Abstract
Rearrangements of the high mobility group protein I-C (HMGI-C) gene, consisting in the loss of the carboxyl-terminal tail, have been frequently detected in benign human tumors of mesenchymal origin. We have previously demonstrated that transgenic (TG) mice carrying a truncated HMGI-C construct (HMGI-C/T) exhibit a giant phenotype together with a predominantly abdominal/pelvic lipomatosis. Here, we report that HMGI-C/T TG mice develop natural killer (NK)-T/NK cell lymphomas starting from 12 months of age. We found an increased expression of IL-2 and IL-15 proteins and their receptors in these lymphomas, and we demonstrate that HMGI-C/T protein positively regulates their expression in vitro. Therefore, the HMGI-C/T-mediated chronic stimulation of the IL-2/IL-15 pathway could be responsible for the onset of NK-T/NK cell lymphomas in HMGI-C/T TG mice.
The high mobility group protein I-C (HMGI-C) protein belongs to the HMGI protein family (1), which also includes the HMG-I and HMG-Y proteins. HMGI proteins bind to the minor groove of AT-rich DNA sequences, thereby inducing a bend within the DNA (2, 3). These proteins cannot stimulate initiation of transcription, but they can enhance promoter binding of transcription factors (3–5).
The full activity of HMGI-Y proteins requires their two abilities: to bind directly to DNA and to enhance the binding of different transcription factors to DNA by protein–protein interaction (3, 4). A similar mechanism of action has been reported for the HMGI-C protein (5). The HMGI proteins may regulate the interaction of transcription factors with the basal transcription machinery, both positively and negatively, thereby controlling the transcriptional activity of several genes including IL-4 (6), and IL-2 receptor α-chain (IL-2 Rα) (7).
The HMGI proteins have three separate DNA binding domains referred to as “AT-hook” motifs and a carboxyl-terminal region that contains a highly acidic tail (1). Chromosomal translocations of 12q13–15, involving the HMGI-C gene, have been frequently detected in benign human tumors of mesenchymal origin (8, 9). These chromosomal alterations lead to rearrangements and disordered expression of the HMGI-C gene. The HMGI-C modifications consist of the loss of the carboxyl-terminal tail and its fusion with ectopic sequences (8, 9). We have previously shown that the truncation of HMGI-C, rather than its fusion with other genes, is responsible for cell transformation (10). In fact, both the chimeric and truncated (deprived of the acidic tail) HMGI-C genes are able to neoplastically transform the murine fibroblast line NIH 3T3 (10), and the acquisition of ectopic sequences does not increase the transforming ability of the truncated form of HMGI-C.
On this basis, we generated transgenic (TG) mice carrying a truncated HMGI-C (referred to as HMGI-C/T mice). These mice developed a giant phenotype together with a drastic expansion of the retroperitoneal and s.c. white adipose tissues (11).
Here, we report that HMGI-C/T TG mice develop natural killer (NK)-T/NK cell lymphomas starting from 12 months of age. We also demonstrate that both HMGI-C and its truncated form can stimulate the IL-2 and IL-15 pathways. Increased expression of these cytokines and their receptors in lymphoid tissues chronically stimulates T cell (mainly the NK-T cell subset) growth, eventually leading to the development of NK-T/NK cell lymphomas.
Materials and Methods
Mice.
TG mice for the HMGI-C truncated gene have been previously described (11). They were maintained under specific pathogen-free conditions in our Laboratory Animal Facility (Thomas Jefferson University, Philadelphia, PA) according to institutional guidelines. As wild-type (WT) control animals, we used the offspring mice derived from the chimeric HMGI-C/T mice that did not inherit the transgene.
Histopathological Analysis.
For light microscopy, tissues were fixed by immersion in 10% formalin and embedded in paraffin by standard procedures. Five-micrometer sections were stained with hematoxylin-eosin. The antibodies used in immunohistochemistry were purchased from Dako.
Tissue Sampling and Flow Cytometric Analysis.
Lymphocytes extracted from spleens, thymus, and lymph nodes from WT and TG mice were blocked with normal rat and goat IgG (Santa Cruz Biotechnology), stained with the fluorochrome-conjugated mAbs, and analyzed by flow cytometry (Coulter and Becton Dickinson) using the XL II System (Becton Dickinson). All of the antibodies used in fluorescence-activated cell sorter (FACS) analysis were anti-mouse monoclonals purchased from PharMingen.
For cytospin analysis, 7 × 104 cells were spun onto microscope slides, air-dried, and stained with the May-Giemsa-Grunwald method.
Reverse Transcription (RT)-PCR Analyses.
Tissues from TG animals were rapidly dissected, frozen on dry ice, and stored at −80°C until use. Total RNA was extracted as reported (11). The PCR amplification was performed as described (11, 12). The primers used to amplify IL-15, IL-2, IL-15 Rα, and the IL-2 Rα mRNAs were described (12). A set of primers specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ref. 11) was used as an internal control for the amount of cDNA tested. The PCR products were separated on agarose gels, blotted, and hybridized with specific oligonucleotide probes labeled with [γ-32P]ATP.
Protein Extraction, Western Blotting, and Antibodies.
Tissue samples from WT and TG mice were lysed and analyzed by Western blot following standard techniques. The primary antibodies against IL-15, IL-2, IL-2 Rα, IL-15 Rα, c-Kit, Janus kinase 3 (Jak-3), and pTyr were obtained from Santa Cruz Biotechnology. The anti-actin antibody was from Sigma and the anti-neuronal cell adhesion molecule (anti-N-CAM) and anti-NK1.1 antibodies were from PharMingen.
Expression Vectors.
The plasmids used in this study were the full-length of HMG-Y cDNA and the full length (pHMGI-C) and the truncated version of HMGI-C cloned in the pRC-CMV vector (10) or in the pcDNA-3 myc-HIS-tagged vector. The IL-15 promoter (13) and the IL-2 Rα promoter region (−394 to +108) were cloned in the pGL-3 reporter vector (Promega). pCMV-β-gal was purchased from Invitrogen.
Cell Lines and Transfections.
The 293 and A459 cell lines were grown in DMEM (GIBCO/BRL) supplemented with 10% heat inactivated FCS (GIBCO/BRL). Cells were transfected with the FuGENE-6 (Roche, Indianapolis, IN) with each pGL-3 construct with or without pHMGI-C, pHMGI-C/T, or pHMG-Y expression vectors. In some experiments, pCMVp65 NF-κB was used with or without the HMGI vectors. Cells were harvested 48 h posttransfection, and lysates were analyzed for luciferase activity. Transfection efficiency was normalized by using the β-galactosidase activity. All of the assays were performed in triplicate and were repeated in at least three independent experiments.
Production of Recombinant Proteins.
The full-length or the truncated version of HMGI-C and HMG-Y cDNAs was cloned in the pET2c (Novagen). BL21/DE3 cells transformed with each vector were grown in LB, induced with isopropyl β-d-thiogalactoside (IPTG), sonicated, and purified by using the His-Trap purification Kit (Amersham Pharmacia) following the manufacturer's instructions.
Electrophoretic Mobility-Shift Assay (EMSA).
The oligonucleotides used in EMSA were: the IL-2 CD28RE and IL-2 CD28RE Mut described previously (14); the IL-2 Rα and IL-2 Rα Mut described before (7); and the IL-15 oligonucleotide spanning from base −84 to −52 of the IL-15 promoter region (13). Recombinant proteins (5–100 ng) were incubated in the presence of radiolabeled oligonucleotide and poly(dCdG) (Sigma) as nonspecific competitor as previously described (7). In some experiments, a 50- or 100-fold excess of specific or nonspecific unlabeled competitor oligonucleotides was added. The DNA–protein complexes were resolved on 5% non-denaturing acrylamide gels and were visualized by exposure to autoradiographic films.
Culture of Fetal Liver Lymphocytes.
Pregnant TG females were killed at day 14.5 postcoitum, and the embryos were collected and analyzed for the presence of the transgene. Fetal livers were excised, and precursor lymphocytes were extracted by mechanical disruption of the liver. Cells were resuspended in RPMI 1640 medium (GIBCO) supplemented with 10% FCS and B & T cell growth supplements (Origen, Austin, Tx) according to the manufacturer's instructions. After 12 days, cells were collected and analyzed for surface antigen expression by FACS analysis.
Results
HMGI-C/T TG Mice Develop NK-T/NK Cell Lymphomas.
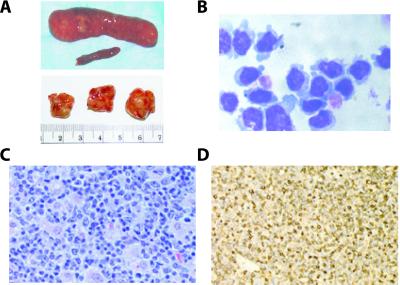
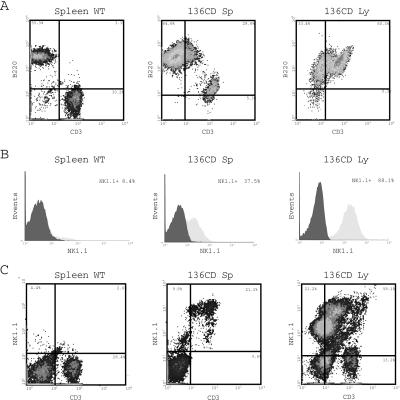
Two lines of TG mice carrying a truncated HMGI-C gene were generated by a previously published embryonic stem cell strategy to obtain high expression levels of the transgene (11). When the HMGI-C/T mice were killed at 1 year of age, autopsy examination revealed that about 35% of the mice derived from both founder lines showed lymphoid pathology. Almost all (8 of 10) of the HMGI-C/T mice killed at 24 months of age displayed similar pathology. In some animals, the size of the lymph nodes was more than 2 cm in diameter with a weight of almost 2 g, and the spleen reached a length of 4.0 cm and a weight of 5 g (Fig. 1A). The histopathological analysis revealed tumors in the mesenteric lymph nodes, spleen, Peyer's patches, and to a lesser extent in the liver, mediastinal lymph nodes, thymus, and kidneys. The lymphomas were categorized as large cell lymphomas with typical histiocytic differentiation patterns or mixed cell lymphomas with a large cell component. These lymphomas were composed totally or partially of immature cells with large nuclei and prominent cytoplasm (Fig. 1C). These features were confirmed by cytospin analysis of the same cases followed by May-Giemsa-Grunwald staining (Fig. 1B). No evidence of lymphoma was found in WT mice younger than 23 months of age. In older control animals, we found very early follicular small cell-type lymphomas with clear B cell plasma cell differentiation (in about 20% of the cases), consistent with previously published data (15). Immunohistochemical analysis showed that four of five large cell lymphomas from the TG mice were CD3 positive (Fig. 1D) and CD20 negative (data not shown) and that the latter was negative for both antigens, indicating the presence of T cell lymphomas. FACScan analysis of lymphocytes isolated from WT or pathological spleen and lymph nodes using the CD3, B220, and CD19 anti-mouse antibodies confirmed the immunohistochemistry data. A double staining of lymphomatous spleen and lymph nodes (Fig. 2A) revealed the presence of a CD3+/B220+/CD19− population. In contrast, only 3–4% of cells were CD3+/B220+ in splenocytes derived from WT animals (Fig. 2A). Because CD3+/B220+ cells isolated from mouse bone marrow could be NK-T/NK cell precursors, which are also positive for the typical NK-T cell surface antigen recognized by the NK1.1 antibody (16), we analyzed the lymphocytes isolated from WT spleens or pathological spleens and lymph nodes for the expression of NK1.1 (Fig. 2B). Although in WT splenocytes the NK1.1-positive cells were about 8% of the total population (Fig. 2B), the percentage of this population increased dramatically in pathological spleens and lymph nodes, reaching up to 80% of total lymphocytes (Fig. 2B). A double staining FACS analysis of lymphocytes with CD3 and NK1.1 antibodies showed that most of the cells from pathological samples were CD3+/NK1.1+ (Fig. 2C), indicating the presence of T/NK lymphocytes. Up-regulation of NK1.1 antigen and other markers of the NK cells such as N-CAM and c-Kit in spleens and lymph nodes derived from the HMGI-C/T mice were confirmed by using Western blot analysis (data not shown). On the basis of the results presented here, we classified these neoplasias as NK-T/NK cell lymphomas.
Figure 1.
HMGI-C/T TG mice develop large cell T lymphomas. (A Upper) Comparison between normal and pathological spleens. (Lower) Enlargement of pathological lymph nodes in TG mice. (B) Cytospin analysis of pathological lymphocytes extracted from laterocervical lymph node. The large cell phenotype and the presence of azurophilic granules are showed. (C) Hematoxylin-eosin staining of a pathological lymph node revealed the presence of large cells. (D) CD3 stained positively in lymphoid cells. In each case, a representative field is shown.
Figure 2.
HMGI-C/T TG mice develop T/NK cell lymphomas. (A) CD3 and B220 expression on lymphocytes extracted from a WT mouse (Left), a pathological spleen (Center), or a pathological lymph node (Right). The percentage of CD3+, B220+, and CD3+/B220+ is reported in the figure. (B) Expression of NK1.1 antigen in WT splenocytes (Left), TG splenocytes (Center), and pathological lymph node (Right). The percentage of NK1.1 positive cells is reported. (C) CD3 and NK1.1 expression in normal (Left) and pathological spleen (Center) and lymph node (Right).
The Stimulation of the Pathway Shared by IL-2 and IL-15 Accounts for the Development of the NK-T/NK Lymphomas in HMGI-C/T TG Mice.
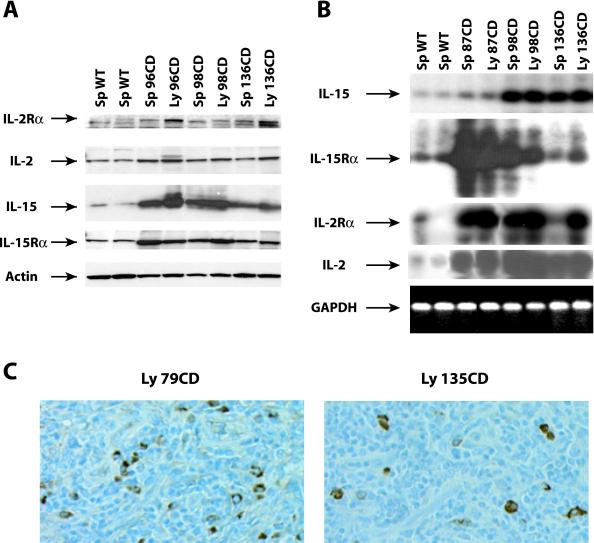
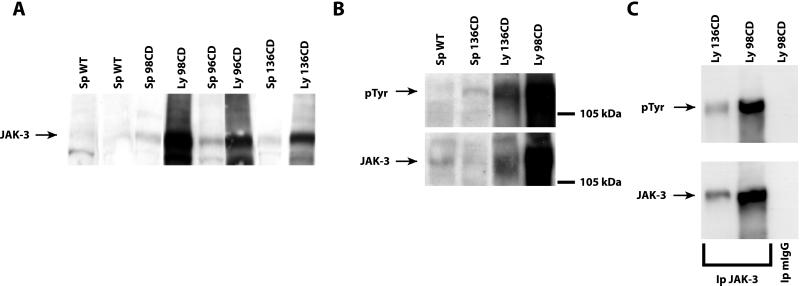
Proliferation of NK-T/NK cells is known to be mainly dependent on the IL-2 and IL-15 pathways (17). Thus, we analyzed the expression of the IL-2, IL-15, and their high affinity receptors (IL-2 Rα and IL-15 Rα) in the lymphomatous tissues. A significant increase was observed in IL-2 (2- to 3-fold increase), IL-15 (3- to 5-fold), IL-2 Rα (2- to 4-fold), and IL-15 Rα (2- to 3-fold) protein levels in the neoplastic tissues in comparison with the normal spleen (Fig. 3A). The up-regulation of IL-15 protein was also confirmed by immunohistochemistry (Fig. 3C). RT-PCR analysis (Fig. 3B) paralleled the protein data, indicating that the induction occurs at the mRNA level, and suggesting that transcriptional control plays a major role in the regulation of the gene expression induced by the HMGI-C/T transgene. The stimulation of the IL-2/IL-15 receptor complex normally results in the activation of Jak-3 kinase (18, 19). We then investigated whether the overexpression of IL-15 and/or IL-2 and their receptors resulted in modification of the Jak-3 status. We found that all of the examined pathological tissues expressed higher Jak-3 protein levels than did normal spleen (Fig. 4A). An increased phosphorylation of Jak-3 was also detected in the lymphomatous tissues (Fig. 4 B and C), indicating that the intracellular pathway induced by IL-2 or IL-15 was also activated in HMGI-C/T TG mice.
Figure 3.
Overexpression of IL-2 and IL-15 and their receptors in NK-T/NK cells derived from HMGI-C/T TG mice. (A) Expression of IL-2 Rα, IL-2, IL-15, and IL-15 Rα proteins in two different control spleens and in six different pathological specimens. The same Western blot was incubated with antibody vs. actin protein to normalize the amount of loaded proteins. (B) RT-PCR analysis of RNA extracted from two different WT spleens and six pathological specimens. The expression of IL-15, IL-15 Rα, IL-2, and IL-2 Rα is shown. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was determined as an internal control of RNA quantity and status. (C) IL-15 expression analyzed by immunohistochemistry in two different pathological lymph nodes.
Figure 4.
Levels and phosphorylation status of Jak-3 protein in WT and pathological specimens. (A) Western blot analysis determining the expression of Jak-3 protein in two different control spleens and in six different pathological specimens. (B) Phosphotyrosinse expression in one WT spleen and three different pathological specimens. The same Western blot was incubated with antibody vs. the Jak-3 protein. (C) Immunoprecipitation of Jak-3 protein in two different lymphomatous samples. The same blot was hybridized with anti-pTyr antibody and then stripped and reprobed with anti Jak-3 Ab. Normal mouse IgGs (mIgG) were used as a negative control.
IL-15 Rather than IL-2 Is the Primary Target of HMGI-C Activity.
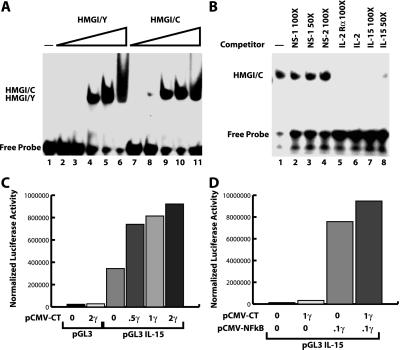
It has been previously reported that HMGI-Y plays a pivotal role in the regulation of IL-2 and IL-2 Rα promoters. Its overexpression enhances the transcription of both of these genes, thereby resulting in an increase of T cell proliferation (7, 14, 20). We examined the possibility that HMGI-C was also able to bind to the promoter region of IL-2 and IL-2 Rα genes. We demonstrated by EMSA that the HMGI-C recombinant protein binds with high affinity to the CD28 responsive element (CD28RE) of the IL-2 promoter and to the −137 to −64 promoter region of IL-2 Rα as it occurs for the HMGI-Y protein (data not shown). To define the functional consequences of this in vitro DNA–protein interaction, we cotransfected a luciferase reporter construct containing the −394 to +108 promoter region of the IL-2 Rα gene with either the HMGI-C or HMGI-C/T expression vectors. We demonstrated that both HMGI-C and HMGI-C/T were able to activate the IL-2 Rα promoter although HMGI-Y protein was more efficient (data not shown). Recent reports emphasize the role of IL-15 rather than IL-2 in the differentiation, survival, and proliferation of NK-T/NK cells (21). The knockout mice involving IL-15, IL-15 Rα, and IL-2 Rβ (which is necessary for the transduction of IL-15 signals in T and NK lymphocytes) developed defects in the NK-T/NK population whereas the absence of IL-2 does not affect this population (22–24). Thus, we evaluated the possibility that HMGI-C could regulate IL-15 transcription. The IL-15 promoter region contains many AT-rich regions to which HMGI-C protein might bind (13). In EMSA experiments, we demonstrated that HMGI-C directly binds to the DNA sequence spanning the −84 to −52 region of the IL-15 promoter (Fig. 5A). This binding was specific, as demonstrated by specific competition with three different oligonucleotides containing the HMGI binding sites (Fig. 5B, compare lanes 1–4 with lanes 5–8). Interestingly, HMGI-C and HMGI-C/T recombinant proteins had a very high affinity for this promoter region, because binding was visible when only 5 or 10 ng of proteins were added (data not shown). To determine whether this binding resulted in increased activity of the IL-15 promoter, we transfected the murine IL-15 promoter cloned in a luciferase reporter vector (13) with increasing amounts of HMGI-C and HMGI-C/T into the A549 lung carcinoma cell line (which has been reported to express the IL-15 protein; ref. 25). The expression of HMGI-C/T enhanced the activity (2- to 3-fold) of the IL-15 promoter in a dose-dependent manner (Fig. 5C). Interestingly, HMGI-C/T protein was more effective than the WT form in enhancing the IL-15 promoter activity (data not shown). Moreover, cotransfection of pCMVp65 NF-κB with HMGI-C/T into the 293 cells resulted in a moderate but reproducible increase of IL-15 promoter activity over the pCMVp65 NF-κB alone (Fig. 5D). As reported previously (13), p65 NF-κB by itself induced a strong activation of the IL-15 promoter.
Figure 5.
HMGI-C/T protein binds to and activates the IL-15 promoter. (A) IL-15 −84 to −52 oligonucleotide recognized by increasing amounts (5, 10, 20, 50, and 100 ng, respectively) of HMGI/Y (lanes 2–6) or HMGI/C recombinant protein (lanes 8–11). (B) In vitro binding of HMGI-C protein to the IL-15 oligonucleotide competed with 30 ng of HMGI/C recombinant protein alone (lane 2) or in presence of two unrelated oligonucleotides (lanes 3 and 4). Specific competition of DNA protein interaction was obtained with a 50- to 100-fold excess of IL-2 Rα (lane 5), IL-2 (lane 6), or IL-15 (lanes 7 and 8) cold oligonucleotides. (C) Luciferase assay using the IL-15 promoter region transfected alone or with increasing concentrations of pCMV-HMGI-C/T vector. (D) Luciferase assay using the IL-15 promoter region transfected alone or with pCMV-NF-κB alone or in combination with pCMV-HMGI-C/T vector. In all cases, a typical experiment is reported.
Overexpression of the HMGI-C/T Protein Increases the Number of NK1.1+ Cells in a Population of Cultured Fetal Liver Lymphocytes.
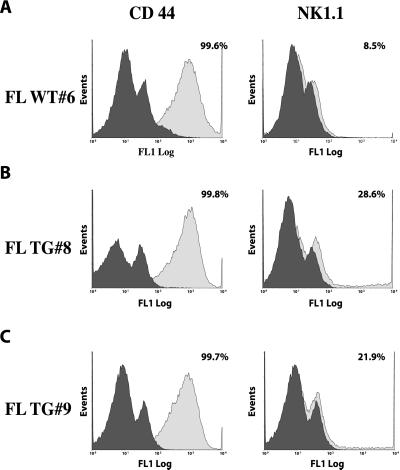
Lu and coworkers (26) reported that fetal liver lymphocytes cultured in the presence of either IL-2 or IL-15 result in the development of a NK1.1 population and that the two cytokines had a synergistic effect in this process. To determine whether the HMGI-C/T overexpression increased the ability of fetal lymphocytes to differentiate into NK-T/NK cells, we cultured fetal liver lymphocytes extracted from WT and TG embryos for 12 days in a mixture of IL-2, IL-4, and IL-6 that allows the growth of both T and B cells. The mixed population was then analyzed for the expression of CD44 (as a positive control) and NK1.1 surface antigens. As shown in Fig. 6, the CD44 antigen is widely expressed in both WT and TG derived fetal lymphocytes. In contrast, a significantly increased number of NK1.1-positive cells was found in lymphocytes derived from the liver of HMGI-C/T mice when compared with the control mice (Fig. 6).
Figure 6.
Increasing expression of NK1.1 antigen on cultured fetal lymphocyte-derived HMGI-C/T TG embryos. CD44 (Left) and NK1.1 (Right) expression on fetal lymphocytes extracted from a WT embryo (A), and two different HMGI-C/T TG embryos (B and C). The percentage of CD44+ and NK1.1+ is reported in the figure.
Discussion
Here, we report that 35% of TG mice carrying a truncated HMGI-C gene develop massive lymph node enlargement and splenomegaly with the presence of immature atypical lymphocytes starting from 12 months. Almost all of the TG mice older than 24 months showed this pathology. Immunohistochemical and FACS analyses showed that these cells expressed the CD3, NK1.1, N-CAM, and c-Kit antigens, suggesting the diagnosis of NK-T/NK cell lymphoma.
In humans, the NK-T/NK cell lymphomas have been only recently classified as an autonomous pathology with a typical surface antigen expression, including CD3ɛ and the CD56 (N-CAM) antigens, and characteristic localization (27). The disease usually starts in the rhinopharyngeal region (nasal NK lymphomas) or in intestinal and cutaneous regions (nasal-type NK lymphomas). In some cases, lymphoma-leukemia pathology was reported with dissemination of the disease to different lymphoid and extra lymphoid organs (28). Accordingly, we found that in mice this pathology was usually localized to the gastrointestinal tract and, in early cases, the localization was primarily observed in Peyer's patches. As in humans, we found in some cases a very aggressive pathology with the involvement of the spleen, the liver, and many other organs, such as lung and pancreas.
An essential role for IL-15 and IL-15 Rα and a contributing role for IL-2 and IL-2 Rα in NK-T/NK cell proliferation has been demonstrated (29–32). IL-15 binds with high affinity to a receptor complex composed of the IL-15 Rα chain and IL-2 Rβ and -γ chains (33). The specificity of the binding is due to the presence of the IL-15 Rα (33). NK and T/NK cells express the shared β and γ chains as well as the high affinity IL-2 and IL-15 Rα subunits. Comparative studies of IL-2 Rα and IL-2/15 Rβ-deficient mice suggest that IL-15 rather than IL-2 might be important for the differentiation of T/NK cells and intraepithelial lymphocytes (23, 24). IFN regulatory factor (IRF-1)-deficient mice lack NK cells because of the inability of their bone marrow stromal cells to elaborate IL-15, suggesting a unique role for IL-15 in NK cell development (34). Finally, IL-15 and IL-15 Rα-deficient mice (24) are generally healthy and lymphopenic and specifically lack NK, T/NK, IELs, and activated CD8+ memory phenotype T cells. Conversely, IL-2 and IL-2 Rα-deficient mice accumulate activated T and B cells and prematurely die of autoimmune disease (35, 36).
Here, we demonstrated that in vivo overexpression of HMGI-C/T protein led to overexpression of IL-2 and IL-15 and their receptors mainly through transcriptional activation. Moreover, in HMGI-C/T TG mice the induction of IL-15 expression was greater than IL-2 expression (3- to 5-fold and 2- to 3-fold, respectively). Accordingly, we demonstrated, by EMSA, that HMGI-C and HMGI-C/T proteins were able to bind to the IL-15 promoter region with greater avidity than did the HMGI-Y protein. In contrast, the promoter regions of IL-2 and IL-2 Rα were bound better by the HMGI-Y than by the HMGI-C protein. According to this result, the TG mice overexpressing the HMGI-Y gene developed lymphomas with a lower frequency and longer latency period (data not shown). Using the IL-15 and IL-2 Rα promoter regions cloned into luciferase reporter vectors, we also demonstrated that HMGI-C/T and HMGI-C stimulate the transcription of IL-15 better than HMGI-Y whereas the latter protein was more effective in the stimulation of the IL-2 Rα promoter. These data strongly suggest that the activation of the IL-15 promoter activity associated with HMGI-C/T overexpression mainly accounts for the onset of NK lymphomas in the HMGI-C/T TG mice. Finally, some pieces of evidence emerging from our studies with HMGI-Y and HMGI-C/T TG mice confirm a central role for the HMGI-C/T protein in the onset of NK/T-NK cell lymphomas in a mouse model. In fact, HMGI-C/T TG mice, generated by the microinjection of the same construct directly into fertilized mouse eggs, resulted in a significantly lower expression of the transgene expression when compared with the embryonic stem cell-mediated strategy (11). Accordingly, the phenotype was less prominent in the first rather than the second mouse line. In fact, in HMGI-C/T TG mice derived from the classical approach, the onset of this pathology is at 15 months of age and a lower proportion of mice develops the lymphopathology (data not shown).
We have shown that the increase in levels of IL-2 and IL-15 proteins and their receptors results in Jak-3 activation. We hypothesized that this increased activity may be responsible for the NK-T/NK expansion. This hypothesis was based on several pieces of experimental evidence. In vivo administration of IL-15 was reported to protect mice from lethal multisystem apoptosis induced by anti-fas antibody (37). Moreover, Caligiuri and coworkers (38) proposed that IL-15 might function as an NK cell survival factor by the maintenance of Bcl-2 protein expression, thereby preventing NK cell apoptosis. TG mice overexpressing human IL-15 showed an increase of NK cells and memory T cells because of the ability of IL-15 to selectively prolong the survival of memory T cells by blocking activation of T cell death mediated by a process involving IL-2 (39). Finally, TG mice overexpressing murine IL-15 showed an early expansion in NK and CD8+ T lymphocytes and developed fatal T/NK lymphocytic leukemia between 3 and 8 months of age (40). Collectively, these data support the emerging notion that IL-2 and IL-15 expression must be balanced to achieve appropriate cellular immunity during immune responses.
Our data obtained by using embryonic liver lymphocytes from TG or WT embryos seem to support our hypothesis. We demonstrated that, in the presence of both B and T cell growth factors, HMGI-C/T embryonic lymphocytes developed with a statistical higher percentage into NK1.1-positive cells when compared with those in the WT mice. This result suggests that overexpression of HMGI-C/T predisposed stem cell lymphocyte precursors to differentiate into NK-T/NK cells and/or protected them from apoptosis. Both of these effects may be accounted for by HMGI-C/T-mediated induction of IL-15 overexpression. Taken as a whole, our data indicate that the HMGI-C/T-mediated induction of IL-15 overexpression leads to NK-T/NK lymphomas in the HMGI-C/T TG mice.
Acknowledgments
We are grateful to J. McCormick from the Flow Cytometry facility of the Kimmel Cancer Center for helping us with FACS analysis. This work was supported by Grants P01CA76259 and P30CA56036 from the National Cancer Institute and by grants from the Associazione Italiana Ricerca sul Cancro (AIRC).
Abbreviations
- HMGI-C
high mobility group protein I-C
- HMGI-Y
high mobility group protein I-Y
- IL-2 Rα
Il-2 receptor α-chain
- IL-15 Rα
Il-15 receptor α-chain
- NK
natural killer
- N-CAM
neuronal cell adhesion molecule
- Jak-3
Janus kinase 3
- TG
transgenic
- WT
wild type
- FACS
fluorescence-activated cell sorter
References
- 1.Manfioletti G, Giancotti V, Bandiera A, Buratti E, Sautiere P, Cary P, Crane-Robinson C, Coles B, Goodwin G H. Nucleic Acids Res. 1991;19:6793–6797. doi: 10.1093/nar/19.24.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodwin G, Bustin M. In: Architecture of Eukaryotic Genes. Kahl G, editor. Weinheim, Germany: VCH; 1988. pp. 187–205. [Google Scholar]
- 3.Thanos D, Maniatis T. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 4.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 5.Mantovani F, Covaceuszach S, Rustighi A, Sgarra R, Heath C, Goodwin G H, Manfioletti G. Nucleic Acids Res. 1998;26:1433–1439. doi: 10.1093/nar/26.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuvpilo S, Schoenberg C, Gerwig R, Heinfling A, Reeves R, Grummt F, Serfling E. Nucleic Acids Res. 1993;21:5694–5704. doi: 10.1093/nar/21.24.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John S, Reeves R B, Lin J X, Child R, Leiden J M, Thompson C B, Leonard W J. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashar H R, Schoenberg Fejzo M, Tkachenko A, Zhou X, Fletcher J A, Weremowicz S, Morton C, Chada K. Cell. 1995;82:57–65. doi: 10.1016/0092-8674(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 9.Schoenmakers E F, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven W J M. Nat Genet. 1995;10:436–443. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 10.Fedele M, Berlingieri M T, Scala S, Chiariotti L, Viglietto G, Rippel V, Bullerdiek J, Santoro M, Fusco A. Oncogene. 1998;17:413–418. doi: 10.1038/sj.onc.1201952. [DOI] [PubMed] [Google Scholar]
- 11.Battista S, Fidanza V, Fedele M, Klein-Szanto A J, Outwater E, Brunner H, Santoro M, Croce C M, Fusco A. Cancer Res. 1999;59:4793–4797. [PubMed] [Google Scholar]
- 12.Hanisch U K, Lyons S A, Prinz M, Nolte C, Weber J R, Kettenmann H, Kirchhoff F. J Biol Chem. 1997;272:28853–28860. doi: 10.1074/jbc.272.46.28853. [DOI] [PubMed] [Google Scholar]
- 13.Azimi N, Tagaya Y, Mariner J, Waldmann T A. J Virol. 2000;74:7338–7348. doi: 10.1128/jvi.74.16.7338-7348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himes S R, Coles L S, Reeves R, Shannon M F. Immunity. 1996;5:479–489. doi: 10.1016/s1074-7613(00)80503-8. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa K, Utsuyama M, Goto H, Kuramoto K. Gerontology. 1984;30:223–233. doi: 10.1159/000212636. [DOI] [PubMed] [Google Scholar]
- 16.Rolink A, ten Boekel E, Melchers F, Fearon D T, Krop I, Andersson J. J Exp Med. 1996;183:187–194. doi: 10.1084/jem.183.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams N S, Klem J, Puzanov I J, Sivakumar P V, Schatzle J D, Bennett M, Kumar V. Immunol Rev. 1998;165:47–61. doi: 10.1111/j.1600-065x.1998.tb01229.x. [DOI] [PubMed] [Google Scholar]
- 18.DiSanto J P. Curr Biol. 1997;7:R424–R426. doi: 10.1016/s0960-9822(06)00208-9. [DOI] [PubMed] [Google Scholar]
- 19.Imada K, Leonard W J. Mol Immunol. 2000;37:1–11. doi: 10.1016/s0161-5890(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 20.Himes S R, Reeves R, Attema J, Nissen M, Li Y, Shannon M F. J Immunol. 2000;164:3157–3168. doi: 10.4049/jimmunol.164.6.3157. [DOI] [PubMed] [Google Scholar]
- 21.Ma A, Boone D L, Lodolce J P. J Exp Med. 2000;191:753–756. doi: 10.1084/jem.191.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki H, Duncan G S, Takimoto H, Mak T W. J Exp Med. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodolce J P, Boone D L, Chai S, Swain R E, Dassopoulos T, Trettin S, Ma A. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy M K, Glaccum M, Brown S N, Butz E A, Viney J L, Embers M, Matsuki N, Charrier K, Sedger L, Willis C R, et al. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoeck M, Kromer W, Gekeler V. Immunobiology. 1998;199:14–22. doi: 10.1016/S0171-2985(98)80060-0. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Patrene K D, Appasamy P M, Herberman R B, Boggs S S. J Exp Hematol. 1999;27:1046–1056. doi: 10.1016/s0301-472x(99)00042-9. [DOI] [PubMed] [Google Scholar]
- 27.Harris N L, Jaffe E S, Stein H, Banks P M, Chan J K, Cleary M L, Delsol G, De Wolf-Peeters C, Falini B, Gatter K C, et al. Blood. 1994;84:1361–1392. [PubMed] [Google Scholar]
- 28.Dunkley S, Gibson J, Mackinlay N, Joshua D. Pathology. 1998;30:157–159. doi: 10.1080/00313029800169136. [DOI] [PubMed] [Google Scholar]
- 29.Carson W E, Fehniger T A, Haldar S, Eckhert K, Lindemann M J, Lai C F, Croce C M, Baumann H, Caligiuri M A. J Exp Med. 1994;180:1395–1403. [Google Scholar]
- 30.Aiba Y, Hirayama F, Ogawa M. Blood. 1997;89:4005–4012. [PubMed] [Google Scholar]
- 31.Vella A T, Dow S, Potter T A, Kappler J, Marrack P. Proc Natl Acad Sci USA. 1998;95:3810–3815. doi: 10.1073/pnas.95.7.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura H, Yajima T, Naiki Y, Tsunobuchi H, Umemura M, Itano K, Matsuguchi T, Suzuki M, Ohashi P S, Yoshikai Y. J Exp Med. 2000;191:157–170. doi: 10.1084/jem.191.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waldmann T A, Tagaya Y. Annu Rev Immunol. 1999;17:19–49. doi: 10.1146/annurev.immunol.17.1.19. [DOI] [PubMed] [Google Scholar]
- 34.Ogasawara K, Hida S, Azimi N, Tagaya Y, Sato T, Yokochi-Fukuda T, Waldmann T A, Taniguchi T, Taki S. Nature (London) 1998;391:700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- 35.Sadlack B, Merz H, Schorle H, Schimpl A, Feller A C, Horak I. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 36.Willerford D M, Chen J, Ferry J A, Davidson L, Ma A, Alt F W. Immunity. 1995;3:521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 37.Bulfone-Paus S, Ungureanu D, Pohl G, Lindner T, Paus R, Ruckert R, Krause H, Kunzendorf U. Nat Med. 1997;3:1124–1128. doi: 10.1038/nm1097-1124. [DOI] [PubMed] [Google Scholar]
- 38.Carson W E, Fehniger T A, Haldar S, Eckhert K, Lindemann M J, Lai C F, Croce C M, Baumann H, Caligiuri M A. J Clin Invest. 1997;99:937–943. doi: 10.1172/JCI119258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konczalik J M, Dubois S, Losi J M, Sabzevari H, Yamada N, Feigenbaum L, Waldmann T A, Tagaya Y. Proc Natl Acad Sci USA. 2000;97:11445–11450. doi: 10.1073/pnas.200363097. . (First Published October 3, 2000; 10.1073/pnas.200363097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fehniger T A, Suzuki K, Ponnappan A, VanDeusen J B, Cooper M A, Florea S M, Freud A G, Robinson M L, Durbin J, Caligiuri M A. J Exp Med. 2001;193:219–232. doi: 10.1084/jem.193.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]