Abstract
Background
Exceptional longevity is associated with substantial heritability. The ε4 allele in Apolipoprotein E and the linked G allele in rs2075650 of TOMM40 have been associated with increased mortality and the ε2 allele with decreased mortality, although inconsistently.
Methods
Offspring from long lived families and spouse controls were recruited at three sites in the US and in Denmark. We used Generalized Estimating Equations to compare the likelihood of carrying risk alleles in offspring (n=2,307) and spouse controls (n=764), adjusting for age, sex, level of education and family membership.
Results
The likelihood of carrying an APOE ε4 allele or a G allele in rs2075650 was lower (OR=0.75, p =.005 and OR=0.70, p = .002) and the likelihood of carrying an APOE ε2 allele was higher (OR= 1.5, p = .007) among family members in the offspring generation than among their spouse controls.
Conclusions
Our findings support the hypothesis that both reduction in the frequency of the ε4 allele and increase in the frequency of the ε2 allele contribute to longevity.
Keywords: Exceptional longevity, familial longevity, offspring, APOE, TOMM40
1. Introduction
Human exceptional longevity is the outcome of a complex interplay between genetic and environmental influences (McGue et al., 1993; Finch and Tanzi, 1997). Exceptional longevity has been noted to cluster in families and is associated with moderate heritability (Herskind et al., 1996; De Benedictis et al., 2001; Mitchell et al., 2001). In a study of centenarians and their siblings, male and female siblings of centenarians were 17 and 8 times more likely, respectively, to survive to 100 years compared with males and females in their birth cohort (Perls et al., 2002). First degree relatives of individuals who lived beyond 95 years were twice as likely to survive to the same age as were relatives of married in controls (Gudmundsson et al., 2000; Kerber et al., 2001).
Several candidate genes have been investigated to determine their effects on life span and apolipoprotein E (APOE) has consistently emerged as a determinant of longevity (Deelen et al., 2011; Nebel et al., 2011, Sebastiani, 2012). APOE has three common alleles, ε2, ε3 and ε4. The ε4 allele of APOE has been associated with early mortality and a decreased frequency of the allele in the oldest old (Schachter et al., 1994; Tan et al., 2004; McKay et al.2011; Soerensen et al., 2012 ). However, the association of the ε4 allele with mortality risk is inconsistent and varies by population (Galinsky et al., 1997; Bader et al., 1998; Lee et al., 2001). Some studies (Sobel et al., 1995; Ewbank, 2002, 2007) reported that the effect of the ε4 allele on mortality risk diminishes with increasing age, but a recent study in the Danish 1905 cohort showed an increased effect of carrying the ε4 alleles with increasing age (Jacobsen et al., 2010). Recent studies have also identified a variant in TOMM40, marked by the single nucleotide polymorphism (SNP) rs2075650, which is associated with longevity, but is close to and in linkage disequilibrium with rs429358, the marker for the APOE ε4 allele (Deelen et al., 2011; Nebel et al., 2011; Sebastiani et al., 2012).
Since longevity is heritable, a family based study design may serve well to address the question of the relation of APOE to longevity. Mortality associated with the presence of a risk allele may lead to reduced frequencies of risk alleles in the parental generation. Thus, it may be more informative to examine if there is a reduced frequency of risk alleles in offspring of people achieving exceptional longevity, where mortality is unlikely to confound the association between genetic risk factors and longevity. We examined the likelihood of carrying an APOE ε4 or ε2 allele and distribution of the TOMM40 SNP rs2075650 in the offspring generation of the Long Life Family Study (LLFS). We hypothesized that offspring of Long Life Family Study members would have a decreased frequency of the APOE ε4 allele, a decreased frequency of the G allele in TOMM40 rs2075650 and an increased frequency of the APOE ε2 allele compared with similarly aged peers not selected for familial longevity.
2. Methods
Long-lived individuals, their siblings and their offspring were recruited for an examination that characterized key intermediate phenotypes of longevity, including major chronic diseases, risk factors, and physical and cognitive function. A referent group consisting of the spouses, primarily of the offspring generation, was also recruited and examined. For this study, we restricted the analysis to the offspring generation and the examined sons/daughters/nieces/nephews (n=2,314) and their spouses (n=773).
2.1 Recruitment
In the United States (US), each field center used Center for Medicare and Medicaid Services (CMS) lists to mail a recruitment brochure to people who were at least 79 years old on January 1, 2005; had no recorded date of death; did not have end-stage renal disease or were not in hospice programs; and lived within 3 hours driving distance of one of the 3 US study centers (Boston University, Columbia University, and the University of Pittsburgh). Mailings were performed in collaboration with CMS and the National Institute on Aging via an Intra-Agency Agreement. Study participants were also recruited from the community using web-based media and newspaper advertisements, mailing lists obtained through local government agencies, purchased public domain lists from various commercial vendors and community presentations at churches and senior centers. The University of Southern Denmark study site first identified individuals who would be aged 90 and above during the study recruitment period through the Danish National Register of Persons, which contains current information on names, including past names such as maiden names for women, addresses, place of birth, marriages, and vital status (Pedersen et al., 2006). Second, using information on the place of birth and the names, parish registers available in regional archives were searched to locate the parents of the elderly individuals in order to identify sibships. Based on the above information, potentially eligible families were identified and contact was made with potential probands to further assess the family’s eligibility for and willingness to participate in the LLFS using criteria parallel to that used in the United States. Recruitment, informed consent and study procedures were approved by the Institutional Review Boards of all participating sites.
2.2 Eligibility and Enrollment
The Family Longevity Selection Score (FLoSS) was developed by the LLFS study in order to rank longevity among families based on birth-year cohort survival probabilities of the proband and siblings (Sebastiani et al., 2009). On screening, a FLoSS of 7 or more was required for a family to be eligible. If a proband’s family was FLoSS eligible, they also had to meet the following criteria: the proband and at least one living sibling were able to give informed consent and willing to participate in the in-person interview and examination, including providing blood for serum and for extraction of DNA; and either the proband or a sibling had a living offspring willing to participate. A minimum of two siblings and one offspring was required for the key triad. Additionally, we recruited and enrolled the spouses of interested individuals, primarily in the offspring generation, to serve as a comparison group ascertained from the same source populations, but not selected for familial longevity. Spouse controls were employed to adjust for characteristics of individuals within a family which are likely to be correlated. Prior to examination, written informed consent was obtained from all enrollees. The Institutional Review Boards at of all of the Field Centers and the Data Management and Coordinating Center (Washington University, St. Louis) in the USA reviewed and approved this project and the regional Institutional Review Board in Denmark reviewed and approved this project.
2.3 Genotyping
Among offspring and their married-in controls, we were able to genotype APOE for 2,307 of 2,371 (97.3%) offspring of probands and their siblings and 764 of 793 (96.3%) spouse controls. We were able to genotype the TOMM40 SNP rs2075650 for 2,274 (95.9%) offspring and 746 (94.1%) married-in controls. Of the 3,071 participants in the analysis, 983 (32.0%) were Danish.
Genotyping of APOE polymorphisms was based on SNPs rs7412 and rs429358 and was performed at the Biomedical Genomics Center (BMGC) at the University of Minnesota. Genotyping was performed using the Taqman genotyping platform and concordance among genotypes for both SNPs was 100% based on 51 blinded duplicates. We also sequenced DNA from 12 samples with different APOE genotypes to assure correct assignment of alleles. Genotyping of the TOMM40 SNP rs2075650 was performed as part of the Long Life Family Study genome wide association study (GWAS) and was typed via the Illumina HumanOmni 2.5 at the Center for Inherited Disease Research at Johns Hopkins University. Participants were classified by the presence or absence of the APOE ε4 and ε2 alleles, and as carrying one or more G alleles at rs2075650.
2.5. Statistical Analysis
Prior to association analysis, we tested all SNPs for Hardy-Weinberg Equilibrium (HWE) using the HAPLOVIEW program (Barrett et al., 2005; Barrett, 2009) and we did not find deviations of genotype frequencies from HWE for any of the SNPs. To minimize population stratification, analysis was restricted to subjects of White ethnicity, resulting in the loss of 8 spouses (1%) and 5 relatives (0.2%) from the analysis. We used logistic regression procedures in Generalized Estimating Equations (GEE) to estimate the likelihood of carrying one or more APOE ε4, one or more APOE ε2 alleles or one or more rs2075650 G alleles in TOMM40. GEE adjusts for the relatedness of the LLFS offspring and controls and for the possibility that the characteristics of family members are correlated by both shared genetics and shared environment, by treating family membership as a cluster. LLFS offspring were compared with spouse controls, adjusting for age, sex, level of education, and family membership (Model A) and then adjusting for age, sex, level of education, family membership and a history myocardial infarction, hypertension, stroke, diabetes and Parkinson’s disease (Model B). In the offspring generation, only 2 people were reported as suffering from dementia, so we did not include dementia as a covariate for these analyses. SNPS in TOMM40 have been shown to be in linkage disequilibrium with the APOE ε4 allele and it has been suggested that rs2075650 may not have an independent effect on longevity (Deelen et al., 2011; Nebel et al., 2011; Sebastiani et al., 2012). To examine this possibility we repeated the analysis for TOMM40 including APOE in the model as a covariate, and also among those with the APOE ε3/ε3 genotype.
3. Results
LLFS offspring and their spouse controls did not differ in mean age at assessment (60.5 vs. 60.9, respectively). The average number of years of education was slightly but significantly higher among LLFS offspring compared with spouse controls (12.6 vs. 12.1)(Table 1). LLFS offspring were also more likely to be female than spouse controls (57.7% vs. 47.3%) and less likely to have a history of hypertension (29.4% vs.33.9%), but did not differ from spouse controls in the frequency of a history of myocardial infarction, history of stroke, diabetes or Parkinson’s disease (Table 1).
TABLE 1.
Demographic Characteristics
| Characteristic | Spouse Controls | Offspring Generation | |
|---|---|---|---|
|
| |||
| Sample Size (N) | 764 | 2,307 | |
|
| |||
| Age (mean ± S.D.) | 60.9 ± 8.6 | 60.5 ± 8.2 | |
|
| |||
| Education (mean ± S.D.)** | 12.1 ± 3.4 | 12.6 ± 3.0 | |
|
| |||
| Sex (n,%)** | |||
| Male | 403 (52.7) | 975 (42.3) | |
| Female | 361 (47.3) | 1332 (57.7) | |
|
| |||
| APOE GENOTYPES (n, %)** | |||
|
| |||
| 2/2 | 5 (0.7) | 15 (0.7) | |
|
| |||
| 2/3 | 79 (10.3) | 360 (15.6) | |
|
| |||
| 2/4 | 17 (2.2) | 40 (1.7) | |
|
| |||
| 3/3 | 482 (63.1) | 1,461 (63.3) | |
|
| |||
| 3/4 | 163 (21.3) | 407 (17.6) | |
|
| |||
| 4/4 | 18 (2.4) | 24 (1.0) | |
|
| |||
| One or More APOE ε4 alleles (n, %)*** | 198 (25.9) | 471 (20.4) | |
|
| |||
| One or More APOE ε2 alleles, (n, %)** | 101 (13.2) | 415 (18.0) | |
|
| |||
| TOMM40 GENOTYPES (n,%)*** | |||
|
| |||
| Sample Size (N) | 746 | 2,274 | |
|
| |||
| GG | 17 (2.3) | 18 (0.8) | |
|
| |||
| GA | 176 (23.6) | 432 (19.0) | |
|
| |||
| AA | 553 (74.1) | 1,824 (80.2) | |
|
| |||
| One or More TOMM40 G alleles*** | 193 (25.9) | 450 (19.8) | |
|
| |||
| Stroke (n,%) | 16 (2.1) | 33 (1.4) | |
|
| |||
| Heart Disease (n, %) | 27 (3.5) | 65 (2.8) | |
|
| |||
| Hypertension (n, %)* | 258 (33.9) | 676 (29.4) | |
|
| |||
| Diabetes (n,%) | 42 (5.5) | 123 (5.4) | |
|
| |||
| Parkinson’s disease | 2 (0.3) | 6 (0.3) | |
p < .05,
p <.01,
p < .001
Compared with spouse controls, the likelihood of carrying one or more ε4 alleles was 25% lower among LLFS offspring (OR= 0.75, 95% CI: 0.6–0.9), adjusting for age, sex, education and family membership and did not change when the additional covariates were added to the model (Table 2, Model A, Model B). When the presence or absence of the G allele in rs2075650 of TOMM40 was added to the model, the relationship between offspring and the likelihood of G allele was attenuated (OR= 0.9, 95% CI 0.7–1.2) (data not shown). Compared with spouse controls, the likelihood of carrying one or more ε2 alleles was 50% higher among offspring (OR= 1.5, 95% CI: 1.1–1.9), adjusting for covariates (Table 2, Model A, Model B).
TABLE 2.
Likelihood of carrying the APOE ε4 allele and APOE ε2 allele by relative group
| Likelihood of the APOE ε4 allele. | No. at Risk | ε4 allele (n,%) | Odds Ratio Model A† | Odds Ratio Model B†† |
|---|---|---|---|---|
| Participants | ||||
| OFFSPRING GENERATION | 2,307 | 471 (20.4) | 0.75 (0.6–0.9) | 0.75 (0.6–0.9) |
| SPOUSE CONTROLS | 764 | 198 (25.9) | 1.0 (reference) | 1.0 (reference) |
| Likelihood of the APOE ε2 allele. | No. at Risk | ε2 allele (n,%) | Odds Ratio Model A† | Odds Ratio Model B†† |
|---|---|---|---|---|
| Participants | ||||
| OFFSPRING GENERATION | 2,307 | 415 (18.0) | 1.5 (1.1–2.0) | 1.5 (1.1–1.9) |
| SPOUSE CONTROLS | 764 | 101 (13.2) | 1.0 (reference) | 1.0 (reference) |
Model A† Adjusted for age, sex, level of education, and family membership.
Model B†† Adjusted for age, sex, level of education, family membership, history of myocardial infarction, hypertension, stroke, diabetes and Parkinson’s disease
We confirmed findings from previous studies that rs2075650 in TOMM40 is in moderate linkage disequilibrium with the APOE ε4 allele (Figure 1). Compared with spouse controls, the likelihood of carrying one or more G alleles was 30% lower among LLFS offspring (OR= 0.70, 95% CI: 0.6–0.9), adjusting for covariates (Table 3, Model A, Model B). When the APOE ε4 allele was added to the model, the relationship between offspring and the likelihood of G allele was attenuated (OR= 0.8, 95% CI 0.6–1.1) (data not shown). Among those with the APOE ε3/ε3 genotype, the likelihood of carrying one or more G alleles remained 30% lower in offspring than in controls (OR= 0.7, 95% CI, 0.5–1.05), but the difference was not statistically significant.
Figure 1.
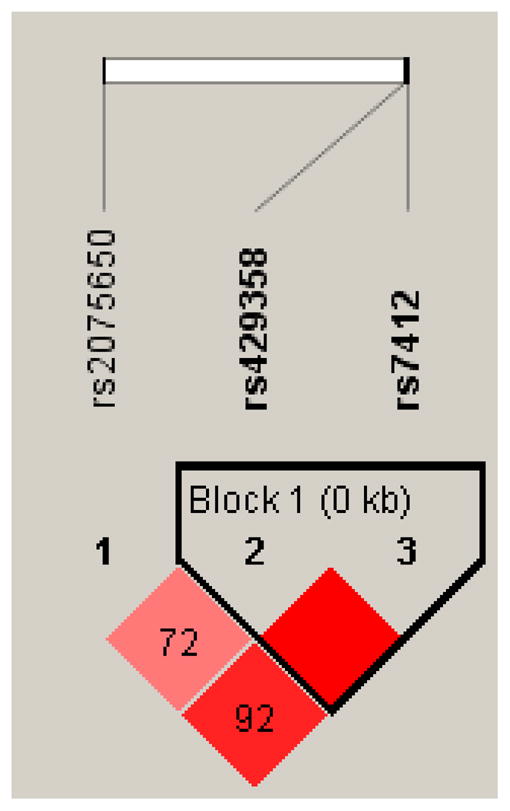
Linkage disequilibrium patterns for SNPs in APOE and TOMM40
Table 3.
Likelihood of carrying the rs2075650 G allele in TOMM40 by relative group and among those with the APOE e3/e3 genotype
| Likelihood of the G allele. | No. at Risk | G allele (n,%) | Odds Ratio Model A† | Odds Ratio Model B†† |
|---|---|---|---|---|
| All Participants | ||||
| OFFSPRING GENERATION | 2,274 | 450 (19.8) | 0.7 (0.6–0.9) | 0.7 (0.6–0.9) |
| SPOUSE CONTROLS | 746 | 193 (25.9) | 1.0 (reference) | 1.0 (reference) |
| Participants with the APOE ε3/ε3 genotype | ||||
| OFFSPRING GENERATION | 1439 | 86 (6.0) | 0.7 (0.5–1.05) | 0.7 (0.5–1.08) |
| SPOUSE CONTROLS | 467 | 39 (8.4) | 1.0 (reference) | 1.0 (reference) |
Model A† Adjusted for age, sex, level of education, and family membership.
Model B†† Adjusted for age, sex, level of education, family membership, history of myocardial infarction, hypertension, stroke, diabetes and Parkinson’s disease.
4. Discussion
Overall, we found that the likelihood of carrying an APOE ε4 allele was lower and the likelihood of carrying an APOE ε2 allele was higher among LLFS family members in the offspring generation than among similarly aged spouse controls. These findings support the hypotheses that the absence of the ε4 allele and presence of the ε2 allele are associated with longevity. Adjustment for common age-related, potentially confounding conditions, including stroke, heart disease, hypertension, diabetes and Parkinson’s disease did not change the pattern of results.
Previous studies have reported lower APOE ε4 allele frequencies in older compared with younger cohorts (Davignon et al., 1988; Kervinen et al., 1994; Rebeck et al., 1994; Schachter et al., 1994; van Bockxmeer, 1994), suggesting that the ε4 allele increases risk for mortality. In this study, we found a lower APOE ε4 allele frequency in a relatively young cohort of offspring of long-lived individuals, before substantial mortality has occurred: thus the reduced frequency of the ε4 allele is the oldest old may not be due to early mortality but to a heritable lower frequency of the risk allele, reducing risk for APOE related disease. Previous studies have suggested that the APOE ε2 allele is associated with longevity, although inconsistently (Schachter et al., 1994; Louhija et al., 1995; Hirose et al, 1997). We also observed a significantly higher frequency of the ε2 allele in offspring of long-lived individuals, consistent with an association of the ε2 allele and longevity in the parent generation. In some studies of older cohorts, however, including studies of Canadian British, and Italian elders, and a randomly chosen Han Chinese population, the frequency of the ε2 allele was similar in the older and younger cohorts, and the frequency of the ε4 allele was not consistently lower in the older cohort (Davignon et al., 1988; Galinsky et al., 1997; Bader et al., 1998; Jian-Gang et al., 1998). In the multi-ethnic WHICAP cohort, mortality risks associated with APOE genotype differed significantly by ethnic group (Lee et al., 2001). These differences underscore the importance of taking ethnicity into account when interpreting associations of genetic data with specific phenotypes and suggest that the impact of APOE risk alleles is likely to be influence by other genetic and environmental factors.
We also found that the frequency and likelihood of carrying a G allele in rs2075650 of TOMM40 was lower among offspring in the Long Life Family Study, compared with the likelihood in married-in controls. Recent genome wide association studies in German, Dutch, Danish and USA Caucasian cohorts of long lived individuals identified rs2075650 in TOMM40 as associated with longevity (Deelen et al., 2011; Nebel et al., 2011; Sebastiani et al., 2012), but also close to and in linkage disequilibrium with the rs429358, the APOE ε4 allele, and the investigators suggested that rs2075650 may not have an independent effect on longevity. Consistent with these analyses, we found that addition of the ε4 allele to our models, attenuated the association between offspring and the likelihood of carrying the G allele. When the analysis was restricted to those with the APOE ε3/ε3, genotype the association of offspring with the likelihood of carrying the G allele was not attenuated, but failed to reach statistical significance. Replication of this association in other long lived cohorts will be needed to elucidate these results.
The limitations of the study include relatively small sample sizes and restriction of the analysis to Caucasians. It would be of interest to examine a wide range of populations of differing ethnicity and ancestry. Additionally, there is some selection bias amongst the older generation of the LLFS for cognitively intact subjects due to the inclusion criteria.
5. Conclusions
Our findings support the hypothesis that reduction in the frequency of the ε4 allele, an increase in the frequency of the ε2 allele contribute to longevity. The decreased allelic frequency of the ε4 allele, increased frequency of the ε2 allele and decreased frequency of the rs2075650 G allele in TOMM40 in LLFS family members compared with controls, and similarities or discrepancies in these associations in other studies suggests the need for follow-up functional studies to better understand the role of these genetic factors and their interactions with other genetic and environmental factors in survival to extreme old age.
Acknowledgments
LLFS: Sponsored by the National Institute on Aging (NIA cooperative agreements U01-AG023712, U01-AG23744, U01-AG023746, U01-AG023749, and U01-AG023755). The Danish 1905-cohort is funded by NIH/NIA, P01 AG08761. The Danish Aging Research Center is funded by the VELUX Foundation.
Footnotes
Disclosure Statement. All authors have contributed to this work and approved the contents of this manuscript. There are no actual or potential conflicts of interest and noauthor has any financial interest in the study or its findings.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bader G, Zuliani G, Kostner GM, Fellin R. Apolipoprotein E polymorphism is not associated with longevity or disability in a sample of Italian octo- and nonagenarians. Gerontology. 1998;44:293–299. doi: 10.1159/000022030. [DOI] [PubMed] [Google Scholar]
- Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Davignon J, Gregg RE, Sing CF. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- De Benedictis G, Tan Q, Jeune B, Christensen K, Ukraintseva SV, Bonafe M, Franceschi C, Vaupel JW, Yashin AI. Recent advances in human gene-longevity association studies. Mech Ageing Dev. 2001;122:909–920. doi: 10.1016/s0047-6374(01)00247-0. [DOI] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van der Breggen R, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank DC. Mortality differences by APOE genotype estimated from demographic synthesis. Genet Epidemiol. 2002;22:146–155. doi: 10.1002/gepi.0164. [DOI] [PubMed] [Google Scholar]
- Ewbank DC. Differences in the Association Between Apolipoprotein E Genotype and Mortality Across Populations. 2007. pp. 899–907. [DOI] [PubMed] [Google Scholar]
- Finch CE, Tanzi RE. Genetics of aging. Science. 1997;278:407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- Galinsky D, Tysoe C, Brayne CE, Easton DF, Huppert FA, Dening TR, Paykel ES, Rubinsztein DC. Analysis of the apo E/apo C-I, angiotensin converting enzyme and methylenetetrahydrofolate reductase genes as candidates affecting human longevity. Atherosclerosis. 1997;129:177–183. doi: 10.1016/s0021-9150(96)06027-3. [DOI] [PubMed] [Google Scholar]
- Gudmundsson H, Gudbjartsson DF, Frigge M, Gulcher JR, Stefansson K. Inheritance of human longevity in Iceland. European journal of human genetics: EJHG. 2000;8:743–749. doi: 10.1038/sj.ejhg.5200527. [DOI] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- Hirose N, Homma S, Arai Y, Kawamura M, Hasegawa H, Ishida H, Shimizu K, Osono Y, Homma A, Nakamura Y. Tokyo Centenarian Study. 4. Apolipoprotein E phenotype in Japanese centenarians living in the Tokyo Metropolitan area. Nippon Ronen Igakkai Zasshi. 1997;34:267–272. doi: 10.3143/geriatrics.34.267. [DOI] [PubMed] [Google Scholar]
- Jacobsen R, Martinussen T, Christiansen L, Jeune B, Andersen-Ranberg K, Vaupel JW, Christensen K. Increased effect of the ApoE gene on survival at advanced age in healthy and long-lived Danes: two nationwide cohort studies. Aging Cell. 2010;9:1004–1009. doi: 10.1111/j.1474-9726.2010.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian-Gang Z, Yong-Xing M, Chuan-Fu W, Pei-Fang L, Song-Bai Z, Nui-Fan G, Guo-Yin F, Lin H. Apolipoprotein E and longevity among Han Chinese population. Mech Ageing Dev. 1998;104:159–167. doi: 10.1016/s0047-6374(98)00067-0. [DOI] [PubMed] [Google Scholar]
- Kerber RA, O’Brien E, Smith KR, Cawthon RM. Familial excess longevity in Utah genealogies. The Journals of Gerontology Series A, Biological sciences and medical sciences. 2001;56:B130–139. doi: 10.1093/gerona/56.3.b130. [DOI] [PubMed] [Google Scholar]
- Kervinen K, Savolainen MJ, Salokannel J, Hynninen A, Heikkinen J, Ehnholm C, Koistinen MJ, Kesaniemi YA. Apolipoprotein E and B polymorphisms--longevity factors assessed in nonagenarians. Atherosclerosis. 1994;105:89–95. doi: 10.1016/0021-9150(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Lee JH, Tang MX, Schupf N, Stern Y, Jacobs DM, Tycko B, Mayeux R. Mortality and apolipoprotein E in Hispanic, African-American, and Caucasian elders. Am J Med Genet. 2001;103:121–127. doi: 10.1002/ajmg.1528. [DOI] [PubMed] [Google Scholar]
- Louhija J, Miettinen HE, Kontula K, Tikkanen MJ, Miettinen TA, Tilvis RS. Aging and genetic variation of plasma apolipoproteins. Relative loss of the apolipoprotein E4 phenotype in centenarians. Arterioscler Thromb. 1994;14:1084–1089. doi: 10.1161/01.atv.14.7.1084. [DOI] [PubMed] [Google Scholar]
- McGue M, Vaupel JW, Holm N, Harvald B. Longevity is moderately heritable in a sample of Danish twins born 1870–1880. J Gerontol. 1993;48:B237–244. doi: 10.1093/geronj/48.6.b237. [DOI] [PubMed] [Google Scholar]
- McKay GJ, Silvestri G, Chakravarthy U, Dasari S, Fritsche LG, Weber BH, Keilhauer CN, Klein ML, Francis PJ, Klaver CC, Vingerling JR, Ho L, De Jong PT, Dean M, Sawitzke J, Baird PN, Guymer RH, Stambolian D, Orlin A, Seddon JM, Peter I, Wright AF, Hayward C, Lotery AJ, Ennis S, Gorin MB, Weeks DE, Kuo CL, Hingorani AD, Sofat R, Cipriani V, Swaroop A, Othman M, Kanda A, Chen W, Abecasis GR, Yates JR, Webster AR, Moore AT, Seland JH, Rahu M, Soubrane G, Tomazzoli L, Topouzis F, Vioque J, Young IS, Fletcher AE, Patterson CC. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. Am J Epidemiol. 2011;173:1357–1364. doi: 10.1093/aje/kwr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Hsueh WC, King TM, Pollin TI, Sorkin J, Agarwala R, Schaffer AA, Shuldiner AR. Heritability of life span in the Old Order Amish. Am J Med Genet. 2001;102:346–352. doi: 10.1002/ajmg.1483. [DOI] [PubMed] [Google Scholar]
- Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, Wittig M, Ellinghaus D, Flachsbart F, Wichmann HE, Meitinger T, Nikolaus S, Franke A, Krawczak M, Lathrop M, Schreiber S. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132:324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Gotzsche H, Moller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53:441–449. [PubMed] [Google Scholar]
- Perls TT, Wilmoth J, Levenson R, Drinkwater M, Cohen M, Bogan H, Joyce E, Brewster S, Kunkel L, Puca A. Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci U S A. 2002;99:8442–8447. doi: 10.1073/pnas.122587599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck GW, Perls TT, West HL, Sodhi P, Lipsitz LA, Hyman BT. Reduced apolipoprotein epsilon 4 allele frequency in the oldest old Alzheimer’s patients and cognitively normal individuals. Neurology. 1994;44:1513–1516. doi: 10.1212/wnl.44.8.1513. [DOI] [PubMed] [Google Scholar]
- Schachter F, Faure-Delanef L, Guenot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- Sebastiani P, Hadley EC, Province M, Christensen K, Rossi W, Perls TT, Ash AS. A family longevity selection score: ranking sibships by their longevity, size, and availability for study. Am J Epidemiol. 2009;170:1555–1562. doi: 10.1093/aje/kwp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Hoh J, Perls TT. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Louhija J, Sulkava R, Davanipour Z, Kontula K, Miettinen H, Tikkanen M, Kainulainen K, Tilvis R. Lack of association of apolipoprotein E allele epsilon 4 with late-onset Alzheimer’s disease among Finnish centenarians. Neurology. 1995;45:903–907. doi: 10.1212/wnl.45.5.903. [DOI] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Tan Q, Thinggaard M, Kleindorp R, Beekman M, Suchiman HE, Jacobsen R, Mcgue M, Stevnsner T, Bohr VA, De Craen AJ, Westendorp RG, Schreiber S, Slagboom PE, Nebel A, Vaupel JW, Christensen K, Christiansen L. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age (Dordr) 2012 doi: 10.1007/s11357-011-9373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Christiansen L, Christensen K, Kruse TA, Bathum L. Apolipoprotein E genotype frequency patterns in aged Danes as revealed by logistic regression models. Eur J Epidemiol. 2004;19:651–656. doi: 10.1023/b:ejep.0000036784.64143.26. [DOI] [PubMed] [Google Scholar]
- van Bockxmeer FM. ApoE and ACE genes: impact on human longevity. Nature Genet. 1994;6:4–5. doi: 10.1038/ng0194-4. [DOI] [PubMed] [Google Scholar]


