Summary
What are the molecular properties of proteins that fall on the radar of protein quality control (PQC)? Here we mutate the E. coli’s gene encoding dihydrofolate reductase (DHFR), and replace it with bacterial orthologous genes to determine how components of PQC modulate fitness effects of these genetic changes. We find that chaperonins GroEL/ES and protease Lon compete for binding to molten globule intermediate of DHFR, resulting in a peculiar symmetry in their action: Over-expression of GroEL/ES and deletion of Lon both restore growth of deleterious DHFR mutants and most of the slow-growing orthologous DHFR strains. Kinetic steady-state modeling predicts and experimentation verifies that mutations affect fitness by shifting the flux balance in cellular milieu between protein production, folding and degradation orchestrated by PQC through the interaction with folding intermediates.
Introduction
Much is known about how cells respond to external stresses like temperature and oxidation stress (Vabulas et al., 2010). However, despite a considerable effort, our understanding of how cells respond to a “genetic shock”, i.e., mutations, is still lacking. The major difficulty in projecting the effects of protein mutations on fitness landscape stems from the involvement of the intracellular environment, a milieu far more complex than the equilibrium conditions of a test tube. Indeed, mutations can cause misfolding and aggregation of destabilized proteins, leading to reduced fitness (Booth et al., 1997; Dobson, 2006; Wang and Moult, 2001), while components of the proteostasis machinery can potentially mitigate or further aggravate these unfavorable outcomes. The extent to which PQC determines the fitness effects of mutations remains unclear.
Considerable efforts have established how prominent components of PQC work, including the chaperonins, GroEL/ES, and the protease, Lon (Hartl et al., 2011; Horwich and Fenton, 2009; Parsell et al., 1994). Using E. coli to overexpress non-endogenous mutant proteins from various organisms, Tokuriki and Tawfik showed that overexpression of GroEL/ES promoted the folding of highly destabilized mutant proteins (Tokuriki and Tawfik, 2009a) In vitro. It should be noted that GroEL/ES only acts on a fraction of the E. coli proteome (Hartl et al., 2011), and it is unknown whether this observation also applies to mutant forms of the endogenous proteins. Like GroEL/ES, Lon has been connected to protein misfolding (Gur and Sauer, 2008; Powers et al., 2012).
While many in vivo studies have established genetic associations between the components of PQC and a phenotype (Fares et al., 2002; Maurizi et al., 1985; Munavar et al., 2005; Queitsch et al., 2002), several important questions remain: 1) How does PQC mediate the fitness effect of mutations? 2) Do the fitness effects of mutations correlate with the thermodynamic stability of proteins? 3) What are the key elements of PQC that detect and respond to structural changes in a mutated protein and how do they interact in the process of a PQC response?
There are two major hurdles to addressing these questions. Firstly, it is difficult to pinpoint the specific mutations and the corresponding changes in a protein’s structural and thermodynamic states that evoke the PQC response in vivo. Secondly, it is hard to know which specific components of the PQC machinery act on these mutation-induced states.
In the current work, we overcome these difficulties by introducing chromosomally incorporated mutations in the endogenously expressed E. coli gene folA (encoding the essential protein DHFR) without perturbing the endogenous promoter, thus preserving the endogenous expression levels. We rationally chose the set of mutations directed at sites in the protein that would produce a broad range of structurally destabilizing mutations while ensuring a minimal change in activity (see Supplemental Experimental Procedure and ref (Bershtein et al., 2012)). In addition we explore the fitness effects of more dramatic genetic changes in the gene encoding DHFR by replacing it with bacterial orthologs.
We evaluated the In vivo response to mutations and orthologous replacements in the folA gene by perturbing the concentrations of the key components of the PQC system and monitoring how the perturbations affected the bacterial growth rate (i.e. fitness) of the mutant strains. Specifically, major non-essential ATP-dependent proteases were knocked-out, and the abundance of the chaperonins was increased. The analysis showed that both Lon protease and GroEL/ES chaperonins act on the compact molten globule-like equilibrium folding intermediates of DHFR(Ionescu et al., 2000; Ptitsyn, 1995). Based on these findings, we developed a simple quantitative “triage”-like (Gottesman et al., 1997; Powers et al., 2012; Wickner et al., 1999) kinetic model where cytoplasm is viewed as an active steady-state medium under constant energy and material flow, rather than an equilibrated solution of proteins’ thermodynamic states. The model predicts that the effect of mutations on protein abundance and fitness in active cytoplasm is fundamentally different from that in equilibrium In vitro.
Results
Mutant DHFR proteins interact with both branches of the PQC system
Each strain was transformed with a plasmid carrying GroEL/ES genes (Table S1) to achieve a multi-fold overexpression of GroEL in the cytoplasm (10-fold over-expression from the plasmid was previously reported(Guisbert et al., 2004)). Comparison of the growth rates under endogenous (GroELo) and over-expressed (GroEL+) levels revealed a dramatically different impact of GroEL on different folA mutants (Figures 1, 2). While wt folA strain grew similarly under both GroELo and GroEL+ regimes, strains with severely and moderately compromised growth rates showed dramatic improvement upon GroEL/ES overexpression, some even back to wt level (Figure 1A,B, Figure 2A right panel). Conversely, for most high-fitness mutant strains, GroEL+ resulted in a slower growth (Figure 2A right panel). Although the most pronounced effect of the GroEL/ES over-expression was observed at 42°C, similar pattern was also seen at lower (30°C and 37°C) temperatures (Figure 2A right panel). Since the analyzed mutant strains differ from each other solely by the mutations in the coding region of the folA gene, the differential effect of the GroEL/ES overexpression on the strains’ fitness is expected to be the result of a specific GroEL - mutant DHFR interaction.
Figure 1. GroEL/ES over-expression and lon deletion rescue a near-lethal phenotype of a DHFR mutant.
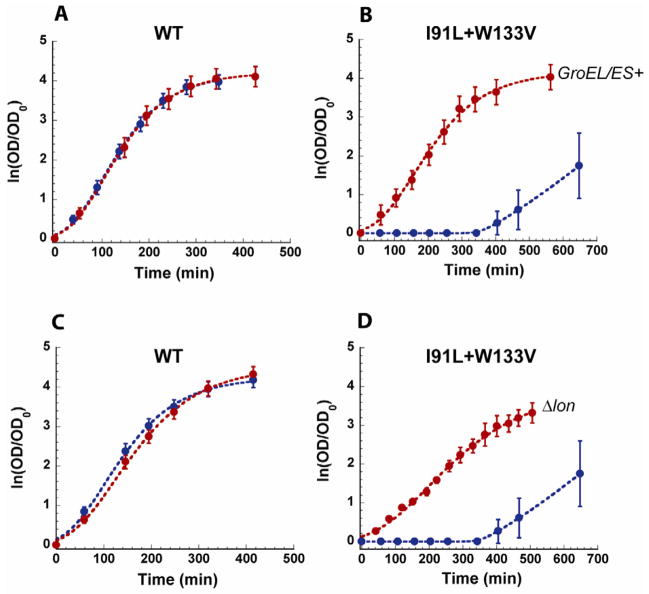
Growth curves of wt and mutant (I91L+W133V) DHFR strains at 42°C. (A, B) Strains with (red) and without (blue) GroEL/ES over-expression. (C, D) Strains carrying lon gene (blue) or lon deletion (red). Error bars represent s.d. of 16 independent measurements.
Figure 2. The effect of GroEL/ES over-expression and lon deletion on the growth rates of DHFR mutant strains.
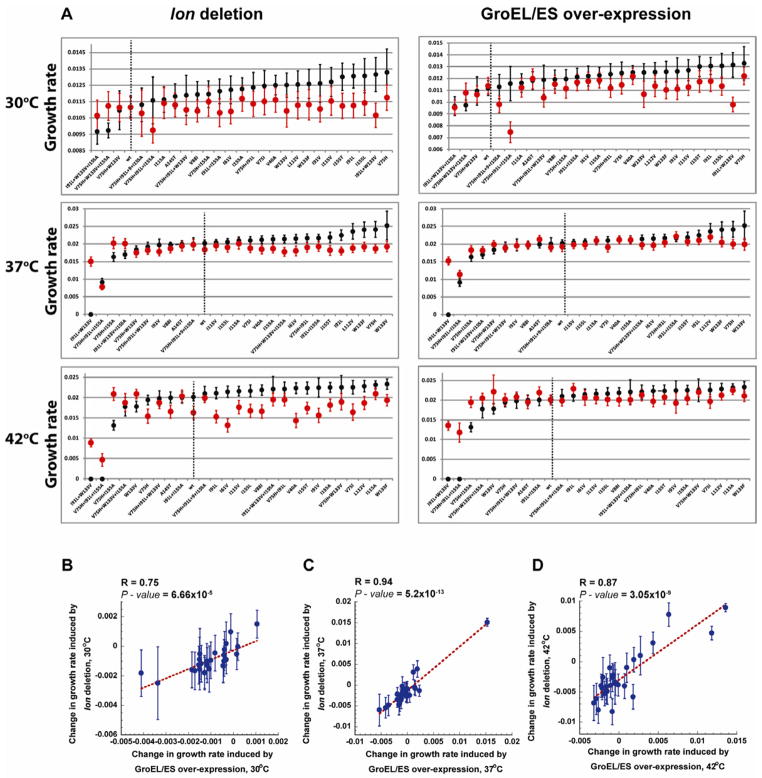
(A) Growth rates of the DHFR mutant strains at 30 °C, 37 °C and 42°C were sorted from the lowest to the highest (black). Growth rates obtained with GroEL/ES over-expression (right) and lon deletion (left) are shown in red. (See also Table S1). Dashed line indicates the wt DHFR strain. Error bars represent s.d. of 16 independent measurements. (B – D) Change in the growth rates of DHFR mutants upon lon deletion and GroEL/ES over-expression is linearly correlated at all temperatures tested (30°C, 37°C, and 42°C). Change in growth rate is calculated by subtracting the growth rates under endogenous conditions from the growth rates upon lon deletion or GroEL/ES over-expression. Error bars for changes in the growth rate were calculated by summation of the relative s.d. of endogenous and lon deletion or GroEL/ES over-expression values. R, and P-values are calculated by ANOVA.
Next, we investigated whether variations in the proteolytic activity of ATP-dependent proteases in vivo might also result in a change in fitness of the folA mutant strains. To this end, three major, but non-essential, ATP-dependent proteases were knocked-out - one at a time: (i) ClpP – the protease component of ClpAP, ClpAPX, and ClpXP complexes, (ii) HslV - the protease component of HslUV, and (iii) Lon protease (see Experimental Procedures). Intervention at the protease level produced an intriguingly mixed picture in which ClpP and HslV deletion did not act in a mutant-specific manner (Figure S1), whereas Lon protease deletion turned out to be very specific to folA mutant strains. Specifically, Δlon mutants rescued the same slow growing strains as the GroEL overexpression, while both Δlon and GroEL+ were mildly deleterious for most mutant strains that grew faster than wt (Figure 1C,D, Figure 2A left panel). Furthermore, changes in the growth rates caused by Δlon were highly correlated with changes in the growth rates caused by GroEL+ for all 26 folA mutant strains at all temperatures tested (Figure 2B–D). Our observations that the effects of Δlon and GroEL+ on growth rates are correlated for all folA mutants is consistent with the “triage” model that advocates a stoichiometric partitioning between folding and degradation branches of the PQC (Gottesman et al., 1997). It is worth noting that despite the strong similarities between Δlon and GroEL+ fitness effects, we also observed a notable difference in that Δlon was mildly deleterious to wt growth rates at higher temperatures (Figure 1A,C, Figure 2A left panel), whereas GroEL+ appeared neutral (Figures 1 and 2). Such behavior is expected, given the central role of the Lon protease in the maintenance of regulatory(Gonzalez et al., 1998; Nishii et al., 2002) and misfolded proteins(Chung and Goldberg, 1981; Gur et al., 2011). Additionally, Lon deletion is known to be deleterious (Torrescabassa and Gottesman, 1987). Altogether, the observed differential fitness effects of Δlon on various folA mutants in the cytoplasm points to a specific interaction between Lon protease and various mutant forms of DHFR.
Chaperonins and Lon protease compete for the intermediate folding state of DHFR
We hypothesized that GroEL and Lon both recognize the same physical state of the protein and compete for it in the cytoplasm. But what are the characteristics of such a state? There was no correlation between the fitness effects induced by Δlon or GroEL+ and the thermodynamic stability (Gibbs free energy difference between folded and unfolded states, ΔG) (see (Bershtein et al., 2012)) of the corresponding DHFR mutant proteins (Figure S2A,B). This fact suggests that the physical state that is recognized by Lon and GroEL/ES might relate to an equilibrium folding intermediate rather than the folded or fully unfolded states of the protein. A clear candidate is a compact molten globule (MG) intermediate, or I-state (Ptitsyn, 1995), which is known to populate wt E. coli DHFR at high temperature (Ionescu et al., 2000). MG-like equilibrium folding intermediate represents an ensemble of compact conformations with exposed hydrophobic surface, making it potentially prone to interact with GroEL (Clark and Frieden, 1999; Martin et al., 1991), be recognized by Lon(Gur and Sauer, 2008; Koodathingal et al., 2009), and form aggregates (Booth et al., 1997).
We used a fluorescent hydrophobic probe bis-1-Anilinonaphthalene-8-sulfonate (bis-ANS) to determine, In vitro, the population of the intermediate states in a broad range of temperatures for wt and the 19 mutants that we were able to purify (all the single and three double mutants). Bis-ANS is a standard probe to detect equilibrium and compact folding kinetic intermediates (Goldberg et al., 1990; Jones et al., 1994; Lindgren et al., 2005). We observed a significant variation in the population of I intermediates between mutants in a broad range of temperatures (Figure 3A,B, Figure S3, Table S2, and Experimental Procedures), yet no correlation can be found between bis-ANS fluorescence and overall stability ΔG (Figure S2C). Furthermore, there was no correlation between bis-ANS fluorescence and the propensity of the mutant DHFR proteins to form oligomeric states at the elevated temperatures (Figure S2D), albeit oligomerization was previously shown to correlate with thermodynamic stability(Bershtein et al., 2012). Consistent with previous reports (Ionescu et al., 2000), wt DHFR protein is characterized by a very low bis-ANS fluorescence in the 25–50°C range with a sharp peak around 60°C (Figure 3A). In fact, most mutant DHFR proteins have a well-defined apparent mid-transition temperature, Tmapp, for thermal transition to the MG state (Figure S3). Such behavior is expected–during thermal unfolding, proteins are known to populate molten globule state because hydrophobic interactions strengthen with temperature (Ptitsyn, 1995; Tanford, 1978). Comparison of the bis-ANS derived Tmapp values with the Tmapp values inferred from the Differential Scanning Calorimetry (DSC) analysis for the same set of the mutant DHFR proteins reveals a strong linear correlation, thus reaffirming the role of bis-ANS as a folding intermediate reporter in our system ((Bershtein et al., 2012) and Figure 3C).
Figure 3. Detection of folding intermediate by bis-ANS fluorescence.
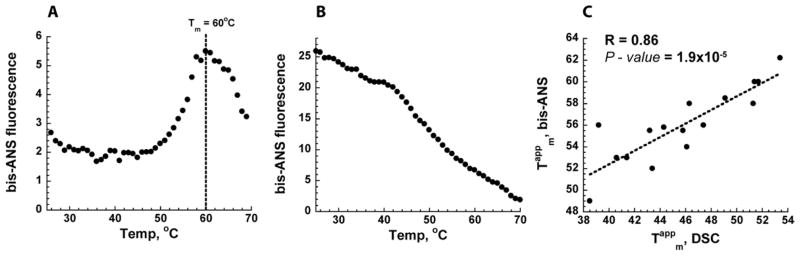
Bis-ANS fluorescent probe was used in vitro to monitor the population of folding intermediates during thermal unfolding of the purified DHFR proteins (25°C –70°C) (see Experimetal Procedures). Panels A and B show representative behavior, the data for all mutants are in Figure S3. (A) Wild-type DHFR protein is characterized by a very low fluorescent signal up to 50°C and a sharp transition through a folding intermediate around 60°C (Tm, mid-transition temperature). (B) V75H+I155A DHFR mutant exhibits a very pronounced folding intermediate already at room temperature (25°C) that tends to disappear with temperature increase due to unfolding. (C) Tmapp values of the DHFR proteins derived from bis-ANS profiling during thermal unfolding (see also Figure S3) is correlated with Tmapp values derived from the thermal unfolding measured by Differential Scanning Calorimetry (Bershtein et al., 2012).
We note that only a small group of the mutant DHFR proteins exhibited high bis-ANS fluorescence at room temperature (Figure 3B, Figure S3), while most mutants exhibit an elevated population of the I-state only at higher temperatures (Figure S3). Strikingly, the slow growing mutants whose growth is significantly rescued by GroEL+ and/or Δlon in vivo (Figure 2) belong to this second group of mutants (W133V, V75H+I155A, I91L+I155A). We hypothesize that this is probably true for the remaining mutants with the impaired fitness, which we were not able to purify (I91L+W133V, V75H+I91L+I155A, V75H+W133V+I155A). Another characteristic feature of the DHFR mutants in this subset is a monotonic decrease in the bis-ANS signal with increasing temperature (Figure 3B, Figure S3). This drop in bis-ANS fluorescence with temperature increase can be explained either by thermal unfolding (molten-globule carrying exposed hydrophobic patches unfolds into a random coil that does not bind bis-ANS any longer), or by aggregation (that screens the hydrophobic areas from bis-ANS). To distinguish between these scenarios, we measured the aggregation propensity, using the Proteostat dye as an aggregation reporter, for several representative DHFR proteins under conditions that are identical to the bis-ANS experiment (see the Supplemental Experimental Procedures). Figure S4 shows that the aggregated fraction does not change substantially within 25°C–70°C temperature range, suggesting that thermal unfolding of the molten globule-like states into a random coil is the most plausible explanation for the observed bis-ANS fluorescence decay with temperature.
E. coli can sustain a substantial drop in DHFR activity without compromising its fitness
An important question is why the strains expressing mutants with populated non-functional molten-globule state even at room temperature can still exhibit near-neutral phenotype at 30°C? For instance, V75H+I155A DHFR mutant shows 8–10 fold increase in bis-ANS fluorescence compared to the wt protein at 30°C (Figure 3B), yet the growth rate of the strain expressing this protein is similar to wt at the same temperature (Figure 2A, Table S1). We address this question by establishing the relation between the abundance of soluble DHFR in the cytoplasm and the bacterial growth rate. To that end we put the folA gene under the IPTG-controllable promoter (see Figure 4A and Experimental Procedures) (Lutz and Bujard, 1997). Under the saturating concentration of IPTG, this strain generates 20–25% of the endogenous DHFR protein level. Decrease in the IPTG concentration results in a tightly regulated drop in the amount of DHFR produced. Growth rates at each IPTG concentration were measured at 30°C, while the DHFR abundances were determined by Western Blot (see Figure 4B, and Supplemental Experimental Procedures). Similar to previous study by (Dykhuizen et al., 1987) on the dependence of fitness on the activity of β-galactozidase, DHFR abundance-fitness relation exhibits the Michaelis-Menten -like behavior. The behavior can be formally expressed as
Figure 4. Dependence of fitness on protein abundance.
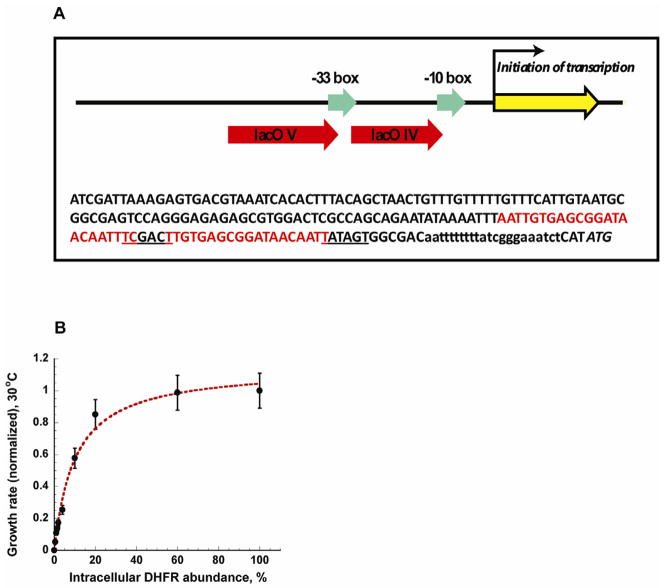
(A) Positions of lacO binding sites introduced upstream of the folA gene to provide IPTG-controllable expression (see Experimental Procedures for a detailed description). Nucleotide sequence represents the non-coding area upstream to folA ORF. Nucleotides in red denote the sequence of the introduced lacO V and IV operators. Underlined sequences represent the −33 and −10 promoter signals. Low case nucleotides indicate the initiation of transcription (+1). The first ATG of the coding sequence is in italic. CAT upstream to the ATG codon was introduced for the cloning purposes. (B) The effect of down-regulation of the wt DHFR expression on growth rate at 30°C. Growth rates of the B3 strain with IPTG-controlled folA expression were measured in the range of IPTG concentrations (see Experimental Procedures) and presented as a function of the corresponding DHFR expression level. Solid line is a fit to Michaelis-Menten-like kinetics (see Experimental Procedures). Error bars represent s.d. of 4 independent experiments.
| (1) |
where [F] is the number of functional DHFR molecules per cell, Gmax is a maximum growth rate achieved at saturation of DHFR, the constant F0 (an analog of the Michaelis-Menten constant KM) is the amount of the expressed DHFR needed to maintain 50% of the fitness. Our analysis (see Figure 4B) shows that best fit to the data is obtained at F0 ≈ 10 constituting roughly 10% of the basal level of DHFR in E. coli (Taniguchi et al., 2010). An important observation is that there is a broad regime at which the drop in the DHFR concentration (approximately up to 20% of the endogenous expression) does not cause appreciable drop in fitness. However, the drop in fitness upon further decrease of DHFR abundance is sharp and approximately linear. This finding implies that fitness-wise cells can sustain up to 80% loss of functional DHFR to a highly populated intermediate state. We measured the catalytic activity of the purified wt and V75H+I155A DHFR proteins (Figure S5, and Supplemental Experimental Procedures), and indeed found that V75H+I155A DHFR mutant retains approximately 20% of the wt activity at 30°C, which is sufficient to sustain the wt-like growth rates.
Our results suggest that GroEL and Lon might both recognize and compete for the I-state of DHFR. In such a scenario, Lon would act to eliminate the aggregation-prone proteins from the cytoplasm, as protein folding intermediates often lead to formation of aggregates (Booth et al., 1997). This view is consistent with the earlier reports that showed an increase in the total protein aggregates upon Lon inactivation (Rosen et al., 2002). The analysis of the DHFR abundance in the insoluble (aggregated) fraction of cell lysates of the mutant DHFR strains further supports this view: We found that lon deletion most strongly affects the abundance of mutants that show highly populated I-state at low temperatures. While no aggregated DHFR can be detected for these mutants under the endogenous Lon levels, Lon depletion resulted in a pronounced insoluble fraction (Figure S6). Furthermore, when lon is deleted, the DHFR insoluble abundance strongly correlates with the bis-ANS fluorescence (Figure 5A, Table S2). This correlation disappears when Lon protease is present at its endogenous levels (Figure 5B).
Figure 5. Protein abundance is correlated with population of folding intermediate (I).
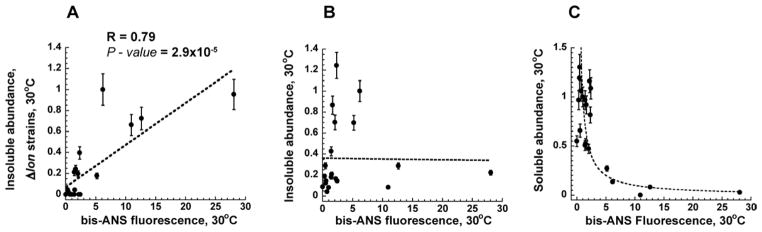
(A) Abundance of the insoluble DHFR fraction detected for all lon deleted DHFR mutant strains is correlated to bis-ANS fluorescence measured in vitro at 30°C. (B) Insoluble fraction of the DHFR proteins in lon+ strains does not correlate with bis-ANS fluorescence (see also Figure S6). Insoluble abundance is normalized against V75H+I91L DHFR strain (arbitrary set to 1). (C) Intracellular protein abundance of the soluble DHFR proteins in the strains grown at 30°C inversely correlates with bis-ANS fluorescence, as predicted by Eq. 3. Abundances are normalized against wt DHFR (arbitrary set to 1). bis-ANS fluorescence is normalized against wt DHFR (arbitrary set to 1), see also Table S2. Error bars respresent s.d. of 3 independent experiments.
A kinetic model of DHFR homeostasis supports the active cytoplasm paradigm
We used these results to develop a simple In vivo kinetic model of the DHFR homeostasis in active cytoplasm (see Graphical Abstract). The model is similar in spirit but not in detail to several earlier models (Kiefhaber et al., 1991) (Bandyopadhyay et al., 2012; Powers et al., 2012; Vendruscolo, 2012; Wickner et al., 1999). The main assumptions of our model are as follows: 1) DHFR is present in the cytoplasm in two interconverting states, F and I, and mutations affect both kinetic kF, kI and equilibrium Keq = kF / kI constants for the F ↔I transitions. 2) GroEL/ES “captures” DHFR in the I-state and releases it in solution in the F state. 3) Lon protease acts on the I state and degrades it but is insensitive to the F state. 4) New DHFR proteins are synthesized in the cytoplasm at a steady state of C molecules per unit time and are released from the ribosome in the F state.
Equilibrium constants for the F ↔ I transitions for each mutant can be obtained from In vitro experiments where bis-ANS fluorescence reports on the equilibrium population of I in solution:
| (2) |
where [F]eq and [I]eq are concentrations of the F and I states of the protein at equilibrium in solution at the conditions of the in vitro ANS fluorescence experiments. C0 is the total concentration of DHFR in equilibrium solution under conditions of the in vitro experiment, ANS is an ANS fluorescence signal, and α is a proportionality constant. If the cytoplasm were an equilibrium solution of proteins, the abundances of DHFR mutants in the F state in cytoplasm would be linearly anti-correlated with their signature ANS fluorescence obtained in In vitro measurements (Eq. 2). On the contrary, under the active cytoplasm assumption, the steady-state abundance of the I-state of DHFR is determined not by its equilibrium with the F state, but rather by the balance between protein production and degradation. The kinetic model predicts that under the steady-state conditions the abundance of folded proteins would depend on the equilibrium constant quite differently than under the equilibrium conditions. Specifically, the model predicts that at low temperature (30°C), when the spontaneous unfolding of F to I is faster than the GroELo F to I turnover, the concentration of F for a DHFR mutant should be proportional to the equilibrium constant Keq for Keq ≥ 1, i.e. inversely proportional to the signature ANS fluorescence signal for a mutant (See Supplemental Experimental Procedures):
| (3) |
When k0Keq ≤ kLon. Here, kLon is a rate constant for degradation of I-state proteins by Lon, k0 is degradation rate of F-state DHFR by other than Lon proteases and A is proportionality constant. Consistent with the active cytoplasm prediction Eq. (3), we found the hyperbolic relation between the soluble DHFR abundance in folA mutant strains and the corresponding in vitro bis-ANS fluorescence of the mutant DHFR (Figure 5C).
Next, we turned to the modeling of the Δlon and GroEL+ effects on fitness. The theory predicts the abundance of folded DHFR [F] in the cytoplasm in steady-state (see Supplemental Experimental Procedures, (Eq. S3)). Substituting this result into (Eq. 1) for growth rates we get:
| (4) |
Where the parameters Gmax, F0, kLon, ku and C are as defined before, and is an effective folding rate that reflects both spontaneous folding and the effects of Lon and GroEL. is the rate constant for GroEL capture of proteins in the I-state; it depends linearly on the concentration of GroEL.
The analysis of the model leads to the following predictions with regards to the impact of PQC on the growth rates of mutant DHFR strains: i) elimination of the Lon protease activity (i.e., making kLon = 0 in Eq. (4)) should result in equalization of the mutants’ growth rates:
| (5) |
i.e. under the Δlon conditions all strains should grow with similar rate.
ii) Unlike the Lon knockout, the GroEL overexpression that leads to a greater k̃F should provide increased growth rates, while maintaining their variation between strains. iii) The GroEL overexpression on the Δlon background should retain homogeneity of growth rates between strains.
Mimicking Horizontal Gene Transfer: Replacement of the folA gene in E. coli with bacterial orthologs
Consistent with the model above, we found that the growth rates of the mutant strains on the Δlon background are indeed more homogenous than on the wt protease background, especially at lower temperatures (Figure 2, left panel). However, the range of growth rates between mutants is somewhat narrow to provide a definitive test of the theory. We sought to broaden it by making the E. coli strains whose folA genes are replaced by orthologs from highly divergent bacteria. Out of 290 available DHFR sequences of the mesophilic bacterial origin we selected 42 sequences whose amino acids identity to the E. coli’s DHFR fell between 29 to 96% (Table S3). To avoid a possible codon-usage related bias, we converted the amino acid sequences of the orthologous DHFRs into the DNA sequence using the codon signature of the E. coli’s folA gene (see Table S4). The synthetic DNA sequences were then used to replace the protein coding part of the chromosomal folA gene, while preserving the integrity of the E. coli’s folA gene regulatory region. Growth rates of the orthologous DHFR strains at 30°C, 37°C, and 42°C fall in a broad range at all temperatures (Figure 6A, Figure 7 and Table S3). Next, we tested the effect of Δlon and GroEL+ on growth rates of the orthologous strains (Figure 6A). Similar to the fitness effects of Δlon and GroEL+ on the folA mutant strains, most of the othologous DHFR strains with impaired growth rates were restored back to the wild-type level, dropping slightly for strains whose growth rates were initially higher than wild-type. The symmetry between the fitness effects of Δlon and GroEL+ was also maintained at all temperatures (Figure 6B).
Figure 6. The effect of GroEL/ES over-expression and lon deletion on the growth rates of DHFR strains with orthologous replacements of the folA gene.
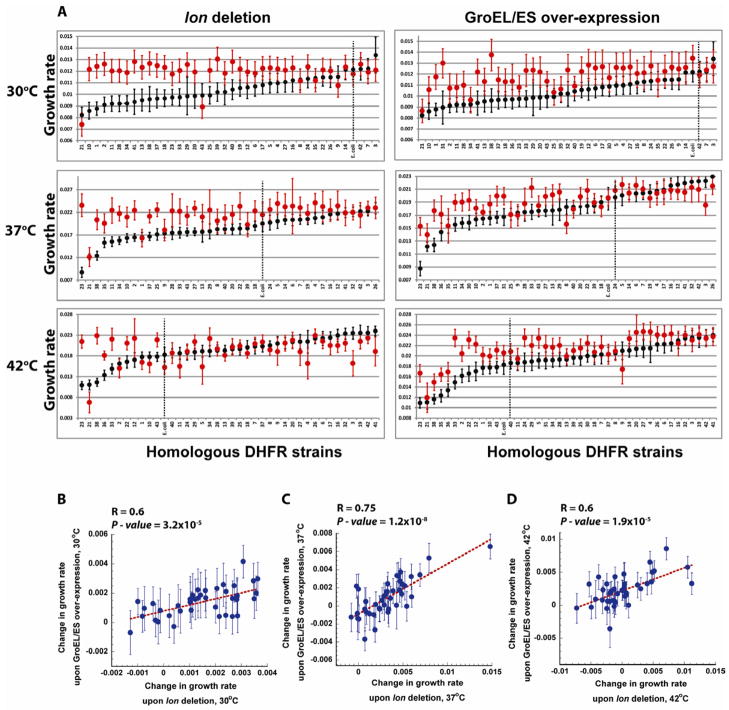
(A) Growth rates of the strains at 30 °C, 37 °C and 42°C were sorted from the lowest to the highest (black). Growth rates obtained with GroEL/ES over-expression (right) and lon deletion (left) are shown in red (see Experimental Procedures and Table S3). Dashed line marks the strain expressing E. coli’s DHFR. Error bars represent s.d. of 16 independent measurements. (B – D) Change in the growth rates of the orthologously replaced DHFR strains upon lon deletion and GroEL/ES over-expression is linearly correlated at all temperatures tested (30°C, 37°C, and 42°C). Change in growth rate is calculated by subtracting the growth rates under endogenous conditions from the growth rates upon lon deletion or GroEL/ES over-expression. Error bars for changes in the growth rate were calculated by summation of the relative s.d. of endogenous and lon deletion or GroEL/ES over-expression values. R, and P-values are calculated by ANOVA.
Figure 7. Distribution of growth rates of DHFR strains with orthologous replacements of the folA gene.
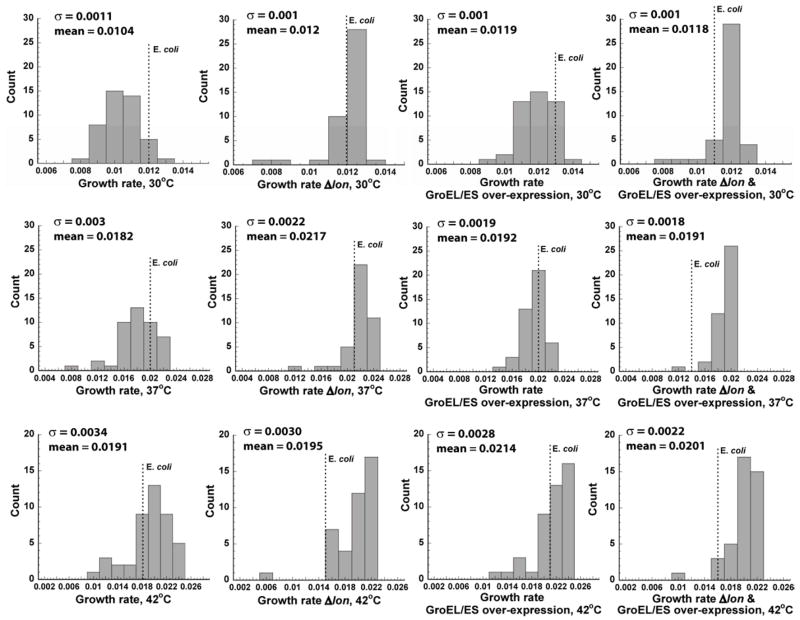
The effect of Δlon, GroEL+, and Δlon with GroEL+ on the distribution of growth rates of the orthologous DHFR strains was measured at 30°C (upper panel), 37°C (middle panel), and 42°C (lower panel). σ is standard deviation. Dashed line indicates the growth rate of the strain expressing E. coli’s DHFR.
Consistent with the model prediction (i), Δlon resulted in a remarkable equalization of the growth rates for all strains (Figure 6A, Figure 7). GroEL/ES overexpression resulted in a much more dispersed distribution of the growth rates, as can be seen from Figure 7, thus confirming the prediction (ii). GroEL+ on the background of Δlon further decreased the dispersion of the growth rates (Figure 7), as postulated by prediction (iii).
Discussion
The view of the cytoplasm as an active medium has been well established (Kiefhaber et al., 1991; Pechmann et al., 2009; Powers et al., 2012; Vabulas et al., 2010; Vendruscolo, 2012; Wickner et al., 1999). In particular, Gierasch and coworkers proposed a general mathematical model for protein homeostasis in E. coli, which considers dynamic interactions of proteins with key players of the PQC system (Powers et al., 2012). Here we reduced the experimentation and mathematical analysis to one protein – DHFR - and specific components of the PQC system, which play a key role in response to mutations. In case of DHFR, growth rate is directly related to the abundance of functional folded proteins in the cytoplasm (Figure 4), so that mutations affect fitness through the “loss of function” effect rather than general misfolding toxicity (Drummond and Wilke, 2008) (Pechmann et al., 2009). Therefore fitness effects of mutations in DHFR can be derived from the model predictions concerning the steady-state amount of folded protein.
Our experiments mimicking horizontal gene transfer of folA genes revealed a significant variability (more than 2-fold) of growth rates between E. coli strains with orthologous folA replacements, whereby the majority of replacements showed slower growth than wild-type E. coli. Remarkably, such diversity is greatly diminished on the Δlon background in a way that matches the theory predictions. Since Lon cannot affect the catalytic activity of DHFR, we conclude that the variability of growth rates under the wt GroEL and Lon backgrounds can be attributed mainly to the variation of F ↔ I interconversion stabilities between the DHFR orthologs from different bacterial species in the environment of the E. coli cytoplasm. This observation brings about an interesting possibility that proteins are thermodynamically and kinetically optimized to fit the environments of their endogenous cytoplasms. It is known that proteomes vary vastly in terms of their average hydrophobic content and charges (pIs) (Nandi et al., 2005). Some aspects of the variation can be attributed to thermal adaptation (Zeldovich et al., 2007a), but other factors such as pH, salinity, and presence of osmolytes (Bandyopadhyay et al., 2012) in the cytoplasm should be considered as well. Another interesting factor that might contribute to a variation of abundances of active orthologous proteins expressed in E. coli is the efficiency of cotranslational folding (Ugrinov and Clark, 2010).
The main insight of this study is that fitness effects of mutations in active cytoplasm are fundamentally different from those in a hypothetical “passive” cytoplasm viewed as a solution of proteins at thermodynamic equilibrium. At equilibrium, mutations result in thermal redistribution between U, I and F forms, while the total amount of protein remains unchanged. In the active cytoplasm mutations also perturb the total amount of protein because they shift the balance between protein synthesis, folding and degradation. Under the Δlon condition, proteins in the I-state do not get degraded or otherwise removed from the cytoplasm (provided that they do not aggregate, which is the case at 30°C). In this case, the abundance of proteins in the folded state under Δlon is governed by the balance between its production in the folded state and degradation in the folded state. The transitions to the I-state do decrease the concentration of the F state. However such decrease is perfectly compensated by the decrease in the F-concentration-dependent degradation rate making the abundance of folded DHFR in the steady-state cytoplasm under the Δlon conditions independent on the F → I transition stability. Therefore, the Δlon condition equalizes the abundances of DHFR in E. coli strains with mutant or orthologous folA replacements and as a consequence, these strains exhibit almost equal growth rates. In the same vein, GroEL+ becomes inefficient under Δlon, because concentration of functional DHFR is already at saturation in the fitness-abundance curve.
In the active cytoplasm folding and unfolding kinetics rates play an important role, along with the equilibrium constants. This conclusion is intuitive. Indeed, any shifts towards the native state provided GroEL must be “memorized” upon the release of the protein into the cytoplasm where it carries out its function. If the protein rapidly equilibrates between its native and nonfunctional states, then all the “work” done by GroEL and burned ATP would be wasted upon release of the protein into cytoplasm. Therefore, for GroEL to be effective, its turnover rate must be comparable or faster than the rate of conversion of F back to I. Importantly, both active (Chakraborty et al., 2010; England et al., 2008; Stan et al., 2007) and passive (Horwich and Fenton, 2009; Tyagi et al., 2011) models of GroEL function will perform equally well under the assumptions of our model, provided that DHFR is released from GroEL in its folded state. Simple caging and release of I states by GroEL (i. e., without converting it into the F form), although protective against aggregation and degradation by Lon and other I-specific proteases, will not affect the concentration of the folded (functional) F fraction at the steady-state, and, therefore, the fitness of the organism.
An important factor that our model currently omits is the aggregation of misfolded proteins. Some mutants which exhibit high bis-ANS fluorescence at room temperature were indeed shown to aggregate at higher temperatures ((Bershtein et al., 2012), Figure S4)). This factor probably introduces an additional variation in the growth rates observed at 42°C, even under the Δlon regime, while at 30°C the theoretical prediction that Δlon should lead to equalizing the growth rates holds with considerable accuracy. It is also possible that the steady-state conditions might be violated due to the feedback mechanisms that can lead to up- or down regulation of protein expression in response to mutations in the ORF. While this possibility is not considered in our present model, it can be included in its future extension.
Admittedly, the growth curve of a bacterial culture measured by changes in the optical density reflects on a total biomass production, and as such is oblivious to possible changes in cell morphology (shape, volume, etc) that may be induced by mutations or growth conditions(Ahmad et al., 1998). However, the difference in growth rates between the mutant strains serves as a good quantitative reporter on the role that cellular proteostasis machinery plays in shaping the fitness effects of mutations. We are currently working to extend the characterization of fitness effects by including the morphological effects of mutations. While we were able to pinpoint GroEL and Lon as important players in the PQC response to mutations, we cannot rule out that there could be other yet unidentified cellular factors that come into play in response to mutations in complex cellular context. However, our minimalistic model is successful in predicting the consequences of genetic changes in the PQC system, suggesting that the mechanisms identified in this work do play an important role.
The distribution of fitness effects of mutations is the key component of most evolutionary models. It is either postulated a’priori (Hartl and Clark, 2007; Wilke, 2005) or derived from a biophysical model that posits a certain relationship between the molecular properties of proteins (e.g. stability, abundance, etc.) and organismal fitness (Drummond and Wilke, 2008; Heo et al., 2011; Tokuriki and Tawfik, 2009b; Wylie and Shakhnovich, 2011; Zeldovich et al., 2007b; Zeldovich et al., 2007c). However, current biophysical multiscale models have two important limitations: 1) they usually view the cytoplasm as an equilibrium solution. 2) they assume unfolding transitions to be two-state between folded and unfolded states. Our study suggests how these assumptions could be improved. In particular, our analysis highlights the crucial importance of folding intermediates in cellular milieu suggesting a potential shift in paradigm in the studies of evolution of protein stability.
Experimental Procedures
Knock-out of protease-encoding genes
Genes encoding ATP-dependent proteases (Lon, HslUV, and ClpP – the protease components of ClpAP, ClpAPX, and ClpXP) were deleted using homologous recombination enhanced by lambda Red, essentially as described (Datsenko and Wanner, 2000).
Analysis of folding intermediates (I)
The relative fraction of the folding intermediate in a range of temperatures was determined in vitro by thermal unfolding of the DHFR proteins in the presence of bis-1- Anilinonaphthalene-8-sulfonat (bis-ANS) followed by a fluorescence signal. To this end, 2 μM of a DHFR protein (10 mM K-phoshate buffer pH8.2, 0.2 mM EDTA, 1 mM beta-mercaptoethanol, 750 μM NADPH) in the presence of 12 μM bis-ANS were heated from 25°C to 70°C at 0.2°C/min. The fluorescence signal that emanated from bis-ANS was monitored (excitation 395 nm, emission 490 nm) using Cary-eclipse fluorescence spectrophotometer, Varian. Integrals of the fluorescence signal over the entire temperature range, or values at 30°C, 37°C, and 42°C were obtained for each of the purified DHFR proteins and normalized against the wtDHFR signal, which was arbitrary set to 1 (Table S2).
Generation of an IPTG- controllable folA regulatory region on the chromosome
Regulatory region of the folA gene was synthesized (GenScript) with the following modifications: The operator sequence of the lac operon (lacO) was introduced upstream to -33 (position V) and -10 (position IV) promoter signals (see Figure 4A, terminology from (Lutz and Bujard, 1997)). The synthetic sequence was cloned in place of the endogenous folA regulatory region in the pKD13 plasmid. The homologous recombination technique described above was then used to generate MG1655 strain carrying a modified regulatory region of the folA gene. Finally, a Z1 cassette carrying lacI gene under constitutive promoter PlacIq (Lutz and Bujard, 1997) was introduced into the chromosome by P1 transduction to generate B3 strain. Under the saturated IPTG concentration (0.6 mM), B3 strain produced 20–25% of the basal DHFR expression level found in wt MG1655 strain, as determined by Western Blot. Decrease in the IPTG concentration resulted in further drop of DHFR production. When grown from a single colony, B3 strain cannot sustain growth unless supplemented with IPTG.
Relating growth rates of B3 strain to fitness
B3 strain cells were grown overnight at 30°C from a single colony in supplemented M9 medium (0.2% glucose, 1mM MgSO4, 0.1% casamino acids, 0.5 μg /ml thiamine) in the presence of 0.6 mM IPTG. Cells were then washed x3 times in M9 media, diluted 1/100, and grown for 5 hours in the absence of IPTG to decrease the intracellular DHFR pool. Next, cells were diluted 1/10, transferred into 96-well microtiter plates (16 wells per each strain), and grown at 30°C in a range of IPTG concentrations (0.6 mM to 0). OD data were collected at 600nm at 35 min intervals. The resulting curves were fit to a bacterial growth model to obtain growth rate parameters. Since full depletion of the DHFR molecules cannot be achieved under this growth regime, the resulting growth rates were scaled from 1 to 0, whereas 1 was assigned to the growth rate of wt MG1655 strain (basal DHFR expression), and 0 was assigned to the lowest growth rate at 0 IPTG. The IPTG values were converted into protein abundance by Western Blot analysis of the soluble fraction of DHFR extracted from the strains grown under the identical conditions. Protein amounts were scaled against DHFR levels found in wt MG1655 strain (set to 100%). The resulted Protein abundance – growth rate data were fit to Michaelis-Menten-like kinetics, [S]xVmax/([S]+KM), where [S] is the DHFR abundance, Vmax is the maximal growth rate obtained under the basal expression levels of DHFR, and KM is the level of DHFR produced at which growth rate is half of its maximal value, see Eq (1).
Generation of strains expressing orthologous DHFR proteins
The blast analysis against E. coli’s DHFR amino acids sequence amongst meshophilic bacteria produced 290 unique DHFR sequences (Huang et al., 2004). This dataset was used to select 42 sequences with amino acids identity to the E. coli’s DHFR ranging between 29 to 96% (Table S3). To avoid possible codon-usage related bias, the amino acid sequence of the chosen DHFRs was converted into the DNA sequence using the codon signature of the E. coli’s folA gene. Specifically, frequency of each codon was calculated, and codons with the highest score were used for protein – DNA sequence conversion (see Table S4). In addition, each DNA sequence, including the E. coli’s folA, was fused to a tag encoding 6 histidines. The resulted DNA sequences were synthesized (GenScript), and cloned into the pKD13 plasmid downstream to folA’s regulatory region. Using the homologous recombination technique (see above), 42 MG1655 E. coli strains were created in which homologous DHFR-encoding sequences replaced the ORF of the chromosomal folA gene, while maintaining the integrity of folA’s regulatory region.
Analysis of growth curves
Cells were grown overnight at 30°C from a single colony in supplemented M9 medium (0.2% glucose, 1mM MgSO4, 0.1% casamino acids, 0.5 μg/ml thiamine), diluted 1/100, transferred into 96-well microtiter plates (16 wells per each strain), and grown at 30°C, 37°C, and 42°C. OD data were collected at 600nm at 35 min intervals. The resulting curves were fit to a bacterial growth model to obtain growth rate parameters (Zwietering et al., 1990)
Statistics
R and P – values for linear correlations were determined by ANOVA (analysis of variation) test.
Supplementary Material
Highlights.
Proteases (Lon) and chaperonins (GroEL/ES), act on folding intermediates
Fitness effects of Lon knockout and GroEL/ES overexpression are correlated
Stability-changing mutations affect the total abundance of a protein in cytoplasm
A dynamic model predicts fitness effects of mutations and orthologous replacements
Acknowledgments
We thank Jiabin Xu for help with data analysis, Dan Tawfik, Art Horwich, and Stephen McQ. Gould for comments. This work was supported by National 973 Project (No. 2012CB720802) and the Natural Science Foundation of China Project (No. 31171705) (WM), the Key Program of National Natural Science Foundation of China (31130043) (JZ), NIH Grant No GM 068670, and long-term postdoctoral fellowship from the Human Frontiers Science Program (SB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Saxena K, Kasturia N, Dalal V, Bhatt N, Rajkumar A, Maity S, Sengupta S, Chakraborty K. Chemical chaperones assist intracellular folding to buffer mutational variations. Nature chemical biology. 2012;8:238–245. doi: 10.1038/nchembio.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Shakhnovich EI. Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. Proc Natl Acad Sci U S A. 2012;109:4857–4862. doi: 10.1073/pnas.1118157109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DR, Sunde M, Bellotti V, Robinson CV, Hutchinson WL, Fraser PE, Hawkins PN, Dobson CM, Radford SE, Blake CC, et al. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature. 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Chung CH, Goldberg AL. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci U S A. 1981;78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AC, Frieden C. The chaperonin GroEL binds to late-folding non-native conformations present in native Escherichia coli and murine dihydrofolate reductases. Journal of molecular biology. 1999;285:1777–1788. doi: 10.1006/jmbi.1998.2403. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein aggregation and its consequences for human disease. Protein Pept Lett. 2006;13:219–227. doi: 10.2174/092986606775338362. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Dean AM, Hartl DL. Metabolic flux and fitness. Genetics. 1987;115:25–31. doi: 10.1093/genetics/115.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England JL, Lucent D, Pande VS. A role for confined water in chaperonin function. Journal of the American Chemical Society. 2008;130:11838–11839. doi: 10.1021/ja802248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares MA, Ruiz-Gonzalez MX, Moya A, Elena SF, Barrio E. Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature. 2002;417:398. doi: 10.1038/417398a. [DOI] [PubMed] [Google Scholar]
- Goldberg ME, Semisotnov GV, Friguet B, Kuwajima K, Ptitsyn OB, Sugai S. An early immunoreactive folding intermediate of the tryptophan synthease beta 2 subunit is a ‘molten globule’. FEBS Lett. 1990;263:51–56. doi: 10.1016/0014-5793(90)80703-l. [DOI] [PubMed] [Google Scholar]
- Gonzalez M, Frank EG, Levine AS, Woodgate R. Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: in vitro degradation and identification of residues required for proteolysis. Genes Dev. 1998;12:3889–3899. doi: 10.1101/gad.12.24.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Wickner S, Maurizi MR. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- Guisbert E, Herman C, Lu CZ, Gross CA. A chaperone network controls the heat shock response in E. coli. Genes Dev. 2004;18:2812–2821. doi: 10.1101/gad.1219204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Biran D, Ron EZ. Regulated proteolysis in Gram-negative bacteria--how and when? Nat Rev Microbiol. 2011;9:839–848. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. 4. Sinauer Associates, Inc; 2007. [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Heo M, Maslov S, Shakhnovich E. Topology of protein interaction network shapes protein abundances and strengths of their functional and nonspecific interactions. Proc Natl Acad Sci U S A. 2011;108:4258–4263. doi: 10.1073/pnas.1009392108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- Huang SL, Wu LC, Liang HK, Pan KT, Horng JT, Ko MT. PGTdb: a database providing growth temperatures of prokaryotes. Bioinformatics. 2004;20:276–278. doi: 10.1093/bioinformatics/btg403. [DOI] [PubMed] [Google Scholar]
- Ionescu RM, Smith VF, O’Neill JC, Jr, Matthews CR. Multistate equilibrium unfolding of Escherichia coli dihydrofolate reductase: thermodynamic and spectroscopic description of the native, intermediate, and unfolded ensembles. Biochemistry. 2000;39:9540–9550. doi: 10.1021/bi000511y. [DOI] [PubMed] [Google Scholar]
- Jones BE, Jennings PA, Pierre RA, Matthews CR. Development of nonpolar surfaces in the folding of Escherichia coli dihydrofolate reductase detected by 1-anilinonaphthalene-8-sulfonate binding. Biochemistry. 1994;33:15250–15258. doi: 10.1021/bi00255a005. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T, Rudolph R, Kohler HH, Buchner J. Protein aggregation in vitro and in vivo: a quantitative model of the kinetic competition between folding and aggregation. Biotechnology (N Y) 1991;9:825–829. doi: 10.1038/nbt0991-825. [DOI] [PubMed] [Google Scholar]
- Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren M, Sorgjerd K, Hammarstrom P. Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophys J. 2005;88:4200–4212. doi: 10.1529/biophysj.104.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Langer T, Boteva R, Schramel A, Horwich AL, Hartl FU. Chaperonin-mediated protein folding at the surface of groEL through a ‘molten globule’-like intermediate. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- Maurizi MR, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. Journal of bacteriology. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munavar H, Zhou Y, Gottesman S. Analysis of the Escherichia coli Alp phenotype: heat shock induction in ssrA mutants. Journal of bacteriology. 2005;187:4739–4751. doi: 10.1128/JB.187.14.4739-4751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S, Mehra N, Lynn AM, Bhattacharya A. Comparison of theoretical proteomes: identification of COGs with conserved and variable pI within the multimodal pI distribution. BMC Genomics. 2005;6:116. doi: 10.1186/1471-2164-6-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii W, Maruyama T, Matsuoka R, Muramatsu T, Takahashi K. The unique sites in SulA protein preferentially cleaved by ATP-dependent Lon protease from Escherichia coli. Eur J Biochem. 2002;269:451–457. doi: 10.1046/j.0014-2956.2001.02668.x. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Levy ED, Tartaglia GG, Vendruscolo M. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc Natl Acad Sci U S A. 2009;106:10159–10164. doi: 10.1073/pnas.0812414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Powers DL, Gierasch LM. FoldEco: A Model for Proteostasis in E. coli. Cell reports. 2012;1:265–276. doi: 10.1016/j.celrep.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn OB. Molten globule and protein folding. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Rosen R, Biran D, Gur E, Becher D, Hecker M, Ron EZ. Protein aggregation in Escherichia coli: role of proteases. FEMS Microbiol Lett. 2002;207:9–12. doi: 10.1111/j.1574-6968.2002.tb11020.x. [DOI] [PubMed] [Google Scholar]
- Stan G, Lorimer GH, Thirumalai D, Brooks BR. Coupling between allosteric transitions in GroEL and assisted folding of a substrate protein. Proc Natl Acad Sci U S A. 2007;104:8803–8808. doi: 10.1073/pnas.0700607104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009a;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009b;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Torrescabassa AS, Gottesman S. Capsule Synthesis in Escherichia-Coli K-12 Is Regulated by Proteolysis. Journal of bacteriology. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi NK, Fenton WA, Deniz AA, Horwich AL. Double mutant MBP refolds at same rate in free solution as inside the GroEL/GroES chaperonin chamber when aggregation in free solution is prevented. FEBS letters. 2011;585:1969–1972. doi: 10.1016/j.febslet.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrinov KG, Clark PL. Cotranslational folding increases GFP folding yield. Biophys J. 2010;98:1312–1320. doi: 10.1016/j.bpj.2009.12.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2:a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M. Proteome folding and aggregation. Curr Opin Struct Biol. 2012;22:138–143. doi: 10.1016/j.sbi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Human mutation. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol Biol. 2005;5:44. doi: 10.1186/1471-2148-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CS, Shakhnovich EI. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA Sequence Determinants of Thermophilic Adaptation. PLoS Comput Biol. 2007a;3:e5. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Chen P, Shakhnovich BE, Shakhnovich EI. A First-Principles Model of Early Evolution: Emergence of Gene Families, Species, and Preferred Protein Folds. PLoS Comput Biol. 2007b;3:e139. doi: 10.1371/journal.pcbi.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Chen P, Shakhnovich EI. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci U S A. 2007c;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwietering MH, Jongenburger I, Rombouts FM, van’t Riet K. Modeling of the bacterial growth curve. Applied and environmental microbiology. 1990;56:1875–1881. doi: 10.1128/aem.56.6.1875-1881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


