Abstract
Information about the basic biological properties of human rhinovirus-C (HRV-C) viruses is lacking due to difficulties with culturing these viruses. Our objective was to develop a cell culture system to grow HRV-C. Epithelial cells from human sinuses (HSEC) were differentiated at air–liquid interface (ALI). Differentiated cultures supported 1–2 logs growth of HRV-C15 as detected by quantitative RT-PCR. Two distinguishing features of HRVs are acid lability and optimal growth at 33–34 °C. We used this system to show that HRV-C15 is neutralized by low pH (4.5). In contrast to most HRV types, replication of HRV-C15 and HRV-C41 was similar at 34 and 37 °C. The HSEC ALI provides a useful tool for quantitative studies of HRV-C replication. The ability of HRV-C to grow equally well at 34 °C and 37 °C may contribute to the propensity for HRV-C to cause lower airway illnesses in infants and children with asthma.
Keywords: Human rhinovirus-C, Air–liquid interface, Differentiated sinus epithelium, qRT-PCR, Temperature sensitivity, pH sensitivity
Highlights
► A system was developed to grow differentiated sinus epithelial cells at air–liquid interface. ► These cells were used as a culture system for HRV-A and HRV-C viruses. ► Replication kinetics for HRV-C15 are similar to those of HRV-A16. ► Like other HRV types, HRV-C15 is neutralized by low pH (4.5). ► In contrast to most HRV-A and HRV-B, HRV-C types grew equally well at 34–37 °C.
Introduction
Human rhinoviruses (HRV) are the causative agents of common cold and are the most frequent cause of acute respiratory tract infection both in children and in adults (Arden et al., 2006, Johnston et al., 1995, Nicholson et al., 1993). HRV have a tremendous diversity and 160 subtypes have been recognized so far. Historically, HRV were classified into 100 serotypes based on cross-reactivity in neutralization tests (Hamparian et al., 1987). These serotypes were categorized in a variety of ways such as receptor usage into major or minor group (Uncapher et al., 1991), or according to the sensitivity to antiviral agents into group A or B (Andries et al., 1990). Based on the full-length or partial (VP2/VP4 and VP1) coding sequences they were initially classified into A and B genotypes (Ledford et al., 2004, Savolainen et al., 2002). With the use of molecular diagnostic techniques a novel group was discovered in 2006 (Arden et al., 2006) and was designated C species. HRV-C are not emerging viruses (Arden and Mackay, 2010) but had been circulating unnoticed due to the inability to culture them. Sequencing of the complete genomes of A, B and available C types confirmed their assignment into three distinct phylogenetic taxa (Palmenberg et al., 2009).
HRV are associated with a broad range of clinical disease including the common cold, bronchiolitis, pneumonia, wheezing and asthma exacerbation especially in children (Arden et al., 2010, Briese et al., 2008, Miller et al., 2009). There is some evidence that HRV-C may be more virulent than other species (Kaida et al., 2011, Mak et al., 2011). For example, HRV-C may be more likely than HRV-B to cause more severe illnesses in infancy (Lee et al., 2012), and HRV-C may be more likely than other species to cause acute exacerbations of asthma (Bizzintino et al., 2011, Fry et al., 2011). These findings, along with distinct genetic features and use of alternative cell surface receptor(s) (Bochkov et al., 2011), suggest that there may be important differences in the replication cycle of HRV-C viruses compared to other HRV.
In fact, unlike HRV-A and HRV-B species, HRV-C could not be propagated in standard cell culture. We previously reported growth of an HRV-C isolate (HRV-C15) in organ cultures of human sinus epithelium (HSEC) obtained as a byproduct of human sinus surgeries (Bochkov et al., 2011). This system provided an important tool for HRV-C research, however, tissue samples obtained from patients are limited in amount and are difficult to standardize for quantitative research.
To overcome these limitations, we developed a system for growing HRV-C in an air–liquid interface of differentiated human sinus epithelium (HSEC) that has pseudostratified morphology, cilia and goblet cells, and used it to study some of the basic biological properties of HRV-C. Two characteristics that distinguish HRV from human enteroviruses are their instability at low pH and their optimal growth at 33–34 °C (Dimmock and Tyrrell, 1964, Taylor-Robinson and Tyrrell, 1962, Turner, 2007). We used the HSEC ALI system to study the effects of low pH treatment and to determine effects of temperature on replication of two different HRV-C types.
Results
Replication of HRV-C15 in different culture systems for sinus epithelium
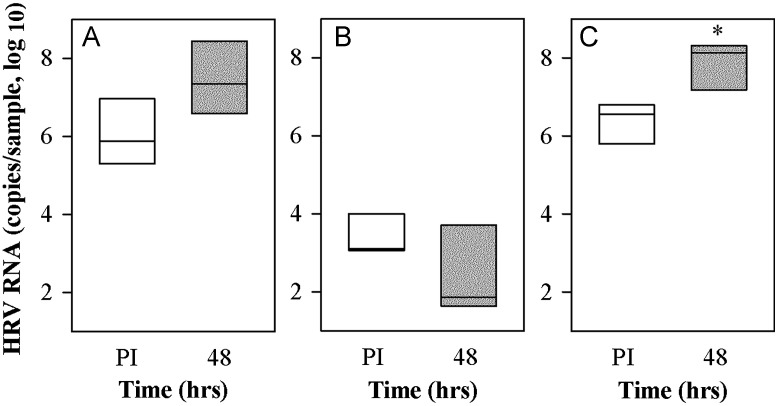
Following inoculation of sinus organ cultures with purified HRV-C15 virions, the mean quantity of cell-associated HRV RNA 48–72 h post-inoculation (PI) increased by 1–1.5 log units (P=0.078; Fig. 2A). However, when these cells were grown in undifferentiated monolayers, viral attachment was lower and remained low over the next 48 h (P=0.244; Fig. 2B). Next, we tested HRV-C15 replication in epithelial cells from human sinus epithelium (HSEC) differentiated under ALI conditions. Approximately two-thirds supported HRV-C15 replication (0.5–2 logs increase in HRV RNA, P<0.05; Fig. 2C). HRV-C15 infection did not cause notable cytopathic effects or changes to ciliary beating (data not shown).
Fig. 2.
Propagation of HRV-C15 in different forms of human sinus epithelium. (A) Pieces of human sinus mucosal tissue were either infected with HRV-C15 (2.8×108 RNA copies) (n=5), (B) dissociated into single cell suspension, passaged, and grown as a monolayer (n=3) and (C) differentiated at ALI (n=3) and then infected with the same dose of HRV-C15. Samples were incubated at 34 °C and tested immediately post-infection (PI) and again 48–72 h later for HRV RNA. *P<0.05.
Comparison of HRV-C15 and HRV-A16 growth in ALI cultures
Following inoculation with HRV-C15 virions (2.0×108 RNA copies), 4.7×105 HRV RNA copies were cell-associated, and 24 h later the RNA had increased to 2.1×107 RNA copies (P=0.002; Fig.3) with slightly less viral RNA after 48 h. After inoculation with the same dose of purified HRV-A16 virions, RNA concentration was greater just after inoculation (1.1×107 RNA copies) and also 24 h later (3.9×108 RNA copies, P=0.002). The growth kinetics of infection and net increase in RNA (HRV-C15 1.6 log increase, HRV-A16 1.5 log increase) among the two viruses were similar.
Fig. 3.
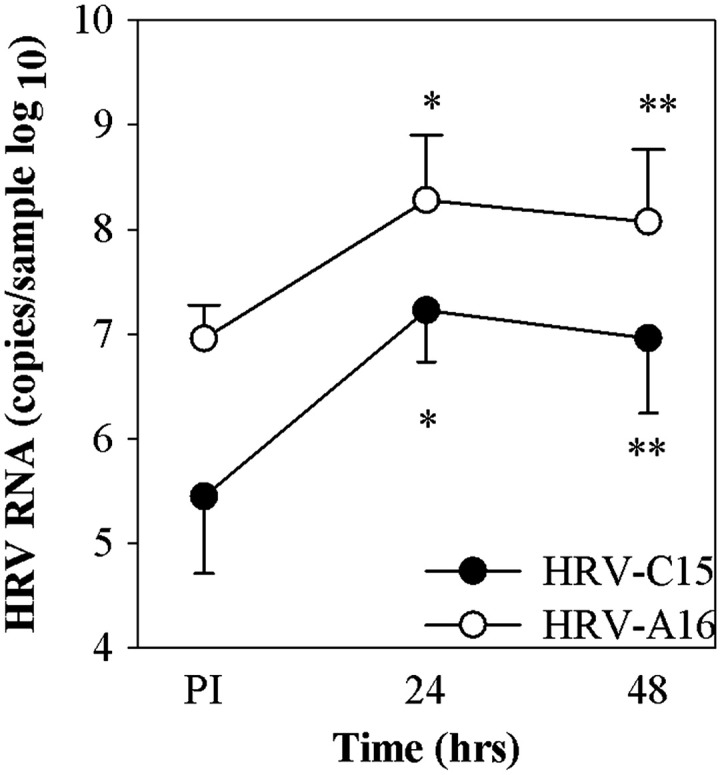
HRV infection of ALI cultures. HSEC ALI cultures (n=6) at 21 days of age were infected with HRV-C15 (2.0×108 RNA copies) or HRV-A16 (3×108 RNA copies) at 34 °C. The cultures for cell-associated virus were collected at 4 h and for recovered virus were incubated for 24–48 h post-infection (PI). *P=0.001, **P<0.005 compared to viral RNA post-inoculation.
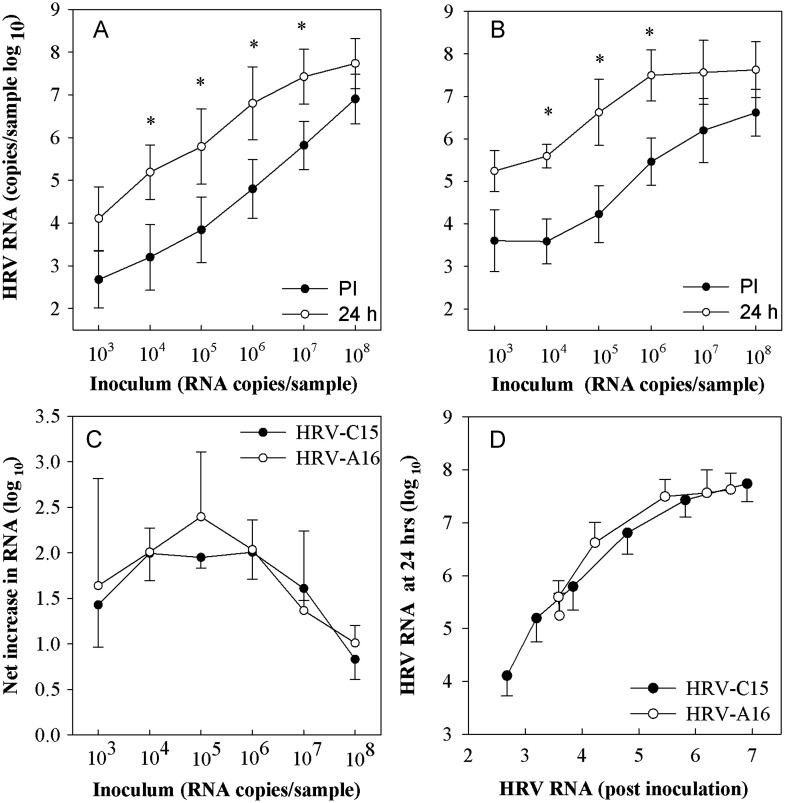
In order to compare dose-related effects on virus replication, HSEC ALI cultures were incubated with 103–108 RNA copies of either HRV-C15 or HRV-A16 virions. For both viruses, there was a 1–2 logs increase in cell-associated HRV RNA over a broad range of inoculating doses (P<0.05; Fig. 4A and B). The peak amplification occurred at the doses of 104–106 RNA copies per culture (Fig. 4C). When normalized for the amount of viral binding, the amounts of output HRV RNA 24 h PI were quite similar (Fig. 4D).
Fig. 4.
Dose response curve of HRV in differentiated HSEC. Differentiated HSEC cultures (n=3) were infected with HRV-C15 or HRV-A16 virions serially diluted (5×108–5×103 RNA copies per well), and cell-associated viral RNA was quantitated immediately post-inoculation (PI) and 24 h later. Panels (A) and (B) illustrate the relationship between input virus, viral binding, and viral yield after 24 h for HRV-C15 and HRV-A16 respectively. These same data were replotted to illustrate the relationship between inoculum dose and net increase in viral RNA (C), and the relationship between viral binding and viral yield for the two viruses (D). *P<0.05.
Optimization of ALI cultures conditions for the amplification of HRV-C15
HRV-C15 replication was not altered by changing the concentration of sodium bicarbonate (0.05%–0.235%), MgCl2 (0.7–30 mM), or altering pH (7.0–7.58). We varied epithelial growth factor (EGF) concentrations (0.8–40 nM) to optimize HSEC cell differentiation and virus propagation. HSEC ALI cultures failed to grow and differentiate in 0.8 nM and 10 nM EGF and 40 nM was the optimal EGF concentration for optimal growth of the HSEC ALI cultures. HRV-C15 replication was similar for EGF concentrations of 20 vs. 40 nM. Adsorption times of 1–4 h produced similar HRV-C15 binding and replication (data not shown).
To test the effects of duration of differentiation on HRV-C15 replication, we cultured HSEC at ALI for 1 month or 2 months and then inoculated them with HRV-C15. Both RNA binding and replication were significantly higher in 2 month ALI cultures compared to those infected after 1 month differentiation (P<0.001, Fig. 5). TER values between 1 and 2 month old cultures were similar (data not shown).
Fig. 5.
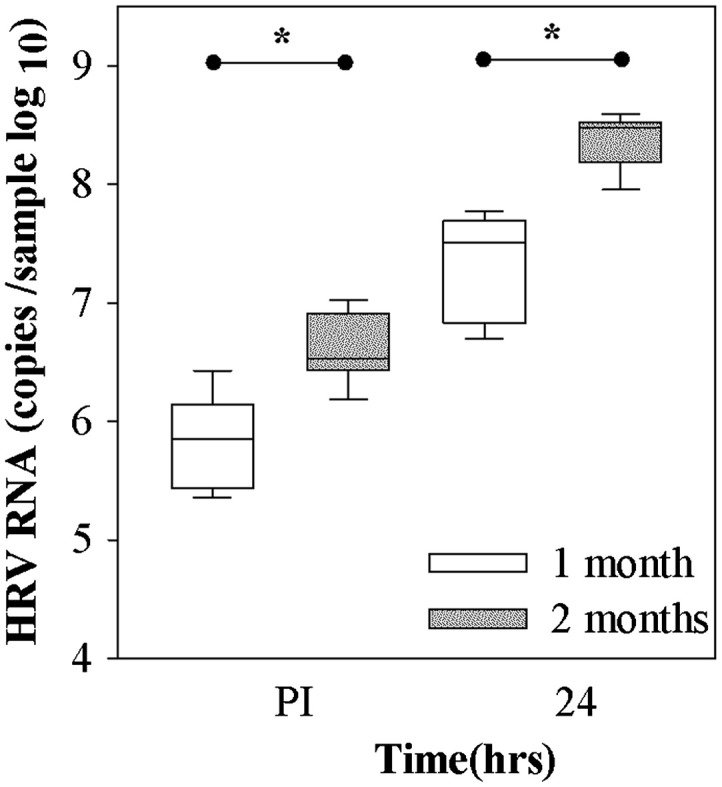
Effect of age of culture on HRV-C15 replication. HSEC cultures (n=3) were allowed to differentiate for one or two months at ALI, and then were infected with HRV-C15 (1×108 RNA copies). Viral replication was measured after 24 h. Differences in the baseline and replication levels of viral RNA between the 1 month and 2 months cultures were determined by the t-test.*P<0.001.
Effects of low pH
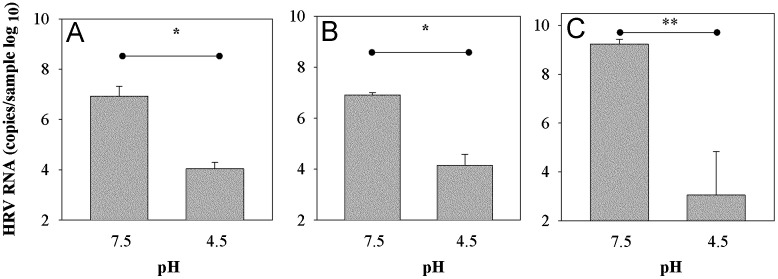
To determine if HRV-C15 infectivity is neutralized by low pH as are other HRV, we treated the HRV-C15 with pH 4.5 and assayed viral RNA replication in sinus organ cultures and in differentiated HSEC ALI. Treatment of HRV-C15 with low pH abrogated replication in organ culture (P<0.05; Fig. 6A) and ALI culture (P<0.05; Fig. 6B). These effects were similar to those of low pH on HRV-A16 (P=0.004; Fig. 6C).
Fig. 6.
Sensitivity of HRV-C15 to low pH. HRV-C15 was diluted 1:10 in citrate phosphate buffer (pH=4.5) or left untreated (pH 7.5), and was then used to infect (A) sinus organ cultures (n=3) (B) differentiated cultures of HSEC (n=3). Similar experiments were conducted with HRV-A16 (C). Virus titers were determined 24 h after inoculation. *P<0.05, **P<0.004 by t-test.
Optimum temperature for HRV-C15 growth
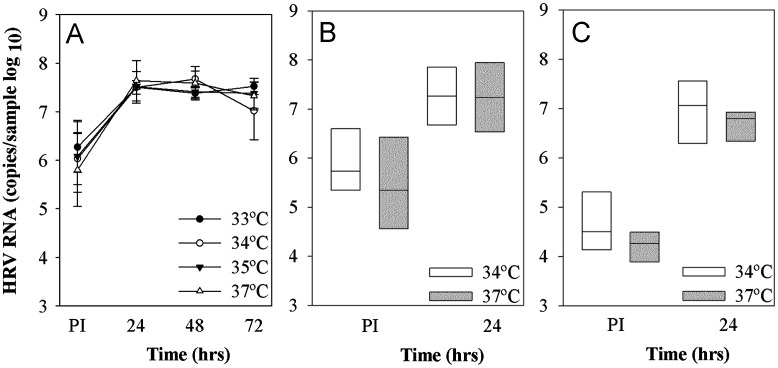
HRV-C15 replication over 24 h was tested at temperatures ranging from 33 to 37 °C. HRV-C15 RNA increased by 1.5 logs after 24 h at all temperatures tested ( Fig. 7A). We repeated the experiments using two different HRV-C (purified C15 and C41 from cell lysates), and found that replication of both viruses was similar at 34 °C and 37 °C (Fig. 7B and C).
Fig. 7.
Effect of temperature on HRV-C replication. (A) Differentiated cultures of HSEC (n=3) were infected with HRV-C15 (1×108 RNA copies), and viral RNA was measured immediately post-inoculation and again after incubation at 33–37 °C for 24, 48 and 72 h. In separate experiments, differentiated HSEC were inoculated with either HRV-C15 (2×108 RNA copies, n=5, panel B) or HRV-C41 (2×107 RNA copies, n=7, panel C) and viral RNA was determined after incubation at either 34 °C or 37 °C for 24 h PI.
Discussion
HRV-C types are of great interest because of their association with childhood pneumonia, wheezing illnesses and the common cold, but not much is known about their biology because of difficulties in culturing these viruses. HRV-C can be cultured in human sinus organ culture, but quantitative research is difficult with this technique. Hence, in this study we developed a system for differentiating sinus epithelium cells in ALI that supported efficient HRV-C replication. Using this system, we demonstrated that HRV-C, like other HRV, is neutralized by low pH, but that HRV-C15 and C41 differ from most HRV-A and HRV-B types in replicating equally well at 33–34 °C and 37 °C.
HRV-C15 grows well in organ culture and in cells differentiated at ALI, but not in sinus epithelial cells grown in a monolayer. The paucity of cell-associated RNA immediately after inoculation indicates that the virus does not bind well to the undifferentiated epithelial cells in the monolayer. This finding indicates that the either the receptor is absent, or else the cells are lacking a cofactor that promotes cellular binding. When the cultures are re-differentiated, both binding and replication are restored, possibly due to re-expression of the receptor.
In the differentiated cells, both HRV-C15 and HRV-A16 replicated well over a broad range of doses (104 RNA copies–107 RNA copies). Peak viral replication was achieved with inoculum size between 104 and 106 RNA copies per sample. These findings suggest that relatively low-dose inoculation produces the best signal for studies of virus replication. If the goal is maximum yield, such as production of viral stocks, more input virus would be desirable.
For any given concentration of input virus, binding of HRV-A16 was greater than that of HRV-C15. This could be due to increased expression of the receptor for HRV-A16 (i.e. ICAM-1), or perhaps a higher affinity of the HRV-A16 to its receptors as compared to HRV-C15. Molecular modeling studies of HRV-C15 have predicted a loss of mass at the 5-fold axis of symmetry, analogous to an area associated with receptor binding in HRV-A and HRV-B (Palmenberg et al., 2009). These changes could contribute to differences in characteristics of HRV-C15 binding to epithelial cells. Notably, replication of the two different strains was nearly identical once the data were normalized for virus binding, suggesting that replication processes distal to binding are similar.
The HSEC-ALI system for HRV-C replication has several advantages over the organ culture. First, it allows for expansion of HSEC before differentiation, thus providing more cells from the same donor for testing of multiple conditions. Notably, stocks of undifferentiated cells can be cryopreserved for use on multiple days. In addition, the number of cells per well is uniform, and the cultures consist almost entirely of epithelial cells which provides a more uniform substrate for viral replication. Another group published the growth of HRV-C15 in ALI system that agrees with our findings (Hao et al., in press). These studies together demonstrate that the HSEC-ALI system is advantageous for quantitative studies of HRV-C biology, pathogenic mechanisms, and pre-clinical tests of anti-viral drugs.
Growth of HRV-C15 in HSEC ALI cultures also enabled studies of basic characteristics of the HRV-C types. HRVs are rendered noninfectious by low pH, which causes a conformational change in virus capsid resulting in a loss of VP4 and uncoating of the virus (Giranda et al., 1992). This is a defining property that distinguishes HRVs from the closely related enteroviruses. Our results demonstrate that HRV-C strains, like HRV-A and HRV-B, are sensitive to low pH.
One other notable characteristic is that most HRV grow optimally at 33–34 °C and have reduced replication at 37 °C (Schroth et al., 1999, Taylor-Robinson and Tyrrell, 1962, Tyrrell and Parsons, 1960, Papadopoulos et al., 1999). Although the mechanism of temperature sensitivity is not completely understood, there has been a speculation that the lower GC content of HRV compared to other enteroviruses is an adaptation for replication at lower temperatures (McIntyre et al., 2010). Overall, HRV-C genomes have a higher G+C content (43%) than HRV-A and HRV-B (37% and 38%) and are closer to HEV (ranging from 42 to 48%) (Linsuwanon et al., 2011); this has led to speculation that HRV-C might grow well at core body temperature. Our results support this theory, since both HRV-C15 and another clinical isolate (HRV-C41) grew equally well at 34 °C vs 37 °C. Additional isolates of HRV-C and other species need to be tested in the future to determine if the ability to grow well at 37 °C is a general and distinct property of the C species.
This culture system has some advantages and some limitations that should be considered. The ALI culture system yields polarized cell layers that are morphologically similar to airway epithelium in vivo. This system, unlike cell lines (e.g. MRC-5, HeLa and WI-38) that are often used for HRV culture, supported the growth of all three species of HRV, and could therefore enable studies of comparative biology. One limitation of our studies is that viral titer is inferred from the quantity of viral RNA and not by the progeny virus particles produced since plaque assays for HRV-C strains await development. Previous studies have demonstrated excellent correlation between HRV measured by plaque assay and by qPCR (Mosser et al., 2005). Addition of retinoic acid and steroids in the ALI medium is essential for development and differentiation of the HSEC epithelium. However, presence of steroids in the culture medium could affect antiviral responses and in turn virus replication. Further studies are needed to decipher the role of such hormones and growth factors on HRV replication in differentiated HSEC.
To summarize, we developed a culture system in which clinical HRV-C strains can be grown and tested. This model system serves as an important tool for studying HRV-C infections and provides useful insights into the biological properties of HRV-C strains. Ability to grow at a higher temperature may have clinical implications, and could contribute to a greater propensity for some HRV-C strains to cause lower airway syndromes such as wheezing illnesses and exacerbations of asthma.
Materials and methods
Viruses
HRV-A16 and HRV-C15 were clinical isolates that were cloned and viruses were produced by transfecting viral RNA into Hela cells as previously described (Bochkov et al., 2011). After three freezes, thaw cycles the cell lysate was treated with RNaseA (10 μg/ml) for 10 min at room temperature to get rid of free RNA that is present outside the virions. This procedure was included so that viral RNA measurements would reflect the quantity of viruses and not unpackaged viral RNA. The viruses were purified by pelleting through a 30% sucrose cushion in a Beckman SW 41 rotor (40,000 rpm, 16 °C, 130 min) (Lee et al., 1995, Wang et al., 1998). Pelleted virus was resuspended in PBS with 0.01% BSA and stored at −70 °C. HRV-C41 was a clinical isolate produced by inoculating nasal secretions into sinus organ culture as previously described (Bochkov et al., 2011). The cultures were frozen and thawed thrice to release the virions and centrifuged at low speed to get rid of the cellular debris and frozen at −70 °C till used.
RNA extraction and quantitative (q) RT-PCR
Total RNA was extracted from sinus tissue or ALI cultures using the RNeasy Mini kit (Qiagen). HRV RNA concentrations were determined by qRT-PCR as previously described (Bochkov et al., 2011).
Sinus organ cultures
Sinus tissue was obtained from residual surgical specimens from individuals undergoing sinus surgery (Bochkov et al., 2011) and the protocol was approved by the University of Wisconsin-Madison Human Subjects Committee. All samples were washed thrice with PBS, cut in 4×4 mm2 pieces and then tested for HRV, adenovirus, influenza, parainfluenza, enterovirus, respiratory syncytial virus, metapneumovirus, coronavirus and bocavirus by multiplex PCR (Lee et al., 2007) before proceeding with experiments.
Cultures of human sinus epithelium
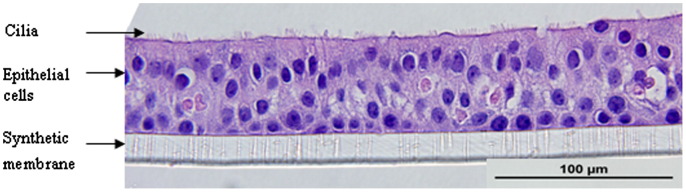
To obtain sinus epithelial cells, the tissue was washed with PBS and digested by pronase (Roche, Basel, Switzerland) and DNase (Sigma–Aldrich, St.Louis, MO) (Schroth et al., 1999). Epithelial cells were seeded in 75 cm2 CellBind® flasks (Costar; Corning Inc., Corning, NY) and cultured in Bronchial Epithelial Growth Medium (BEGM, Lonza, Walkersville, MD). At 90% confluence, the cells were dissociated with trypsin and seeded either as monolayer cultures or for differentiation at ALI. The cells obtained at P-0 and P-1 were frozen in liquid nitrogen for future use. For monolayers, cells were seeded into 12-well tissue culture plates at a density of 1×105 cells/ml. For differentiated cultures, cells were seeded at a density of 1.5×105 cells/cm2 in BEGM (Lonza, Walkersville, MD) onto the apical surface of 1.13 cm2 Transwell polycarbonate inserts (Costar 0.4 μm pore size, 10 μm membrane thickness; Corning Inc., Corning, NY) coated with human placental collagen Type VI (Sigma–Aldrich, St.Louis, MO) in a 12-well plate. The cells were fed basally with BEGM and incubated at 37 °C for first 24 h. Media was removed from the apical surface and the basal medium was changed to BEGM and Dulbecco's modified Eagle's medium (DMEM) in a 1:1 ratio supplemented with the following additives: insulin, 2.5 μg/ml; transferrin, 5 μg/ml; recombinant epithelial growth factor, 0.25 ng/ml; hydrocortisone, 0.25 μg/ml; epinephrine, 0.25 μg/ml; triiodothyronine, 3.25 ng/ml; retinoic acid, 0.05 ng/ml; bovine pituitary extract, 1% (v/v); gentamycin, 50 μg/ml (“ALI” medium) (Gray et al., 1996). The medium was replaced daily for the first seven days and every other day thereafter. The cultures were allowed to mature for at least 21 days until ciliary movement was visualized. The cultures exhibited trans-epithelial resistance (TER) values from 800–1200 Ω/cm2 at 21 days consistent with formation of tight junctions. The cultures on average contained 1×106 cells/well at 30 days of age and formed a pseudo-stratified epithelium (Fig. 1). Tissues from a total of 13 donors were tested, and all except one differentiated in the ALI conditions.
Fig. 1.
Photomicrograph of human sinus epithelium after differentiation at ALI (Haemotoxylin & Eosin, 600× magnification).
HRV inoculation
Once the epithelial cells had differentiated, the medium was removed and cells were washed thrice with PBS. Aliquots of HRV-C15, HRV-C41 or HRV-A16 were diluted in BEGM/0.01% BSA to a desired concentration and a 100 μl inoculum was added either to the apical surface of the ALI culture, to the monolayer or to the dissociated cells in suspension. The cultures were gently shaken (15 min, RT) and then incubated with virus at 34 °C for 1–4 h. The cultures were washed at the apical surface thrice with PBS (Ca and Mg free) and samples to assess cell-associated virus were collected in 350 μl RLT buffer (Qiagen). The remaining samples were fed basally with 1 ml of ALI medium while dissociated cells were plated in a 12-well plate and incubated (34–37 °C) for 24–72 h. After aspirating the medium, cells were collected in 350 μl of RLT buffer (Qiagen) for RNA extraction and qRT-PCR analysis.
Acid lability of HRV-C15
Purified HRV-C15 and HRV-A16 were diluted 1:10 in 0.15 M citrate buffer (pH=4.5) for 15 min at room temperature. The pH treated and untreated viruses (1×108 RNA copies) were used to infect different pieces of sinus organ cultures or ALI cultures.
Statistical analysis
Student's t-test was used to analyze viral replication between two groups (in the Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7) using SigmaPlot 11.0 (Systat Inc., San Jose, CA), and P<0.05 was considered significant.
Acknowledgments
This work was supported by the National Institute of Health Grant nos. U19 AI070503 and P01 HL070831.We thank Fue Vang, Tressa Pappas, Kristine Grindle and Rose Vrtis for technical assistance.
References
- Andries K., Dewindt B., Snoeks J., Wouters L., Moereels H., Lewi P.J., Janssen P.A. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J. Virol. 1990;64:1117–1123. doi: 10.1128/jvi.64.3.1117-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., Faux C.E., O'Neill N.T., McErlean P., Nitsche A., Lambert S.B., Nissen M.D., Sloots T.P., Mackay I.M. Molecular characterization and distinguishing features of a novel human rhinovirus (HRV) C, HRVC-QCE, detected in children with fever, cough and wheeze during 2003. J. Clin. Virol. 2010;47:219–223. doi: 10.1016/j.jcv.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., Mackay I.M. Newly identified human rhinoviruses: molecular methods heat up the cold viruses. Rev. Med. Virol. 2010;20:156–176. doi: 10.1002/rmv.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzintino J., Lee W.M., Laing I.A., Vang F., Pappas T., Zhang G., Martin A.C., Khoo S.K., Cox D.W., Geelhoed G.C., McMinn P.C., Goldblatt J., Gern J.E., Le Souef P.N. Association between human rhinovirus C and severity of acute asthma in children. Eur. Respir. J. 2011;37:1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov Y.A., Palmenberg A.C., Lee W.M., Rathe J.A., Amineva S.P., Sun X., Pasic T.R., Jarjour N.N., Liggett S.B., Gern J.E. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat. Med. 2011;17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Renwick N., Venter M., Jarman R.G., Ghosh D., K+¦ndgen S., Shrestha S.K., Mette Hoegh A., Casas I., Adjogoua E.V. Global distribution of novel rhinovirus genotype. Emerg. Infect. Dis. 2008;14:944–947. doi: 10.3201/eid1406.080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmock N.J., Tyrrell D.A.J. Some physico-chemical properties of rhinoviruses. Br. J. Exp. Pathol. 1964;45:271. [PMC free article] [PubMed] [Google Scholar]
- Fry A.M., Lu X., Olsen S.J., Chittaganpitch M., Sawatwong P., Chantra S., Baggett H.C., Erdman D. Human rhinovirus infections in rural Thailand: epidemiological evidence for rhinovirus as both pathogen and bystander. PLoS One. 2011;6:e17780. doi: 10.1371/journal.pone.0017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giranda V.L., Heinz B.A., Oliveira M.A., Minor I., Kim K.H., Kolatkar P.R., Rossmann M.G., Rueckert R.R. Acid-induced structural changes in human rhinovirus 14: possible role in uncoating. Proc. Natl. Acad. Sci. USA. 1992;89:10213–10217. doi: 10.1073/pnas.89.21.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T.E., Guzman K., Davis C.W., Abdullah L.H., Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1996;14:104. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- Hamparian V.V., Colonno R.J., Cooney M.K., Dick E.C., Gwaltney J.M., Jr, Hughes J.H., Jordan W.S., Jr, Kapikian A.Z., Mogabgab W.J., Monto A. A collaborative report: rhinoviruses – extension of the numbering system from 89 to 100. Virology. 1987;159:191. doi: 10.1016/0042-6822(87)90367-9. [DOI] [PubMed] [Google Scholar]
- Hao, W., Bernard, K., Patel, N., Ulbrandt, N., Feng, H., Svabek, C., Wilson, S., Stracener, C., Wang, K., Suzich, J.A., Infection and propagation of human rhinovirus C in human airway epithelial cells. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- Johnston S.L., Pattemore P.K., Sanderson G., Smith S., Lampe F., Josephs L., Symington P., Toole S.O., Myint S.H., Tyrrell D.A.J. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ. 1995;310:1225. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida A., Kubo H., Takakura K.I., Togawa M., Shiomi M., Kohdera U., Iritani N. Molecular epidemiology of human rhinovirus C in patients with acute respiratory tract infections in Osaka city. Jpn. J. Infect. Dis. 2011;64:488–492. [PubMed] [Google Scholar]
- Ledford R.M., Patel N.R., Demenczuk T.M., Watanyar A., Herbertz T., Collett M.S., Pevear D.C. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J. Virol. 2004;78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.M., Grindle K., Pappas T., Marshall D.J., Moser M.J., Beaty E.L., Shult P.A., Prudent J.R., Gern J.E. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J. Clin. Microbiol. 2007;45:2626–2634. doi: 10.1128/JCM.02501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.M., Lemanske R.F., Evans M.D., Vang F., Pappas T., Gangnon R., Jackson D.J., Gern J.E. Human rhinovirus species and season of infection determine illness severity. Am. J. Respir. Crit. Care Med. 2012 doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.M., Wang W., Rueckert R.R. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes. 1995;9:177–181. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- Linsuwanon P., Payungporn S., Suwannakarn K., Chieochansin T., Theamboonlers A., Poovorawan Y. Complete coding sequence characterization and comparative analysis of the putative novel human rhinovirus (HRV) species C and B. Virol. J. 2011;8:5. doi: 10.1186/1743-422X-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak R.K., Tse L.Y., Lam W.Y., Wong G.W., Chan P.K., Leung T.F. Clinical spectrum of human rhinovirus infections in hospitalized Hong Kong children. Pediatr. Infect. Dis. J. 2011;30:749–753. doi: 10.1097/INF.0b013e31821b8c71. [DOI] [PubMed] [Google Scholar]
- McIntyre C.L., McWilliam Leitch E.C., Savolainen-Kopra C., Hovi T., Simmonds P. Analysis of genetic diversity and sites of recombination in human rhinovirus species C. J. Virol. 2010;84:10297–10310. doi: 10.1128/JVI.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.K., Edwards K.M., Weinberg G.A., Iwane M.K., Griffin M.R., Hall C.B., Zhu Y., Szilagyi P.G., Morin L.L., Heil L.H. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 2009;123:98–104. doi: 10.1016/j.jaci.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser A.G., Vrtis R., Burchell L., Lee W.M., Dick C.R., Weisshaar E., Bock D., Swenson C.A., Cornwell R.D., Meyer K.C. Quantitative and qualitative analysis of rhinovirus infection in bronchial tissues. Am. J. Respir. Crit. Care Med. 2005;171:645–651. doi: 10.1164/rccm.200407-970OC. [DOI] [PubMed] [Google Scholar]
- Nicholson K.G., Kent J., Ireland D.C. Respiratory viruses and exacerbations of asthma in adults. Br. Med. J. 1993;307:982–986. doi: 10.1136/bmj.307.6910.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A.C., Spiro D., Kuzmickas R., Wang S., Djikeng A., Rathe J.A., Fraser-Liggett C.M., Liggett S.B. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos N.G., Sanderson G., Hunter J., Johnston S.L. Rhinoviruses replicate effectively at lower airway temperatures. J. Med. Virol. 1999;58:100–104. doi: 10.1002/(sici)1096-9071(199905)58:1<100::aid-jmv16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Savolainen C., Blomqvist S., Mulders M.N., Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- Schroth M.K., Grimm E., Frindt P., Galagan D.M., Konno S.I., Love R., Gern J.E. Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1999;20:1220. doi: 10.1165/ajrcmb.20.6.3261. [DOI] [PubMed] [Google Scholar]
- Taylor-Robinson D., Tyrrell D.A.J. Serotypes of viruses (rhinoviruses) isolated from common colds. Lancet. 1962;279:452–454. doi: 10.1016/s0140-6736(62)91418-6. [DOI] [PubMed] [Google Scholar]
- Turner R.B. Rhinovirus: more than just a common cold virus. J. Infect. Dis. 2007;195:765–766. doi: 10.1086/511829. [DOI] [PubMed] [Google Scholar]
- Tyrrell D.A., Parsons R. Some virus isolation from common colds. III. Cytopathic effects in tissue culture. Lancet. 1960;275:239–242. doi: 10.1016/s0140-6736(60)90168-9. [DOI] [PubMed] [Google Scholar]
- Uncapher C.R., DeWitt C.M., Colonno R.J. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180:814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]
- Wang W., Lee W.M., Mosser A.G., Rueckert R.R. WIN 52035- dependent human Rhinovirus 16: assembly deficiency caused by mutations near the canyon surface. J. Virol. 1998;72:1210–1218. doi: 10.1128/jvi.72.2.1210-1218.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]