Abstract
Persistent forms of synaptic plasticity are widely thought to require the synthesis of new proteins. This feature of long-lasting forms of plasticity largely has been demonstrated using inhibitors of general protein synthesis, such as either anisomycin or emetine. However, these drugs, which inhibit elongation, cannot address detailed questions about the regulation of translation initiation, where the majority of translational control occurs. Moreover, general protein synthesis inhibitors cannot distinguish between cap-dependent and cap-independent modes of translation initiation. In the present study, we took advantage of two novel compounds, 4EGI-1 and hippuristanol, each of which targets a different component of the eukaryotic initiation factor (eIF)4F initiation complex, and investigated their effects on long-term potentiation (LTP) at CA3-CA1 synapses in the hippocampus. We found that 4EGI-1 and hippuristanol both attenuated long-lasting late-phase LTP induced by two different stimulation paradigms. We also found that 4EGI-1 and hippuristanol each were capable of blocking the expression of newly synthesized proteins immediately after the induction of late-phase LTP. These new pharmacological tools allow for a more precise dissection of the role played by translational control pathways in synaptic plasticity and demonstrate the importance of multiple aspects of eIF4F in processes underlying hippocampal LTP, laying the foundation for future studies investigating the role of eIF4F in hippocampus-dependent memory processes.
Keywords: late-phase long-term potentiation, RNA helicase, eukaryotic initiation factor 4E, cap-dependent translation
the importance of protein synthesis in synaptic plasticity was first demonstrated in the rodent hippocampus in the dentate gyrus of behaving rats (Krug et al. 1984) and in brain slice preparations using puromycin (Stanton and Sarvey 1984). Subsequently, the role of protein synthesis and its regulation have been studied extensively using pharmacological, genetic, and electrophysiological approaches (Frey et al. 1996; Frey and Morris 1997; Nguyen et al. 1994; Scharfman et al. 2001). Protein synthesis-dependent forms of synaptic plasticity are believed to be the cellular substrates for cognitive phenomena, including memory consolidation. Therefore, understanding the mechanisms underlying protein synthesis-dependent forms of synaptic plasticity is critical for gaining insights about brain function at the molecular level.
Synaptic translation is regulated via both extra- and intracellular signals and can occur by two distinct pathways (Hoeffer and Klann 2009; Raught et al. 2000). The most prevalent mechanism involves the recruitment of ribosomes to the 5′-end of the mRNA template (Pestova et al. 2001). This process is enhanced by the presence of the mRNA 5′ “cap” motif (7-methylguanosine GTP cap) and regulated by the activities of multiple eukaryotic initiation factors (eIFs). One eIF central to cap-based translational initiation is eIF4G, a multifunctional scaffolding protein that recruits the 5′ mRNA cap-binding protein eIF4E and the RNA helicase eIF4A, an RNA dead box helicase, that together with eIF4G and eIF4E, forms the initiation factor eIF4F (Raught et al. 2000). RNA helicases play a pivotal role in translation by providing enzymatic activity that reduces or modifies RNA secondary structure, thereby affecting the efficiency of ribosomal processing. The combined activities of the eIF4F complex are a central factor in the translation of capped mRNAs. The second pathway for protein synthesis uses an alternative method of ribosome recruitment to the substrate mRNA, via an internal ribosome entry site (IRES), bypassing the requirement for capped mRNA (Pestova et al. 2001). Although both cap-dependent and cap-independent forms of translation rely on some shared initiation factors, several of the required eIFs for IRES-mediated translation differ from those for cap-dependent translation (Morley et al. 2005; Pestova et al. 2001).
A number of compounds are able to block protein synthesis. These inhibitors act at different points along the protein synthesis pathway, from initiation to the incorporation of new amino acids into an elongating peptide chain (for reviews, see Kelly et al. 2000; Tenson and Mankin 2006). The most commonly used protein synthesis inhibitors cannot be used to address at least two questions with respect to protein synthesis and synaptic plasticity. First, protein synthesis inhibitors targeting the ribosome cannot be used to distinguish between the major forms of eukaryotic mRNA translation, cap-dependent and cap-independent/IRES-based protein synthesis. Second, they also cannot be used to directly assess the role of upstream initiation factor regulatory activities, such as eIF4F, in synaptic plasticity. The latter point has been addressed indirectly using compounds targeting molecular signaling upstream of translational initiation. These studies have used compounds that inhibit either mammalian target of rapamycin (mTOR) or ERK (Kelleher et al. 2004; Sharma et al. 2010; Tang et al. 2002). However, these are hub kinases that regulate multiple signaling cascades, so a direct assessment of the role of eIF4F in translation-dependent synaptic plasticity remains largely unexplored.
Compounds that can be used to directly assess eIF4F-related activities are now available. 4EGI-1 is a small-molecule inhibitor of the interaction between eIF4G and eIF4E and, consequently, disrupts the translational activity of eIF4F (Hoeffer et al. 2011; Moerke et al. 2007). eIF4A exists in two forms: a free form and an eIF4F-bound form (eIF4A-eIF4F) (Edery et al. 1983; Grifo et al. 1983). Hippuristanol is a small-molecule inhibitor that significantly blocks eIF4A-eIF4F activity (Bordeleau et al. 2006). Using these two compounds, in the present study, we show here, for the first time, that specific inhibition of eIF4F-mediated activities blocks the expression of protein synthesis-dependent long-term potentiation (LTP) in the Schaffer collateral circuit of the hippocampus.
METHODS
Mouse husbandry.
All mice used in the experiments were male C57BL/6J and were obtained from Taconic Farms (Albany, NY). Mice were maintained on a 12:12-h light-dark schedule with food and water available ad libitum. Mice tested were between 8 and 16 wk of age. All procedures were approved by the Institutional Animal Care and Use Committee of New York University and complied with the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Slice preparation and electrophysiology.
Transverse hippocampal slices (400 μm) were prepared from mice at 2–4 mo of age using a vibratome as previously described (Hoeffer et al. 2008). Slices were maintained at room temperature in a submersion chamber with artificial cerebral spinal fluid (aCSF) containing the following (in mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4, 24 NaHCO3, and 15 glucose bubbled with 95% O2-5% CO2. Slices were incubated for at least 2 h before removal for experiments. For electrophysiology experiments, slices were transferred to recording chambers (preheated to 32°C), where they were superfused with oxygenated aCSF. Monophasic, constant-current stimuli (100 μs) were delivered with a bipolar silver electrode placed in the stratum radiatum of area CA3, and field excitatory postsynaptic potentials (fEPSPs) were recorded in the stratum radiatum of area CA1 with electrodes filled with aCSF (resistance: 2–4 MΩ). Baseline fEPSPs were monitored by delivering stimuli at 0.033 Hz. fEPSPs were acquired, and amplitudes and maximum initial slopes measured, using pCLAMP 10 (Molecular Devices). Slices were allowed to recover in the recording chamber at least 30 min before recordings began. Basal fEPSPs were stable for at least 20 min before the start of each experiment. Late-phase LTP (L-LTP) was induced by two types of high-frequency stimulation (HFS) protocols: 1) 4× HFS, consisting of four 1-s 100-Hz trains, with an intertrain interval of 5 min, delivered at 40–50% of the intensity that evoked maximum fEPSPs, and 2) 2× HFS, consisting of two 1-s 100-Hz trains, with an intertrain interval of 60 s, delivered at 70–80% of the intensity that evoked maximum fEPSPs (Ma et al. 2011). Drug preincubation was performed at room temperature in submersion maintenance chambers containing aCSF saturated with 95% O2-5% CO2. All drugs were prepared as stock solutions in DMSO and then added to the aCSF [4EGI-1 final vehicle concentration: 1% DMSO and 0.5% β-cyclodextrin (Sigma); hippuristanol final vehicle concentration: 1% DMSO]. Drugs were applied for 60 min before HFS and for various times after HFS (20–40 min after HFS, as indicated).
Isolation of N-methyl-d-aspartate receptor-mediated fEPSPs.
Baseline fEPSPs were collected as described above. After a stable baseline had been established, slices were incubated in either vehicle or 4EGI-1 (100 μM) for 60 min. aCSF containing 0 mM MgCl2 and 4 mM CaCl2 was then applied to treated slices to evoke mixed α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid- and N-methyl-d-aspartate (NMDA) receptor-mediated fEPSPs. 6-Cyano-2,3-dihydroxy-7-nitro-quinoxaline at 20 μM then was applied to slices to isolate NMDA receptor-mediated fEPSPs. Confirmation that the remaining fEPSP was mediated by NMDA receptors was obtained with a final application of 100 μM 2-amino-5-phosphonovaleric acid (APV) to the slices.
Immunoprecipitation.
Tissue was homogenized in ice-cold lysis immunoprecipitation buffer as previously described (Hoeffer et al. 2011). Cleared hippocampal homogenate (150–250 μg) was incubated with anti-eIF4G (1:100, Bethyl Laboratories) and gently shaken overnight at 4°C. The antibody-lysate mix was incubated with 75 μl IgG bound to agarose beads (Pierce). The bead-sample slurry was incubated through rocking at 25°C for 2 h (or 4°C overnight). The supernatant was removed and saved, and immunoprecipitates were washed three times in lysis buffer and once in wash buffer containing (in mM) 50 HEPES (pH 7.5), 40 NaCl, and 2 EDTA before resolution with Western blot analysis.
Puromycin protein labeling.
Hippocampal slices were prepared as described above (Hoeffer et al. 2008). Slices then were subjected to pharmacological pretreatment (cycloheximide, 4EGI-1, or hippuristanol) for 60 min at the desired concentration. Proteins were labeled using an adaption of the SuNSET protocol (Hoeffer et al. 2011; Schmidt et al. 2009). At the end of the protein synthesis inhibitor incubation, puromycin (10 μg/ml in vehicle) was added, and slices were incubated for another 60 min. During this incubation time, newly synthesized proteins were end labeled with puromycin. Slices were transferred to oxygenated aCSF in three successive washes of 2 min and then flash frozen on dry ice. Area CA1 was microdissected from the slices, and protein lysates were prepared and blotted. Puromycin-labeled proteins were identified on blots using mouse monoclonal antibody 12D10 (1:5,000 from a 5-mg/ml stock). Because only a small fraction of the brain proteins were labeled, the signal from blots was identified using ECL-Advance. Protein synthesis levels were determined by taking the total lane signal from 250 to 15 kDa and subtracting the signal from an unlabeled protein lane. Comparisons for time points were made as fold changes of the labeled vehicle for the corresponding time point.
Western blot analysis.
Either freshly extracted or treated hippocampal slices were immediately flash frozen on dry ice. Soluble protein extracts were prepared by homogenizing tissue samples in ice-cold buffer [containing 50 mM Tris·HCl (pH 7.5), 150 mM KCl, 1 mM DTT, 1 mM EDTA, 1× complete protease inhibitor mixture III, 1× phosphatase inhibitor mixture I, and 1× phosphatase inhibitor mixture II (Sigma)]. Protein concentration was measured with the BCA assay (Pierce). SDS-PAGE buffer (6×) was added to aliquots of protein (15–20 μg, or 50 μg for puromycin-labeled protein) that were resolved on Novex precast 4–12% gradient gels (Invitrogen), transferred to a polyvinylidene difluoride membrane, and processed for overnight incubation with primary antibodies (see below) followed by secondary antibodies. Membranes were washed, and proteins were detected with enhanced chemiluminescence reagent (ECL+, GE Healthcare) and visualized using a Kodak 4000MM or GE LAS4000 imager to obtain pixel density values for the band of interest. All images were obtained using maximum sensitivity settings with no binning (0- to 65-K signal range). No images analyzed presented saturating signals for the bands of interest (>65-K grayscale value). Band density values were normalized to one of the following: β-actin, GAPDH, or eIF4G (eIF4G for immunoprecipitation experiments). All gels included loading controls of known concentration to allow comparisons across different blots.
Antibodies.
The following antibodies were used in this study: eIF4E monoclonal mouse antibody (1:1000, Abgent), eIF4G1 polyclonal rabbit antibody (1:100, Bethyl Laboratories), eIF4G1 polyclonal mouse antibody (1:1000, R&D Systems), GAPDH rabbit polyclonal antibody (1:1000, Chemicon), and puromycin monoclonal antibody (mouse 12 D 10, 1:5000). For information about the puromcyin monoclonal antibody, see Schmidt et al. (2009). The following secondary antibodies used: goat anti-rabbit IgG-horseradish peroxidase conjugate (1:10,000, Promega) and goat anti-mouse IgG-horseradish peroxidase conjugate (1:10,000, Promega).
Data analysis.
Data are presented as means ± SEM. Statistics were performed using GraphPad software (GraphPad). For comparison between two groups, either repeated-measures ANOVA or a two-tailed independent Student's t-test was used (for times spanning the last 20 min of recording). For comparisons between multiple groups, multiway (n-way) ANOVA was used followed by individual post hoc tests when applicable. Error probabilities of P < 0.05 were considered statistically significant.
RESULTS
4EGI-1 disrupts hippocampal eIF4E/eIF4G interactions but does not affect either basal synaptic transmission or paired-pulse facilitation in area CA1.
We recently used 4EGI-1 to test the role of eIF4F formation and cap-dependent translation in amygdala-dependent associative fear memory (Hoeffer et al. 2011). To confirm that 4EGI-1 would also impair eIF4F formation in hippocampal slices, we incubated hippocampal slices with 4EGI-1 and performed immunoprecipitation experiments to measure eIF4E/eIF4G interactions. We incubated slices with 100 μM 4EGI-1 for 60 min and then isolated eIF4F complexes by immunoprecipitating eIF4G. 4EGI-1 disrupted eIF4F formation, as indicated by significantly reduced eIF4E/eIF4G interactions (Fig. 1A). In agreement with previous findings (Hoeffer et al. 2011; Moerke et al. 2007), 4EGI-1 worked in a dose-dependent manner, with lower concentrations of 4EGI-1 also reducing eIF4F formation but not to the same levels as with 100 μM (data not shown). We proceeded to examine basal synaptic transmission, as measured by synaptic output in response to a stimulatory input in hippocampal slices treated with 4EGI-1. We found that input/output curves were indistinguishable between slices treated with 4EGI-1 and vehicle (Fig. 1B). We next examined whether 4EG1–1 exposure was neurotoxic when applied for extended periods of time. When slices were exposed to either 50 or 100 μM 4EGI-1, we did not observe a significant reduction in basal fEPSPs (Fig. 1C). To determine whether 4EGI-1 treatment altered NMDA glutamate receptor function, we isolated NMDA receptor-mediated fEPSPs in area CA1 prepared from 4EGI-1- and vehicle-treated slices. NMDA receptor-mediated fEPSPs from 4EGI-1-treated slices were indistinguishable from vehicle-treated slices (Fig. 1D). Finally, we examined whether 4EGI-1 affected paired-pulse facilitation (PPF), a form of presynaptic plasticity (Katz and Miledi 1968). PPF was normal in 4EGI-1-treated slices compared with vehicle-treated slices at several interpulse intervals (Fig. 1D). Taken together, these data suggest that 4EGI-1 does not affect either basal synaptic transmission or presynaptic Ca2+-release properties in area CA1 of hippocampal slices.
Fig. 1.

4EGI-1 disrupts eukaryotic initiation factor (eIF)4F formation but does not affect basal synaptic function in the hippocampus. A: representative Western blot of eIF4F levels, as measured by immunoprecipitation (IP) of eIF4G and its interactions with eIF4E in hippocampal slices treated with 100 μM 4EGI-1. Treatment of slices with 50 μM 4EGI-1 also reduced eIF4F levels, but to a lesser exent (data not shown). The blot represents three independent experiments. B: input versus output plot showing that 4EGI-1- and vehicle-treated slices have comparable field excitatory postsynaptic potential (fEPSP) slopes evoked by increasing synaptic stimulation. Open circles, vehicle-treated mice; solid circles, 100 μM 4EGI-1-treated slices. n = 12–17 slices, 5–7 mice/treatment. P > 0.05 by repeated-measures ANOVA. C, bottom: 4EGI-1 treatment did not alter baseline fEPSPs. Both 50 and 100 μM 4EGI-1 did not produce detectable effects on baseline fEPSPs. Representative traces for fEPSPs are shown at the top before treatment (1) and after 60 min of exposure to 4EGI-1 (2). Solid circles, 50 μM 4EGI-1-treated slices; shaded circles, 100 μM 4EGI-1-treated slices. n = 12–15 slices, 8 mice/treatment. P > 0.05 by repeated-measures ANOVA. D: 4EGI-1-treated slices displayed normal N-methyl-d-aspartate (NMDA)-mediated fEPSPs compared with vehicle-treated slices. fEPSPs with artificial cerebrospinal fluid (aCSF) containing 0 mM Mg2+ and 4 mM Ca2+. α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-mediated fEPSPs were determined after 6-cyano-2,3-dihydroxy-7-nitro-quinoxaline (CNQX) treatment (20 μM). NMDA receptor-mediated fEPSPs were determined by sensitivity to APV (100 μM). n = 8–11 slices/condition. P > 0.05 by repeated-measures ANOVA. E: 4EGI-1-treated slices exhibited normal paired-pulse facilitation (PPF) compared with vehicle-treated slices. Percent facilitation, determined by the ratio of the second fEPSP to the first fEPSP, is shown at interpulse intervals from 10 to 300 ms. n = 15 vehicle-treated slices and 16 EGI-1-treated slices, 8 mice/treatment. P > 0.05 by repeated-measures ANOVA.
Disruption of eIF4E/eIF4G interactions with 4EGI-1 impairs protein synthesis-dependent forms of LTP.
Having established that 4EGI-1 did not impact either basal synaptic transmission or presynaptic facilitation, we next tested the idea that eIF4F formation was required for protein synthesis-dependent forms of synaptic plasticity. We examined LTP at Schaffer collateral synapses in area CA1 of the hippocampus. We began with an examination of the forms of LTP that did not rely on protein synthesis to determine whether perturbing eIF4F would alter more transient forms of synaptic plasticity. To do this, we induced early-phase LTP (E-LTP) with a single train of HFS. Compared with vehicle controls, slices treated with 4EGI-1 did not show any differences in E-LTP (Fig. 2A), suggesting that eIF4F is not required for protein synthesis-independent LTP. We next examined protein synthesis-dependent LTP, or L-LTP, which was induced with four spaced trains of HFS. L-LTP in slices treated with 4EGI-1 decreased substantially during the course of the 3-h experiment compared with L-LTP in control slices (Fig. 2B). Similar results were obtained when L-LTP was elicited with a protocol of two trains of HFS (Tsokas et al. 2007) (Fig. 2C). These results indicate that the inhibition of eIF4F impairs long-lasting forms of LTP in area CA1 that normally require protein synthesis. Taken together, these data suggest that a 60-min pretreatment with 4EGI-1 does not affect either basal synaptic transmission or presynaptic Ca2+-release properties in treated slices.
Fig. 2.

4EGI-1 impairs protein synthesis-dependent LTP. A: a single train of high-frequency stimulation (HFS) resulted in similar levels of early-phase long-term potentiation (E-LTP) in vehicle- and 4EGI-1-treated slices. n = 11 vehicle-treated slices and 8 4EGI-1-treated slices, 5–8 mice/treatment. P > 0.05 by repeated-measures ANOVA. B: 4EGI-1 blocked late-phase long-term potentiation (L-LTP) elicited by four trains of HFS compared with vehicle-treated slices. n = 13 vehicle-treated slices and 15 EGI-1-treated slices, 7–9 mice/treatment. P < 0.01 by repeated-measures ANOVA. C: 4EGI-1 blocked L-LTP induced by two trains of HFS compared with vehicle-treated slices. n = 6 vehicle-treated slices and 8 EGI-1-treated slices, 5–6 mice/treatment. P < 0.05 by two-tailed Student's t-test. Representative traces for fEPSPs are shown at the top of A–C before LTP-inducing stimulation (1) and near the end of the recording period (2) for each experiment.
Inhibition of eIF4A activity does not affect either basal synaptic transmission or PPF.
In addition to mRNA cap binding via eIF4E, the eIF4F complex also contains eIF4A, an RNA helicase critical for cap-dependent translation (Raught et al. 2000). To investigate the role of eIF4A in protein synthesis-dependent LTP, we used hippuristanol (Bordeleau et al. 2006) to block eIF4A activity in the hippocampus. We first examined basal synaptic transmission and found that input/output curves were indistinguishable between slices treated with hippuristanol (10 μM) and vehicle (Fig. 3A). In addition, when slices were exposed to 10 μM hippuristanol for 60 min, there was no significant alteration in fEPSPs (Fig. 3B). Finally, PPF was similar in hippuristanol-treated slices and vehicle-treated slices at several interpulse intervals (Fig. 3C). Taken together, these findings indicate that inhibition of eIF4A activity with hippuristanol does not affect either basal synaptic transmission or presynaptic Ca2+-release properties in area CA1 of hippocampal slices.
Fig. 3.

Inhibition of eIF4A helicase activity does not affect either basal synaptic transmission or presynaptic facilitation. A: input versus output plot showing that hippuristanol- and vehicle-treated slices have comparable fEPSP slopes evoked by increasing synaptic stimulation. Open circles, vehicle-treated slices; solid circles, 10 μM hippuristanol-treated slices. n = 9–12 slices, 6–8 mice/treatment. P > 0.05 by repeated-measures ANOVA. B, bottom: hippuristanol treatment did not alter baseline fEPSPs. Hippuristanol (10 μM) did not produce detectable effects on baseline fEPSPs. Representative traces for fEPSPs are shown at the top before treatment (1) and after 60 min of exposure to hippuristanol (2). n = 5 vehicle-treated slices and 5 hippuristanol-treated slices, 5 mice/treatment. C: hippuristanol-treated slices exhibited normal PPF compared with vehicle-treated slices. Percent facilitation, determined by the ratio of the second fEPSP to the first fEPSP, is shown at interpulse intervals from 10 to 300 ms. n = 10 vehicle-treated slices and 10 hippuristanol-treated slices, 5 mice/treatment. P > 0.05 by repeated-measures ANOVA.
eIF4F-mediated RNA helicase activity is required for protein synthesis-dependent LTP.
Because inhibition of eIF4A activity did not affect either basal synaptic transmission or presynaptic facilitation, we next tested whether eIF4A activity was required for protein synthesis-dependent forms of LTP. Using a strategy similar to what we used in experiments with 4EGI-1 (Fig. 2), we first examined E-LTP in slices treated with hippuristanol. We found that E-LTP was indistinguishable in slices treated with hippuristanol compared with slices treated with vehicle (Fig. 4A). We next examined the expression of protein synthesis-dependent L-LTP. Similar to experiments with 4EGI-1, L-LTP induced with four trains of HFS in hippuristanol-treated slices was significantly inhibited, returning to baseline ∼2 h after the last train of HFS (Fig. 4B). Similar results were obtained when L-LTP was elicited with trains of HFS (Fig. 2C). These results demonstrate that another eIF4F-mediated function, eIF4A RNA helicase activity, is required for L-LTP. Combined with the experiments with 4EGI-1 (Fig. 2), these results indicate that multiple components of eIF4F are required for long-lasting forms of LTP in hippocampal area CA1 that normally require protein synthesis.
Fig. 4.
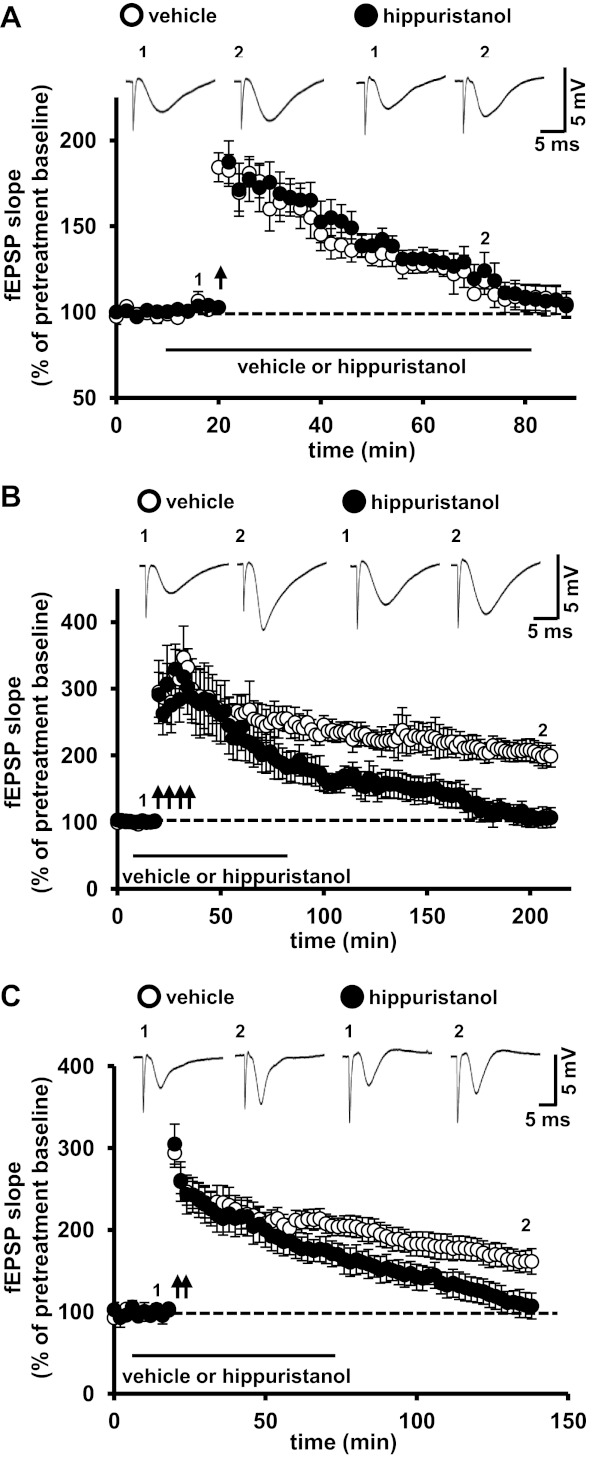
eIF4A activity is required for protein synthesis-dependent LTP. A: a single train of HFS resulted in similar levels of E-LTP in vehicle- and hippuristanol-treated slices. n = 11 vehicle-treated slices and 8 hippuristanol-treated slices, 5–8 mice/treatment. P > 0.05 by repeated-measures ANOVA. B: hippuristanol blocked L-LTP elicited by four trains of HFS compared with vehicle-treated slices. n = 10 vehicle-treated slices and 11 hippuristanol-treated slices, 8 mice/treatment. P < 0.01 by repeated-measures ANOVA. C: hippuristanol blocked L-LTP induced by two trains of HFS. n = 10 vehicle-treated slices and 10 hippuristanol-treated slices, 8 mice/treatment. P < 0.05 by two-tailed Student's t-test. Representative traces for fEPSPs are shown at the top of A–C before LTP-inducing stimulation (1) and near the end of the recording period (2) for each experiment.
Multiple components of eIF4F are required for the increased protein synthesis associated with L-LTP.
Previously, we showed that eIF4F levels increase in response to L-LTP-inducing HFS and after learning (Banko et al. 2005; Hoeffer et al. 2011). However, these studies did not provide direct evidence linking L-LTP-inducing stimulation to eIF4F and protein synthesis. Therefore, we determined whether increased protein synthesis occurs after L-LTP-inducing stimulation and, if so, whether the increases could be blocked by inhibition of eIF4F. First, we investigated whether either 4EGI-1 or hippuristanol inhibited basal protein synthesis in hippocampal slices. To measure protein synthesis in hippocampal slices, we incubated slices with subinhibitory concentrations of puromycin to effectively end-label newly synthesized peptides with a puromycin molecule (Hoeffer et al. 2011; Schmidt et al. 2009). Hippocampal proteins were labeled in the presence of either vehicle or eIF4F inhibitors. We found that preincubation of slices with either 4EGI-1 or hippuristanol inhibited protein synthesis (Fig. 5A). We also observed that neither 4EGI-1 nor hippuristanol inhibited protein synthesis as effectively as cycloheximide, a general protein synthesis inhibitor (Fig. 5A). We next asked whether induction of L-LTP was correlated with increased protein synthesis and, if so, whether eIF4F was required for the increase. We incubated hippocampal slices with eIF4F inhibitors in a manner identical to what was used for the LTP experiments (Figs. 2 and 4) and then labeled newly synthesized proteins as outlined in the schematic shown in Fig. 5B. Using four trains of HFS to induce L-LTP, we observed a significant increase in newly synthesized proteins compared with control slices (Fig. 5C). We also observed that both 4EGI-1 and hippuristanol attenuated the L-LTP-associated increase in protein synthesis (Fig. 5C). These data provide evidence demonstrating that multiple components of eIF4F are involved in the synthesis of new proteins in response to stimulation that induces protein synthesis-dependent LTP.
Fig. 5.
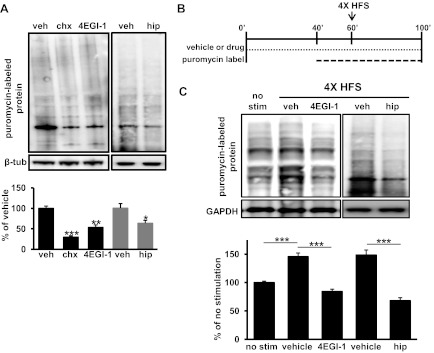
Blockade of eIF4F function impairs L-LTP-induced protein synthesis. A: new protein synthesis was measured by SuNSET. Puromycin labeling of newly synthesized proteins showed that protein synthesis was inhibited by cycloheximide (chx), 4EGI-1, and hippuristanol (hip). Total protein lysate was visualized after 60-min incubation with vehicle (veh) and either 20 μg/ml cycloheximide, 100 μM 4EGI-1, or 10 μM hippuristanol followed by a 60-min puromycin-labeling chase. Newly synthesized protein after inhibitor treatment was measured by the total lane signal at 250–10 kDa compared with the vehicle control (cycloheximide: 33.5%, 4EGI-1: 59.2%, hippuristanol: and 66.0%). n = 6 for vehicle treatment, 4 for cycloheximide treatment, 6 for EGI-1 treatment, and 3 for hippuristanol treatment. Data for vehicle, cycloheximide, and 4EGI-1 treatment were averaged from four independent experiments and two for hippuristanol treatment. *P < 0.05, **P < .01, and ***P < .001 by two-tailed Student's t-test. B: schematic representation of inhibitior treatment and puromycin-labeling scheme for protein synthesis measurements in hippocampal slices after four trains of HFS. Slices were pretreated with either vehicle or drug (100 μM 4EGI-1 or 10 μM hippuristanol) for 40 min, and puromycin label was then added to the bath for 20 min. After 60 min of vehicle/drug incubation, slices were stimulated with four trains of HFS. Forty min after the initial HFS, slices were harvested for protein extraction. C: four trains of HFS induced protein synthesis in area CA1 of the hippocampus that was dependent on eIF4F. Protein synthesis induced by HFS was blocked by either 4EGI-1 (81.3% of unstimulated control) or hippuristanol (68.4% of control). The signal for each treatment was averaged against the no-stimulation control. Western blots from four independent experiments were averaged for vehicle, cycloheximide, and 4EGI-1 treatment and two were averaged for hippuristanol treatment. n = 11 for vehicle treatment, 9 for 4EGI-1 treatment, and 3 for hippuristanol treatment. ***P < 0.001 by two-tailed Student's t-test.
DISCUSSION
In this study, we used two distinct small-molecule inhibitors targeting different components of eIF4F to explore its role in persistent, protein synthesis-dependent forms of synaptic plasticity. We found that blockade of either eIF4E/eIF4G interactions or eIF4A helicase activity did not impair either basal synaptic transmission or transient forms of synaptic plasticity at Schaffer collateral synapses in area CA1 of the hippocampus. However, we discovered that disrupting either component of eIF4F interfered with the expression of long-lasting, protein synthesis-dependent forms of LTP at this synapse. Finally, we demonstrated that both 4EGI-1 and hippuristanol ablate increases in protein synthesis that are triggered by L-LTP-inducing stimulation. This study provides additional evidence for the important role of protein synthesis in persistent forms of synaptic modification. Importantly, these findings provided more detailed information about the role of eIF4F long-lasting synaptic plasticity and provide a description of new tools that are available for examining translational control in synaptic plasticity and memory formation.
Although it is generally accepted that protein synthesis is required to maintain long-term functional synaptic changes, the role of protein synthesis in synaptic plasticity has been questioned (Routtenberg 2008; Rudy 2008). The vast majority of studies establishing a role for protein synthesis in long-lasting plasticity and long-term memory have used general inhibitors of protein synthesis, such as anisomycin and cycloheximide, which target ribosomal functions, such as ribosomal subunit binding or substrate translocation (Barbacid and Vazquez 1975; Frey et al. 1988; Krug et al. 1984; Nguyen and Kandel ; Obrig et al. 1971; Stanton and Sarvey 1984; Tenson and Mankin 2006). Most of these compounds are known to have multiple off-target effects, which have been seized upon by critics as potential experimental confounds (Adams 2003; Edwards and Mahadevan 1992; Flexner and Goodman 1975; Hazzalin et al. 1998). Although we cannot exclude the possibility that 4EGI-1 and hippuristanol may also have side effects, they do not impact either stress responses that increase ERK activation or alterations in DNA and RNA synthesis that have been shown with more general protein synthesis inhibitors (Bordeleau et al. 2006; Hoeffer et al. 2011; McMahon et al. 2011). Thus, the characterization of selective small-molecule inhibitors of cap-dependent protein synthesis such as 4EGI-1 and hippuristanol should allow for more precise pharmacological inhibition of synaptic translation that circumvent some of the potential experimental confounds of inhibitors used in previous studies.
A previous study used hippuristanol to examine translation-dependent plasticity in interneurons in the hippocampus, but synaptic plasticity at Schaffer collateral synapses in area CA1 was not studied (Ran et al. 2009). Thus, whether components of eIF4F are required for L-LTP has not been examined. A role for eIF4F in protein synthesis-dependent synaptic plasticity has been inferred from studies using either mTORC1 inhibitors or pharmacogenetic manipulation of mTOR signaling to examine L-LTP (Cammalleri et al. 2003; Hoeffer et al. 2008; Tang et al. 2002; Tsokas et al. 2005). However, at least one study using similar approaches to examine L-LTP in the dentate gyrus in vivo did not identify a role for mTORC1 in the regulation of eIF4F (Panja et al. 2009). In addition, a recent study has cast doubt on the phosphoregulation of eIF4F-binding protein 2, suggesting that protein synthesis-dependent synaptic plasticity in the brain uses a mechanism independent of mTORC1 phosphorylation of eIF4F-binding protein 2 and, thus, eIF4F (Bidinosti et al. 2010). Thus, the findings detailed here demonstrating that directly impeding eIF4E/eIF4G interactions and eIF4A helicase activity provide directs evidence for the critical role of eIF4F in the manifestation of LTP.
Treatment of slices with either 4EGI-1 or hippuristanol produced a comparable decrease in puromycin-labeled, newly synthesized proteins in slices given LTP-inducing HFS, but the level of blockade was less than what was seen in unstimulated slices treated with either inhibitor (compare Fig. 5, C with A). This suggests that blockade of L-LTP-induced protein synthesis with 4EGI-1 and hippuristanol was incomplete but sufficient to disrupt L-LTP (Figs. 2, B and C, and 4, A and C). Another interesting possibility is that that blockade of eIF4F-mediated translation reveals other modes of translation induced by L-LTP. Interestingly, both 4EGI-1 and hippuristanol can distinguish between cap-dependent and cap-independent translation (Bordeleau et al. 2006; Moerke et al. 2007), raising the possibility that IRES-mediated translation supports some aspects of protein synthesis-dependent synaptic plasticity. In our experiments, it is likely that eIF4A bound to eIF4G is the critical target of hippuristanol blockade because its affinity for eIF4A as part of the eIF4F complex is much greater than that eIF4A in its free form (Oberer et al. 2005) and it has been previously shown that hippuristanol is a potent inhibitor of eIF4A activity in the eIF4F complex (Bordeleau et al. 2006). Finally, we also observed that the distinctive routes of eIF4F inhibition resulted in qualitatively different L-LTP blockade (Figs. 2 and 4), suggesting that the different eIF4F-regulated activities contribute differentially to the expression of LTP. Thus, our findings suggest that the multiple components of the eIF4F complex are integral to L-LTP and also point to the eIF4F complex as an important molecular marker for additional types of long-lasting changes in neuronal function.
A promising field of investigation has emerged exploring the causal link between dysregulation of protein synthesis and deficits cognitive function as well as in neurological disorders (Auerbach et al. 2011; Ehninger et al. 2009; Hoeffer et al. 2008, 2011; Kwon et al. 2006). These studies have improved our understanding of pathophysiology in cognitive disorders in humans in addition to the molecular sequelae triggered by patterns of activity that induce synaptic plasticity in the normal brain. In several instances, synaptic plasticity and cognitive deficits in animal models of human disorders have been reversed using inhibitors of protein synthesis (Ehninger et al. 2008; Zhou et al. 2009). Although these studies have demonstrated an essential proof of principle, the critical requirement for protein synthesis for nearly all aspects of cellular function makes it unlikely that such generalized approaches could be reasonably translated to the treatment of human disorders. Critical to the advancement of new therapies based on these studies is the identification of novel compounds that can be used to more precisely target translational control machinery to modulate specific pools of neuronal translation (i.e., either a dendritic spine or a lipid raft-associated signaling complex), specific substrates (i.e., IRES-bearing mRNAs), or regulate translational responsiveness to synaptic stimulation. The characterization of inhibitors such as 4EGI-1 and hippuristanol represents a step in that direction, and future efforts at developing similar compounds will hopefully lead to new treatments for human neurological disorders rooted in aberrant neuronal protein synthesis.
GRANTS
This work was supported by National Institute of Neurological Disorders and Stroke Grants NS-034007 and NS-047384 (to E. Klann).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: C.A.H., P.P., J.P., and E.K. conception and design of research; C.A.H., E.S., T.M., E.C.A., A.M.W., and H.W. performed experiments; C.A.H., E.S., T.M., E.C.A., A.M.W., and H.W. analyzed data; C.A.H., E.S., T.M., E.C.A., H.W., P.P., J.P., and E.K. interpreted results of experiments; C.A.H., E.S., and T.M. prepared figures; C.A.H., E.S., and T.M. drafted manuscript; C.A.H., E.S., T.M., E.C.A., A.M.W., H.W., and E.K. edited and revised manuscript; C.A.H., E.S., T.M., E.C.A., A.M.W., H.W., P.P., J.P., and E.K. approved final version of manuscript.
REFERENCES
- Adams JM. Ways of dying: multiple pathways to apoptosis. Genes Dev 17: 2481–2495, 2003 [DOI] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 480: 63–68, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Poulin F, Hou L, DeMaria CT, Sonenberg N, Klann E. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci 25: 9581–9590, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M, Vazquez D. Ribosome changes during translation. J Mol Biol 93: 449–463, 1975 [DOI] [PubMed] [Google Scholar]
- Bidinosti M, Ran I, Sanchez-Carbente MR, Martineau Y, Gingras AC, Gkogkas C, Raught B, Bramham CR, Sossin WS, Costa-Mattioli M, DesGroseillers L, Lacaille JC, Sonenberg N. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol Cell 37: 797–808, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220, 2006 [DOI] [PubMed] [Google Scholar]
- Cammalleri M, Lutjens R, Berton F, King AR, Simpson C, Francesconi W, Sanna PP. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA 100: 14368–14373, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Humbelin M, Darveau A, Lee KA, Milburn S, Hershey JW, Trachsel H, Sonenberg N. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem 258: 11398–11403, 1983 [PubMed] [Google Scholar]
- Edwards DR, Mahadevan LC. Protein synthesis inhibitors differentially superinduce c-fos and c-jun by three distinct mechanisms: lack of evidence for labile repressors. EMBO J 11: 2415–2424, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, de Vries PJ, Silva AJ. From mTOR to cognition: molecular and cellular mechanisms of cognitive impairments in tuberous sclerosis. J Intellect Disabil Res 53: 838–851, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med 14: 843–848, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexner LB, Goodman RH. Studies on memory: inhibitors of protein synthesis also inhibit catecholamine synthesis. Proc Natl Acad Sci USA 72: 4660–4663, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol 490: 703–711, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res 452: 57–65, 1988 [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature 385: 533–536, 1997 [DOI] [PubMed] [Google Scholar]
- Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem 258: 5804–5810, 1983 [PubMed] [Google Scholar]
- Hazzalin CA, Le Panse R, Cano E, Mahadevan LC. Anisomycin selectively desensitizes signalling components involved in stress kinase activation and fos and jun induction. Mol Cell Biol 18: 1844–1854, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Cowansage KK, Arnold EC, Banko JL, Moerke NJ, Rodriguez R, Schmidt EK, Klosi E, Chorev M, Lloyd RE, Pierre P, Wagner G, LeDoux JE, Klann E. Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc Natl Acad Sci USA 108: 3383–3388, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Klann E. Biology of the NMDA Receptor. Boca Raton, FL: CRC, 2009, p. 103–122 [Google Scholar]
- Hoeffer CA, Tang W, Wong H, Santillan A, Patterson RJ, Martinez LA, Tejada-Simon MV, Paylor R, Hamilton SL, Klann E. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron 60: 832–845, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B, Miledi R. The role of calcium in neuromuscular facilitation. J Physiol 195: 481–492, 1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Govindarajan A, Jung HY, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 116: 467–479, 2004 [DOI] [PubMed] [Google Scholar]
- Kelly A, Mullany PM, Lynch MA. Protein synthesis in entorhinal cortex and long-term potentiation in dentate gyrus. Hippocampus 10: 431–437, 2000 [DOI] [PubMed] [Google Scholar]
- Krug M, Lossner B, Ott T. Anisomycin blocks the late phase of long-term potentiation in the dentate gyrus of freely moving rats. Brain Res Bull 13: 39–42, 1984 [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron 50: 377–388, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Hoeffer CA, Wong H, Massaad CA, Zhou P, Iadecola C, Murphy MP, Pautler RG, Klann E. Amyloid β-induced impairments in hippocampal synaptic plasticity are rescued by decreasing mitochondrial superoxide. J Neurosci 31: 5589–5595, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon R, Zaborowska I, Walsh D. Noncytotoxic inhibition of viral infection through eIF4F-independent suppression of translation by 4EGi-1. J Virol 85: 853–864, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, Halperin JA, Wagner G. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128: 257–267, 2007 [DOI] [PubMed] [Google Scholar]
- Morley SJ, Coldwell MJ, Clemens MJ. Initiation factor modifications in the preapoptotic phase. Cell Death Differ 12: 571–584, 2005 [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science 265: 1104–1107, 1994 [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Kandel ER. A macromolecular synthesis-dependent late phase of long-term potentiation requiring cAMP in the medial perforant pathway of rat hippocampal slices. J Neurosci 16: 3189–3198, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 19: 2212–2223, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrig TG, Culp WJ, McKeehan WL, Hardesty B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J Biol Chem 246: 174–181, 1971 [PubMed] [Google Scholar]
- Panja D, Dagyte G, Bidinosti M, Wibrand K, Kristiansen AM, Sonenberg N, Bramham CR. Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J Biol Chem 284: 31498–31511, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran I, Laplante I, Bourgeois C, Pepin J, Lacaille P, Costa-Mattioli M, Pelletier J, Sonenberg N, Lacaille JC. Persistent transcription- and translation-dependent long-term potentiation induced by mGluR1 in hippocampal interneurons. J Neurosci 29: 5605–5615, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras AC, NS Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor, 2000, p. 245–294 [Google Scholar]
- Routtenberg A. The substrate for long-lasting memory: if not protein synthesis, then what? Neurobiol Learn Mem 89: 225–233, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy JW. Is there a baby in the bathwater? Maybe: some methodological issues for the de novo protein synthesis hypothesis. Neurobiol Learn Mem 89: 219–224, 2008 [DOI] [PubMed] [Google Scholar]
- Scharfman HE, Smith KL, Goodman JH, Sollas AL. Survival of dentate hilar mossy cells after pilocarpine-induced seizures and their synchronized burst discharges with area CA3 pyramidal cells. Neuroscience 104: 741–759, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277, 2009 [DOI] [PubMed] [Google Scholar]
- Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci 30: 694–702, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton PK, Sarvey JM. Blockade of long-term potentiation in rat hippocampal CA1 region by inhibitors of protein synthesis. J Neurosci 4: 3080–3088, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA 99: 467–472, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Mankin A. Antibiotics and the ribosome. Mol Microbiol 59: 1664–1677, 2006 [DOI] [PubMed] [Google Scholar]
- Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, Iyengar R, Landau EM, Blitzer RD. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci 25: 5833–5843, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P, Ma T, Iyengar R, Landau EM, Blitzer RD. Mitogen-activated protein kinase upregulates the dendritic translation machinery in long-term potentiation by controlling the mammalian target of rapamycin pathway. J Neurosci 27: 5885–5894, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Blundell J, Ogawa S, Kwon CH, Zhang W, Sinton C, Powell CM, Parada LF. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J Neurosci 29: 1773–1783, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]


