrium simulation84. Subsequent nucleophilic attack by the 3′ O− on the NTP was estimated at 17 kcal/mol, thus lowering the highest activation barrier by 2 kcal/mol. Notably, regardless of the deprotonation pathway, the dissociation of the PPi was determined to be the rate-limiting step, with activation energy of 23–24 kcal/mol84. These calculations did not take into account the exit of PPi in the protonated form, nor the potential assisting role played by the positively charged residues in the secondary channel (the likely exit route for PPi), both of which can dramatically alter the numerical values of the activation barrier. The models used in these simulations were also quite abbreviated, leaving out some catalytically important residues (e.g. Asp837). This would not impact the results of the water-assisted deprotonation of the 3′ OH as much as the simulations of the nucleophilic attack and the PPi dissociation.
In their later work Salahub and colleagues supplemented ReaxFF computations with density functional theory (DFT) simulations in analysis of the RNAPII active site, focusing on the catalytic loop (DFDGD motif) and the Mg2+ coordination85. They have used 3.3 Å structure of the pre-translocated RNAPII elongation complex (EC) (1i6h67) to build models of the catalytic loop in the RNA-free RNAPII, bound to DNA, and the pre-translocated EC; the model of the catalytic loop in the post-translocated form was built using 4.3 Å structure of the UTP-bound EC (1r9s69). DFT simulations were limited to 57, 91, and 108 atom models, whereas ReaxFF calculations were based on 250 atom model of the post-translocated EC, developed previously84, solvated with 400 water molecules85. Through the combination of these approaches Salahub et al. were able to generate an improved equilibrium model of Mg2+ and NTP coordination in the active site of RNAPII in solution85.
Comparing pre- and post-translocated models, the authors noted structural expansion-contraction of the catalytic loop in the EC depending on the presence of the substrate NTP: it becomes more compact upon binding of NTP and expands upon completion of the chemical step, all the while stabilized by an ordered network of H-bonds, connecting carboxylate oxygen of one Asp residue to the backbone amide of the neighboring one85. Notably, a contraction of the catalytic loop was reported by Vassylyev and colleagues, who compared the structures of the apo-holoenzyme RNAP from Thermus thermophilus and the holoenzyme bound by the antibiotics rifabutin and rifapentin; they have linked this contraction to the apparent concomitant loss of catalytic Mg2+ from the active site86 (this interpretation was later challenged by Ebright and colleagues87). Changes in Mg2+ coordination due to the binding of transcription factors GreA/B (bacteria) and TFIIS (eukaryotes) and introduction of new potential Mg2+-coordination partners (both Gre and TFIIS feature conserved acidic residues they deposit into the active site upon binding to RNAP) have been proposed as the mechanism of the “remodeling” of the active site from RNA synthesis to transcript cleavage72,88,89. Structurally similar to Gre cleavage factors, the inhibitory protein Gfh1, was proposed by Darst, Vassylyev and colleagues to coordinate Mg2+ in a catalytically inactive configuration90,91 (for alternate mechanism of Gfh1 action see Tagami et al.47). The central role Mg ions play in all these scenarios, the proposed changes in efficiency and even the direction of the catalyzed reaction potentially caused even by a subtle alteration in Mg2+ coordination92–94 necessitate rigorous exploration of reaction mechanisms involving these cations.
Lately the universality of the two-metal catalysis scheme, that owes its inception and broad acceptance to structural work, has been challenged based on biochemical data and computational simulations95,96, and the presence of His residue (His1085 in yeast RNAPII) in the catalytic site apparently makes RNAP a subject for additional scrutiny regarding the functionality of the second Mg ion (MgB)96.
A comprehensive exploration of the catalytic mechanism of yeast RNAPII by computational means has been carried out by Maria Ramos and colleagues97. In order to arrive at an unbiased reaction mechanism Ramos et al. interrogated four independent hypothetical scenarios, all variations on the general two-metal catalysis52, but each having distinct yet plausible candidates for proton donor and acceptor. What sets this work apart from other computational interrogations of RNAPII mechanism84,85 is the combination of the full-atom Gromacs-based MD simulations with thermodynamic integration (TI) free energy calculations, DFT, and hybrid quantum mechanics/molecular mechanics (QM/MM) methods (DFT:PM3MM). This comprehensive combinatorial approach allowed to create a higher quality starting full atom model (before equilibration the fragments missing in the 3.95 Å structure of NTP-bound EC (2e2h) were rebuilt using 2nvz and 2e2i (all57)), to carry out high level DFT and lower level PM3MM interrogation of catalytic mechanisms on an expanded (relative to earlier studies84,85) set of reactants (GTP substrate, 3′-terminal RNA nt, the catalytic tetrad (Asp481, 483, 485, and 837), MgA and B, His1085, Arg446, 766, 1020, and Lys967), and to calculate free energy activation barriers associated with each mechanism97. In retrospect, the recently illuminated effect of Watson-Crick base pairing on the kcat of NTP condensation81 argues for the importance of incorporation of the templating DNA nt into DFT:PM3MM simulations, however, this was implicitly addressed in the full-atom MD simulation.
Alternate hypothetical mechanisms explored by Ramos and colleagues included the following: 1) acid-base catalysis by the substrate Oα; 2) free OH− hypothesis, 3) MgA-bound OH− hypothesis, and 4) Asp485 acting as general base hypothesis97. All these used a model of 3′ OH weakly coordinated by MgA, as the preliminary DFT simulations showed that strongly coordinated MgA-assisted deprotonation of the 3′ OH led to the non-stationary solutions approximately 17 kcal/mol higher in energy, otherwise the strongly coordinated 3′ OH spontaneously converted into weakly coordinated upon movement towards Pα during the nucleophilic attack97.
The first mechanism involved the deprotonation of the 3′ OH by the Oα of the substrate (substrate-assisted catalysis), acting first as the base, followed by the nucleophilic attack on the Pα and protonation of the PPi by the Oα, acting as the acid. Attractive in its simplicity and conforming to the two-proton transfer hypothesis74, this reaction mechanism by the way of TI yielded a very high activation barrier for the deprotonation step, resulting in the energy barrier between the reactants and the transition state equal to 34.9 kcal/mol (from the bulk turnover data the authors estimated that the highest energy barrier for NTP condensation is ≤ 18.1 kcal/mol)97. Based on TI calculations this mechanism was rejected.
The second reaction mechanism employed free OH− from the bulk solvent as the proton acceptor for deprotonation of the 3′ OH. In estimation by Florian et al., the free energy needed for the OH−-assisted deprotonation of the 3′ OH at the neutral pH was 1.5 – 3 kcal/mol, making hydroxide ion from the bulk solvent an attractive replacement of the substrate acting as a general base98. This is even lower than Zhu and Salahub estimate for the energy barrier in deprotonation of the 3′ OH by bulk water (7 kcal/mol)84. The reaction mechanism according to this scenario comprised transfer of OH− from bulk solvent to the RNAP active site, deprotonation of the 3′ OH by the hydroxide, protonation of the Oβ by His1085 acting as general acid, followed by nucleophilic attack on Pα, and protonated PPi leaving the active site (Fig. 4)97. Entry of hydroxide into the active site at neutral pH appeared to be associated with an energy barrier of 7.5 kcal/mol, the highest energy barrier for the entire reaction mechanism reaching 9.9 kcal/mol – about half of the estimate for the barrier of the rate-limiting step in NTP condensation, and the total free energy change of −5.8 kcal/mol, making this reaction pathway feasible both kinetically and thermodynamically97.
Figure 4. Two-metal catalysis of NTP condensation by yeast RNA polymerase II.
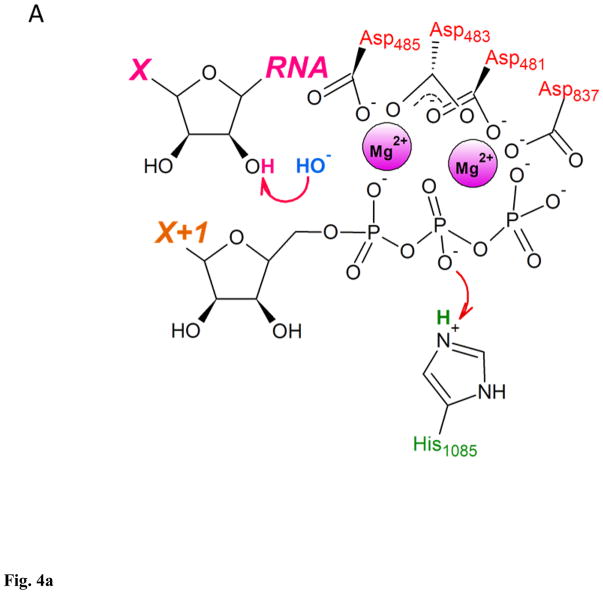
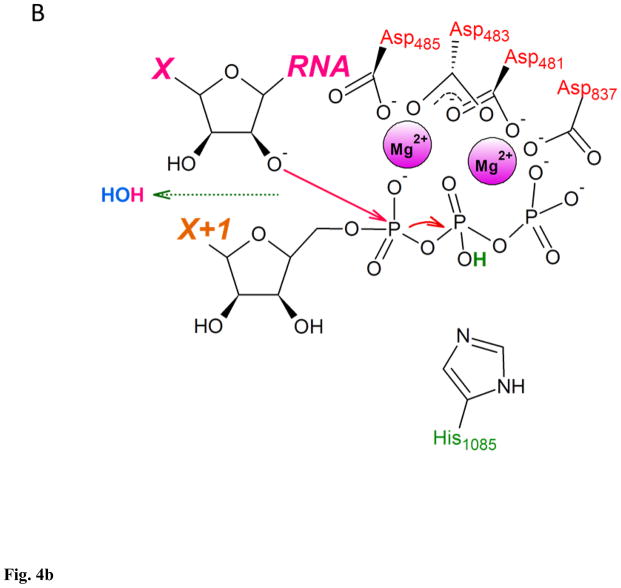
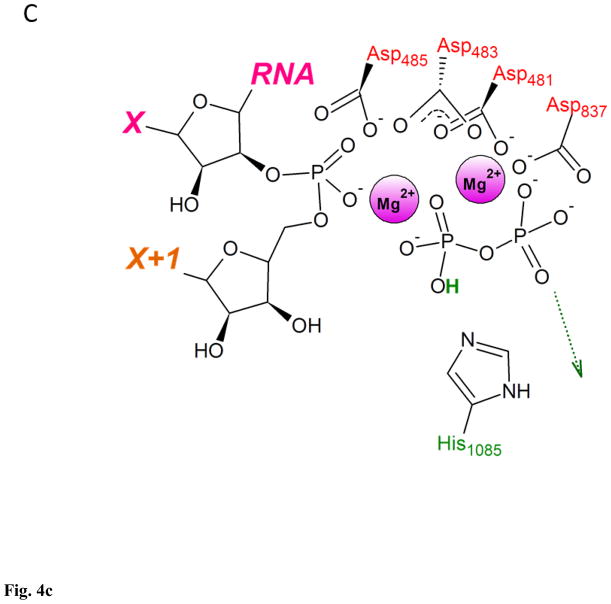
A. RNA 3′ OH is deprotonated by the OH− from the bulk solvent as substrate’s Oβ is protonated by His1085. B. Nucleophilic attack by RNA 3′ O− on substrate’s Pα. C. Substrate in a form of NMP added to the 3′ end of RNA, pyrophosphate in a form of protonated (Mg-PPi)− leaves active site. This figure was adapted from Carvalho et al97, and shows reaction mechanism as a sequence of distinct steps, for clarity and ease of analysis. Proton-inventory experiments reported by Castro et al. indicated that these reaction can take place within the same associative-like transition state82.
The next, third reaction pathway explored by Ramos and colleagues, was similar to the second hypothesis, and included the protonation of the leaving PPi by the His1085, but the deprotonation of the 3′ OH was carried out by MgA-bound OH−, instead of 3′ OH-bound hydroxide transferred from the bulk solvent. Positioning the OH− into the coordination sphere of MgA led to significant rearrangements of the reactants, in particular O2β left Mg2+ coordination sphere, whereas pKa of the O1β increased to the point of spontaneous abstraction of proton from His1085 and MgA coordination geometry switching to tetrahedral. Overall energy of the Mg-coordinated OH− system was 10.8 kcal/mol higher than the same of the system in second hypothesis containing hydroxide unbound from Mg2+. The potential energy surface for the entire reaction shows an energy barrier of 31.7 kcal/mol and the total free energy change of −5.8 kcal/mol97. Being thermodynamically identical to the previous reaction mechanism, this pathway is clearly kinetically disadvantageous, yielding an energy barrier more than 3 times of that for free OH− reaction (pathway 2).
The fourth and the final reaction mechanism investigated by Ramos and colleagues was similar to the second scenario modeled in ReaxFF simulations by Zhu and Salahub84, where deprotonation of the 3′ OH by catalytic residue Asp485 occurs simultaneously with the nucleophilic attack by the 3′ O− on Pα. The activation free energy for this reaction amounted to 26.2 kcal/mol, lower than in mechanisms 1 and 3, but still higher than that of pathway 2 (9.9 kcal/mol) and the estimate for the NTP condensation rate-limiting step (18.1 kcal/mol); the overall free energy change for the reaction was thermodynamically unfavorable at 14.5 kcal/mol97.
The preceding analysis highlights the operational advantage of combining MD simulations with TI, which together allow for more comprehensive ranking of both kinetically and thermodynamically unfavorable reaction pathways. It bears noting that although the NTP condensation along the pathway featuring free OH− from bulk solvent acting as the base, and the His1085 as the general acid emerges as the best candidate, both kinetically and thermodynamically97, alternate, kinetically unfavorable mechanisms may govern the action of “slow” variants of RNAP44, and/or slow nucleotide addition to paused ECs99.
Pyrophosphate release and translocation
The detailed simulations of the NTP condensation and formation of the PPi reviewed above did not consider the mechanism of PPi diffusion from the active site post-chemistry. It is, however, an important mechanistic question, as release of PPi not only had been linked to translocation in the so-called “power-stroke” hypothesis (largely based on the studies of the single-subunit RNAP)100, but is also essential for readying the active site for loading of the next substrate NTP. Multi-subunit RNAPs, including yeast RNAPII, are believed to operate not by the power-stroke mechanism, but to behave instead as isothermal “Brownian ratchet” motors, translocating back and forth along the template with input only of thermal energy32,43,44,101,102. According to this hypothesis PPi release is uncoupled from translocation.
Although consistent with the bulk biochemical, crystallographic, and biophysical data, the isothermal ratchet hypothesis does not provide for an atomic level mechanism of PPi release, nor the order of translocation and release steps. These were investigated by Huang and colleagues by the means of MD103. To this end they had rebuilt missing polypeptides of the 3.95 Å structure of the yeast RNAPII elongation complex (2e2h57) using SWISS-MODEL homology modeling software, and restored the 3′ OH omitted in the GTP-bound complex using 1i6h67 as the template. Starting model for PPi release was generated by breaking Pα-O bond in the GTP in 2e2h. MD simulations were carried out using GROMACS 4.5 software with AMBER03 force field. In order to investigate long scale dynamics from short MD simulations the authors employed Markov State Model to identify macrostates using MSMBuilder software.
Huang and colleagues in the final analysis considered the leaving group as unprotonated (Mg-PPi)2−, believing this to be a more prevalent form at physiological conditions and indicating that in the preliminary simulations differently protonated forms behaved similarly103. The release of the PPi was manually driven (steered) in different directions from the active site towards the pore/secondary channel to simulate potential random diffusion routes.
An immediately important observation made by comparing the pre- and post-chemistry simulations, was that the formation of the PPi lead to significant destabilization of the closed TL. If in pre-chemistry EC fluctuations of the TL tip residues (Gln1078 to Ala1087) were smaller than those averaged over the length of Rpb1 (RMSF of 0.78 Å vs. 1.63 Å), in post-chemistry complex amplitude of the fluctuations of the same residues on average doubled, whereas it remained nearly the same for the reference set103. Notably, formation of the PPi and destabilization of the TL led to loss of interactions between the residue Gln1078 and the substrate 3′ OH, between ribo-discriminating Asn479 and 2′ OH of the substrate, and to the increase in mobility of His1085 and Phe1086. In turn PPi was observed to establish new contacts, importantly, with Rpb1 Lys752, which together with His1085 formed the initial gateway for PPi release (Fig. 5). Breaking of the closed TL contacts with the RNA and the increased destabilization of its tip prompted by PPi formation likely serves as the starting point for conversion of TL into an open configuration and for translocation. Huang and colleagues did stress that they did not observe translocation in their simulation and that in their estimates the time scale of the PPi elimination from the active site (~1.5 μs) is several orders of magnitude shorter than that of NTP condensation, arguing against coupling of PPi release and translocation103.
Figure 5. Hopping pathway of pyrophosphate release from the active site of yeast RNA polymerase II.
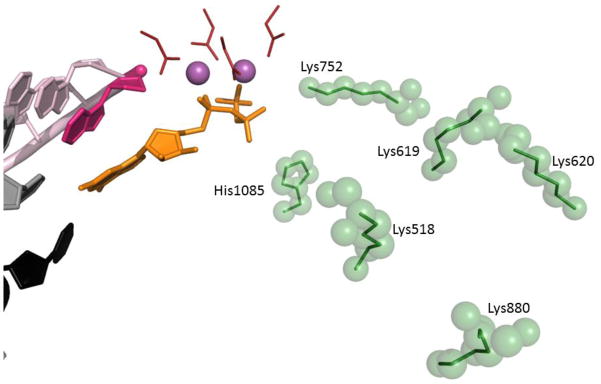
Elements of the active site (from GTP-bound 2e2h) are represented as in Fig. 2. Amino acid residues implicated in facilitating pyrophosphate release are green sticks with green semitransparent spheres.
The simulations of PPi release did indicate a strong coupling between it and the destabilization of the closed TL: not only the formation of the PPi led to an increased mobility of the TL tip residues, but in turn the intrinsic motions of the TL appeared to promote the transition from the active site to the second metastable state (“hopping” position S2, Fig. 5) towards the pore/secondary channel103. The overall trajectory of the PPi diffusion was determined to take a form of hopping between four metastable states, sequentially further removed from the active site (Fig. 5). The gateway state S1 forms post-chemistry, where (Mg-PPi)2− establishes interactions with Lys752 in addition to the pre-existing interaction with His1085. The second state is defined by moving (Mg-PPi)2− further away from the active site, and adding interactions with Lys619, while maintaining the ones with Lys572 and His1085. These are broken during transition to the third state (S3) where (Mg-PPi)2− interacts with Lys619 and Lys518. In the next and last state before release of (Mg-PPi)2− from the pore, it is bound to Lys620 and Lys880. Hopping as the means of facilitated diffusion across long distances is broadly employed in DNA/RNA enzymes, including diffusion of the RNAP itself104,105.
These simulations also highlighted the stimulatory role His1085 plays in PPi release, in addition to its role in NTP binding79 and in chemistry step (as the general acid74,97): in the system where this His residue was substituted by Phe, PPi tended to be trapped in the active site103. Such substitutions can and are being explored genetically and biochemically: Kornberg and colleagues reported the lethality of His1085Phe substitution, and the severe defect in in vitro transcription by the His1085Tyr-substituted RNAPII68. While these experiments emphasized the functional importance of the targeted residue, amino acid substitutions can and do affect more than one parameter, which can influence the outcome of genetic and biochemical assays in addition to catalysis: protein structure, stability, dynamics (allostery), etc. Alchemical manipulations afforded by MD and QM/MM simulations (such as replacement of His by Phe in post-chemistry model) allow exploration of the effects of such substitutions on each isolated step (e.g. PPi release) separately, without affecting others (such as chemistry), hence they can not only complement but also aid in interpretation of complex behaviors, observed in vitro and in vivo.
The interplay between translocation, trigger loop dynamics, and the PPi release was recently explored biochemically by Belogurov and colleagues106. In an elegant experimental set-up, which allowed them to monitor translocation of E. coli RNAP in real time using quenching of fluorophores incorporated into template DNA, combined with the improved fluorescent detection of free PPi, the authors were able to demonstrate that translocation and the release of PPi into solution occur on a similar time scale (t1/2 of 7–12 ms and 6–9 ms, respectively), the former preceding or occurring simultaneously with the latter106. Both steps were delayed relative to the chemistry step. Belogurov and colleagues also demonstrated that acting as a Brownian ratchet, RNAP can nevertheless be biased in the direction of translocation through the opening or closing of the active site by the TL, with active site opening favoring forward translocation106. Taken together with the findings of Da et al.103 linking release of PPi with opening of the TL, one can find features of both the power-stroke100 and the isothermal ratchet44,102 in the post-chemistry action of RNAP.
Concluding remarks
Although it might appear that “the basic mechanism of mRNA synthesis during gene transcription is known” from the structural point of view107, from the studies reviewed herein one can see that the same crystallographic models of RNAPII can be consistent with a number of dramatically different reaction mechanisms, which then have to be ranked kinetically and thermodynamically using complementary methods, such as MD and TI97. In fact, many transcription “mechanisms” proposed to date amount to little more than nominative narratives, enumerating the actual or the conjectural steps of a pathway with no regard to physical parameters (such as potential energy surface, activation barrier, initial velocity for molecular collisions, etc.) which would determine their feasibility. Both the structural and the computational explorations of the basic mechanism of transcription illuminate the complexity of this process, most elements of which still elude full realization as an atomic-resolution physico-chemical model. The details of the nucleotide condensation mechanism and its linkage to TL dynamics are starting to emerge and congeal into a physically tractable mechanism. Basic mechanistic and structural foundations for the large scale dynamics of translocation, back-tracking, arrest, initiation, and termination still await their full elucidation.
Highlights.
NTP condensation by RNA polymerase II occurs via modified two-metal catalysis
Post-chemistry pyrophosphate release takes place in the enzyme pore via hopping mechanism
Pyrophosphate release is linked to opening of the trigger loop
Acknowledgments
This work was supported by the NIH grant GM058750 to E.N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. Journal of molecular biology. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: structural analysis. Journal of molecular biology. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annual review of biochemistry. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer LM, Aravind L. Insights from the architecture of the bacterial transcription apparatus. Journal of structural biology. 2011 doi: 10.1016/j.jsb.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Opalka N, Brown J, Lane WJ, Twist KA, Landick R, Asturias FJ, Darst SA. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukhamedyarov D, Makarova KS, Severinov K, Kuznedelov K. Francisella RNA polymerase contains a heterodimer of non-identical alpha subunits. BMC molecular biology. 2011;12:50. doi: 10.1186/1471-2199-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doherty GP, Fogg MJ, Wilkinson AJ, Lewis PJ. Small subunits of RNA polymerase: localization, levels and implications for core enzyme composition. Microbiology. 2010;156:3532–3543. doi: 10.1099/mic.0.041566-0. [DOI] [PubMed] [Google Scholar]
- 8.Bergsland KJ, Haselkorn R. Evolutionary relationships among eubacteria, cyanobacteria, and chloroplasts: evidence from the rpoC1 gene of Anabaena sp. strain PCC 7120. Journal of bacteriology. 1991;173:3446–3455. doi: 10.1128/jb.173.11.3446-3455.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imashimizu M, Tanaka K, Shimamoto N. Comparative Study of Cyanobacterial and E. coli RNA Polymerases: Misincorporation, Abortive Transcription, and Dependence on Divalent Cations. Genetics research international. 2011;2011:572689. doi: 10.4061/2011/572689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liere K, Weihe A, Borner T. The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. Journal of plant physiology. 2011;168:1345–1360. doi: 10.1016/j.jplph.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Steiner S, Schroter Y, Pfalz J, Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant physiology. 2011;157:1043–1055. doi: 10.1104/pp.111.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray MW, Lang BF, Burger G. Mitochondria of protists. Annual review of genetics. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- 13.Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. Organization and expression of organellar genomes. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2010;365:785–797. doi: 10.1098/rstb.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner M, Thuriaux P, Soutourina J. Structure-function analysis of RNA polymerases I and III. Current opinion in structural biology. 2009;19:740–745. doi: 10.1016/j.sbi.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 15.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nature reviews Genetics. 2011;12:459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 16.Razin SV, Gavrilov AA, Pichugin A, Lipinski M, Iarovaia OV, Vassetzky YS. Transcription factories in the context of the nuclear and genome organization. Nucleic acids research. 2011;39:9085–9092. doi: 10.1093/nar/gkr683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 18.Cramer P. RNA polymerase II structure: from core to functional complexes. Current opinion in genetics & development. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Cramer P. Structure and function of RNA polymerase II. Advances in protein chemistry. 2004;67:1–42. doi: 10.1016/S0065-3233(04)67001-X. [DOI] [PubMed] [Google Scholar]
- 20.Kornberg RD. The molecular basis of eukaryotic transcription. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, Pikaard CS. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Molecular cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nature reviews Molecular cell biology. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 23.Lahmy S, Bies-Etheve N, Lagrange T. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics : official journal of the DNA Methylation Society. 2010;5:4–8. doi: 10.4161/epi.5.1.10435. [DOI] [PubMed] [Google Scholar]
- 24.Landick R. Functional divergence in the growing family of RNA polymerases. Structure. 2009;17:323–325. doi: 10.1016/j.str.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PloS one. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colson P, Yutin N, Shabalina SA, Robert C, Fournous G, La Scola B, Raoult D, Koonin EV. Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of Mimivirus genes. Genome biology and evolution. 2011;3:737–742. doi: 10.1093/gbe/evr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knutson BA, Broyles SS. Expansion of poxvirus RNA polymerase subunits sharing homology with corresponding subunits of RNA polymerase II. Virus genes. 2008;36:307–311. doi: 10.1007/s11262-008-0207-3. [DOI] [PubMed] [Google Scholar]
- 29.Broyles SS, Knutson BA. Poxvirus transcription. Future Virol. 2010;5:639–650. [Google Scholar]
- 30.Hirata A, Murakami KS. Archaeal RNA polymerase. Current opinion in structural biology. 2009;19:724–731. doi: 10.1016/j.sbi.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jun SH, Reichlen MJ, Tajiri M, Murakami KS. Archaeal RNA polymerase and transcription regulation. Critical reviews in biochemistry and molecular biology. 2011;46:27–40. doi: 10.3109/10409238.2010.538662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tagami S, Sekine SI, Yokoyama S. A novel conformation of RNA polymerase sheds light on the mechanism of transcription. Transcription. 2011;2:162–167. doi: 10.4161/trns.2.4.16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Molecular cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 34.Sekine S, Tagami S, Yokoyama S. Structural basis of transcription by bacterial and eukaryotic RNA polymerases. Current opinion in structural biology. 2012;22:110–118. doi: 10.1016/j.sbi.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochimica et biophysica acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annual review of biochemistry. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 37.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nature reviews Molecular cell biology. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brueckner F, Ortiz J, Cramer P. A movie of the RNA polymerase nucleotide addition cycle. Current opinion in structural biology. 2009;19:294–299. doi: 10.1016/j.sbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 39.Cheung AC, Cramer P. A Movie of RNA Polymerase II Transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. The Journal of biological chemistry. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 41.Brueckner F, Armache KJ, Cheung A, Damsma GE, Kettenberger H, Lehmann E, Sydow J, Cramer P. Structure-function studies of the RNA polymerase II elongation complex. Acta crystallographica Section D, Biological crystallography. 2009;65:112–120. doi: 10.1107/S0907444908039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 43.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nature structural & molecular biology. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 44.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 45.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 48.Hein PP, Landick R. The bridge helix coordinates movements of modules in RNA polymerase. BMC biology. 2010;8:141. doi: 10.1186/1741-7007-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinzierl RO. The Bridge Helix of RNA polymerase acts as a central nanomechanical switchboard for coordinating catalysis and substrate movement. Archaea. 2011;2011:608385. doi: 10.1155/2011/608385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan CD, Kornberg RD. A bridge to transcription by RNA polymerase. Journal of biology. 2008;7:39. doi: 10.1186/jbiol99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 53.Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. The EMBO journal. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sosunov V, Zorov S, Sosunova E, Nikolaev A, Zakeyeva I, Bass I, Goldfarb A, Nikiforov V, Severinov K, Mustaev A. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic acids research. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Current opinion in structural biology. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 57.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. The EMBO journal. 2011;30:4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 60.Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGeagh JD, Ranaghan KE, Mulholland AJ. Protein dynamics and enzyme catalysis: insights from simulations. Biochimica et biophysica acta. 2011;1814:1077–1092. doi: 10.1016/j.bbapap.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Read RJ, Adams PD, Arendall WB, 3rd, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011;19:1395–1412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gore S, Velankar S, Kleywegt GJ. Implementing an X-ray validation pipeline for the Protein Data Bank. Acta crystallographica Section D, Biological crystallography. 2012;68:478–483. doi: 10.1107/S0907444911050359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 65.Joosten RP, Joosten K, Cohen SX, Vriend G, Perrakis A. Automatic rebuilding and optimization of crystallographic structures in the Protein Data Bank. Bioinformatics. 2011;27:3392–3398. doi: 10.1093/bioinformatics/btr590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 67.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 68.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Molecular cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 70.Treutlein B, Muschielok A, Andrecka J, Jawhari A, Buchen C, Kostrewa D, Hog F, Cramer P, Michaelis J. Dynamic Architecture of a Minimal RNA Polymerase II Open Promoter Complex. Molecular cell. 2012;46:136–146. doi: 10.1016/j.molcel.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Molecular cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 72.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 73.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Molecular cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 74.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nature structural & molecular biology. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Svetlov V, Vassylyev DG, Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. The Journal of biological chemistry. 2004;279:38087–38090. doi: 10.1074/jbc.C400316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Molecular cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 77.Svetlov V, Nudler E. Jamming the ratchet of transcription. Nature structural & molecular biology. 2008;15:777–779. doi: 10.1038/nsmb0808-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Larson MH, Zhou J, Kaplan CD, Palangat M, Kornberg RD, Landick R, Block SM. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang X, Wang D, Weiss DR, Bushnell DA, Kornberg RD, Levitt M. RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15745–15750. doi: 10.1073/pnas.1009898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yildirim Y, Doruker P. Collective motions of RNA polymerases. Analysis of core enzyme, elongation complex and holoenzyme. Journal of biomolecular structure & dynamics. 2004;22:267–280. doi: 10.1080/07391102.2004.10507000. [DOI] [PubMed] [Google Scholar]
- 81.Kellinger MW, Ulrich S, Chong J, Kool ET, Wang D. Dissecting Chemical Interactions Governing RNA Polymerase II Transcriptional Fidelity. Journal of the American Chemical Society. 2012;134:8231–8240. doi: 10.1021/ja302077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Gotte M, Konigsberg W, Cameron CE. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science. 2011;333:633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu R, Salahub DR. Mechanisms of Nucleotidyl Transfer Catalyzed by the Yeast RNA Polymerase II. AIP Conference Proceedings. 2007;963:104–110. [Google Scholar]
- 85.Zhu R, Janetzko F, Zhang Y, van Duin ACT, Goddard WA, III, Salahub DR. Characterization of the active site of yeast RNA polymerase II by DFT and ReaxFF calculations. Theoretical Chemistry Accounts. 2008;120:479–489. [Google Scholar]
- 86.Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Tahirov TH, Vassylyev DG. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 87.Feklistov A, Mekler V, Jiang Q, Westblade LF, Irschik H, Jansen R, Mustaev A, Darst SA, Ebright RH. Rifamycins do not function by allosteric modulation of binding of Mg2+ to the RNA polymerase active center. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14820–14825. doi: 10.1073/pnas.0802822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. The EMBO journal. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 90.Lamour V, Hogan BP, Erie DA, Darst SA. Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. Journal of molecular biology. 2006;356:179–188. doi: 10.1016/j.jmb.2005.10.083. [DOI] [PubMed] [Google Scholar]
- 91.Symersky J, Perederina A, Vassylyeva MN, Svetlov V, Artsimovitch I, Vassylyev DG. Regulation through the RNA polymerase secondary channel. Structural and functional variability of the coiled-coil transcription factors. The Journal of biological chemistry. 2006;281:1309–1312. doi: 10.1074/jbc.C500405200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maguire ME, Cowan JA. Magnesium chemistry and biochemistry. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2002;15:203–210. doi: 10.1023/a:1016058229972. [DOI] [PubMed] [Google Scholar]
- 93.Cowan JA. Structural and catalytic chemistry of magnesium-dependent enzymes. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2002;15:225–235. doi: 10.1023/a:1016022730880. [DOI] [PubMed] [Google Scholar]
- 94.Sreedhara A, Cowan JA. Structural and catalytic roles for divalent magnesium in nucleic acid biochemistry. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2002;15:211–223. doi: 10.1023/a:1016070614042. [DOI] [PubMed] [Google Scholar]
- 95.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nature structural & molecular biology. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dupureur CM. One is enough: insights into the two-metal ion nuclease mechanism from global analysis and computational studies. Metallomics : integrated biometal science. 2010;2:609–620. doi: 10.1039/c0mt00013b. [DOI] [PubMed] [Google Scholar]
- 97.Carvalho ATP, Fernandes PA, Ramos MJ. The Catalytic Mechanism of RNA Polymerase II. J Chem Theory Comput. 2011;7:1177–1188. doi: 10.1021/ct100579w. [DOI] [PubMed] [Google Scholar]
- 98.Florian J, Goodman MF, Warshel A. Computer simulations of protein functions: searching for the molecular origin of the replication fidelity of DNA polymerases. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6819–6824. doi: 10.1073/pnas.0408173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 101.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maoileidigh DO, Tadigotla VR, Nudler E, Ruckenstein AE. A unified model of transcription elongation: what have we learned from single-molecule experiments? Biophysical journal. 2011;100:1157–1166. doi: 10.1016/j.bpj.2010.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Da LT, Wang D, Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. Journal of the American Chemical Society. 2012;134:2399–2406. doi: 10.1021/ja210656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bratton BP, Mooney RA, Weisshaar JC. Spatial distribution and diffusive motion of RNA polymerase in live Escherichia coli. Journal of bacteriology. 2011;193:5138–5146. doi: 10.1128/JB.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schonhoft JD, Stivers JT. Timing facilitated site transfer of an enzyme on DNA. Nature chemical biology. 2012;8:205–210. doi: 10.1038/nchembio.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic acids research. 2012 doi: 10.1093/nar/gks383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cramer P. Towards molecular systems biology of gene transcription and regulation. Biological chemistry. 2010;391:731–735. doi: 10.1515/BC.2010.094. [DOI] [PubMed] [Google Scholar]


