Abstract
The MHC in humans encodes the most polymorphic genes, the HLA genes, which are critical for the immune system to clear infection. This can be attributed to strong selection pressure as populations moved to different parts of the world and encountered new kinds of infections, leading to new HLA class II alleles. HLA genes also have the highest relative risk for autoimmune diseases. Three haplotypes—HLA-DR2DQ6, DR4DQ8, and DR3DQ2—account for HLA association with most autoimmune diseases. We hypothesize that these haplotypes, along with their multiple subtypes, have survived bottlenecks of infectious episodes in human history because of their ability to present pathogenic peptides to activate T cells that secrete cytokines to clear infections. Unfortunately, they also present self-peptides/mimics to activate autoreactive T cells secreting proinflammatory cytokines that cause autoimmune diseases.
Keywords: autoimmunity, cytokines, inflammation, MHC, rheumatoid arthritis, T cells, Th1/Th2 cells
Among the great apes (human, chimpanzee, gorilla, and orangutan), humans have the longest life span (1). The low life expectancy in nonhuman primates is due to high mortality rates caused by infections. Similarly, the biggest threat to survival of humans is infections caused by microorganisms (eg, viruses, bacteria, fungi, parasites). The MHC in humans, the HLA genes, maps to chromosome 6 and is crucial in fighting infections by encoding molecules responsible for controlling the invading pathogens (2, 3). The MHC is the most dense region of the human genome, encompassing ~4 Mbps, or 0.1% of the genome but containing 6 times more genes (0.6% of total genes). Most of these genes encode for proteins of the immune defense system.
The HLA complex is divided into 3 main regions, the HLA classes I, II, and III (2, 3). Classes I and II genes are the most polymorphic in the human genome (see http://www.ebi.ac.uk/imgt/hla). The high polymorphism in this region can be attributed to strong selection pressure as humans moved to different parts of the world and encountered new infections, leading to diversity in MHC genes through mutation and gene duplication and conversion (4-6). A population with diverse alleles of HLA classes I and II leads to herd resistance to infection and a survival advantage (7, 8).
The HLA class II molecule is composed of two polypeptides chains (α and β). Each α and β chain has 2 domains—a highly conserved α2 and β2 region and a highly polymorphic α1 and β1 domain (2). Since α1 and β1 come together to form the antigen-binding groove, high polymorphism in this region leads to generation of new class II molecules with ability to recognize new epitopes. The class II molecules are expressed on APCs, such as B lymphocytes, macrophages, dendritic cells, endothelial cells, and other organ-specific APCs. Crystal structure shows that class II molecules have a single peptide-binding groove that can accommodate various peptides of 14 to 25 amino acids, depending on charge, stability, and binding affinity (9). The class II region codes genes for three class II molecules DP, DQ, and DR. The DRβ1 genes are the most polymorphic, with almost 450 alleles known at present. The HLA-DR, DP, and DQ genes are in linkage disequilibrium and are inherited en bloc as a haplotype.
Human populations have gone through various infectious episodes in evolution, where severe uncontrolled infections led to a high mortality rate, termed a bottleneck, because only a small percentage survived. Survival of the individuals from these bottlenecks was suggested to be due to the presence of specific HLA class II alleles that were able to mount an effective immune response and clear the infection (4, 5, 10, 11). Thus, only those HLA class II alleles that can overcome a population bottleneck survived and were passed on to the next generation. The HLA class II molecules that survived the episodes went through random mutation or gene conversion, or both, to generate novel alleles that, again, were put through natural selection to generate a better class II allele.
In evolution, HLA alleles offering an advantage during reproductive years have been selected and occur in the population with much higher frequencies. We used the MHC class II prediction model to compare the generation of an antigenic peptide pool between mouse class II (H2-IA) and human HLA-DR molecules. We used myelin oligodendrocyte glycoprotein (MOG) as a self-antigen to predict whether HLA-DR molecules bind this antigen better than mouse class II. Using the Immune Epitope Database Analysis Resource (http://tools.immuneepitope.org), we observed that among the top 100 binders, 79 were HLA-DRB1*1501 epitopes, with 21 predicted to bind mouse class II. We also ran prediction for mouse hepatitis virus as an infectious agent and found similar results, where more than 70% of top binders were HLA-DRB1*1501 epitopes. This was also true for HLA-DRB1*0301and *0401.
Thus, these T-cell epitope prediction data support our hypothesis that during evolution, promiscuous HLA class II alleles with capacity to recognize a large pool of antigens were selected. In humanized mice, the DQ8 molecule linked with diabetes and rheumatoid arthritis (RA) selects a larger pool of VβT cell repertoire than DQ6 and DR molecules (12). A number of studies in various ethnic populations support this hypothesis. The African population is considered the oldest and most diverse compared with other populations, such as whites. In analysis of DRB-DQA-DQB polymorphism through restriction fragment length polymorphism, Olerup et al (13) observed that HLA class II genes were twice as polymorphic in the African population as in whites and suggested that the population bottleneck might have led to generation of a European white population. Another example of population bottleneck and selection of HLA class II is the presence of a restricted set of HLA alleles in Ticuna Indians, which faced severe selection pressure because of novel pathogens introduced by Europeans in the 15th century (14). A recent meta-analysis on the relation between HLA class II polymorphism and hepatitis B virus infection showed that persons carrying the HLA-DR4 allele were significantly associated with clearance of hepatitis B virus infection (15). Similarly, presence of HLA-DRB1*0401 and DRB1*1501 had been linked with increased clearance of hepatitis C virus infection (16).
Thus, HLA-DQ and DR molecules have evolved to present pathogenic peptides to activate a subset of CD4 T cells that secrete specific cytokines to clear infections. Further, the presence of certain HLA-DR and DQ alleles together (linkage disequilibrium) might lead to generation of CD4 T cells with more diverse repertoire as compared to HLA-DR or –DQ alone. We hypothesize that three MHC class II haplotypes DR2 (DRβ1*1501)/DQ6 (DQβ1*0602), DR3 (DRβ1*0301)/DQ2 (DQβ1*0201), and DR4 (DRβ1*0401)/DQ8 (DQβ1*0302) have survived bottlenecks of infectious episodes in the history of mankind and generated new subtypes that are effective in clearance of infections. The presence of these haplotypes at higher gene frequency in every geographical location, ethnic population and racial groups support our hypothesis (17). Unfortunately, the presence of these HLA class II allele(s) also predisposes them to develop autoimmunity because of their ability to activate autoreactive T cells in a small subset of people carrying the haplotype.
CD4 T cell Subsets
Recognition of the MHC peptide complex by CD4 T cells leads to secretion of cytokines. CD4 T cells are differentiated on the basis of the type of cytokines they secrete. Since CD4 T cells help other cells through secretion of cytokine, they are also called helper T cells to differentiate them from cytotoxic CD8 T cells activated by MHC class I peptide complex. Initially, helper T cells were divided into 2 subsets, a Th1 subset secreting IL-2 and IFN-γ responsible for cellular immunity and a Th2 subset secreting IL-4 required for humoral immune response (18, 19). The division was followed by an addition of a subset of regulatory CD4 T cells (Tregs) that secrete IL-10 and TGF-β (20). However, presence of inflammatory diseases such as arthritis and experimental autoimmune encephalomyelitis (EAE) in IFN-γ–deficient mice indicated existence of other helper T cell subsets and led to the discovery of the Th17 subset secreting IL-17 and IL-23 (21).
Recently, other helper T cell subsets have been assigned on the basis of the secretion of IL-9 (Th9) or IL-21 (Tfh-follicular helper) (22). Thus, the current knowledge of T-cell characterization is based on the cytokine secretion pattern of CD4 T cells, and this wide array of cytokines help in various tasks associated with immune function. Since the main function of the MHC molecule is to clear infection through activation of adaptive immune response and the cytokines secreted by the activated CD4 T cells play a major role in this process, it can be argued that MHC molecules control immune response through activation of specific CD4 T cells that determine the cytokine network.
Bacterial pathogens can be divided into 2 major categories on the basis of their postinfection location—intracellular (eg, Mycobacterium tuberculosis, Leishmania major, Cryptosporidium parvum) and extracellular (eg, Borrelia burgdorferi, Klebsiella pneumoniae). To fight these 2 different kinds of infections, 2 different types of helper T cell subsets evolved, called T helper 1 (Th1) and T helper 2 (Th2) cells. Th1 response protects against intracellular infections by secreting IFN-γ (23), which induces cellular immunity and a phagocytic pathway leading to cell lysis (2, 24). IFN-γ activates a number of genes after binding to the IFN-γ receptor, expressed on the majority of immune cells, including macrophages. IFN-γ also activates differentiation of Th1 cells, suppresses differentiation of Th2 and Th17 subsets, and activates NK cells, macrophages, and CD8 T cells.
Activation of macrophages by IFN-γ has an important role in the clearance of microbes as they produce IL-1, TNF-α, IL-6, and IL-8. In contrast, extracellular infections are controlled by the Th17 subset of T cells secreting IL-17 (25, 26). Th17 cytokine, such as IL-17, helps in clearing infections through recruitment of neutrophils to the infected tissue. Nonimmune cells, such as fibroblasts, endothelial cells, airway smooth muscle cells, and epithelial cells, also express IL-17 receptor, and IL-17 induces the proinflammatory mediators IL-1β, IL-6, TNF-α, GM-CSF, G-CSF, NO, and chemokines (CXCL1, CXCL8, CCL2, CCL7, and CCL20). IL-6 is a known activator of Th17 subsets and acts in a positive feedback loop to amplify differentiation of Th17 cells besides activating acute phase protein and complement. IL-17 is also an efficient B cell helper and promotes formation of a germinal center, antibody production, and isotype class switching (21). The IL17-induced humoral immune response plays an important part in neutralization and clearance of extracellular bacteria (27).
During viral infection, IFN-γ–secreting CD4 Th1 cells have showed an important role in the generation of effective CTL response, as well as humoral immune response (28). Besides directly suppressing viral replication, IFN-γ activates various pathways, including antigen presentation/MHC expression, which results in generation of an effective adaptive immune response. Importance of IFN-γ in clearance of viral infection can be highlighted with the fact that most viruses encode for the proteins that prevent induction of IFN-γ or its signaling.
Although recent studies have showed induction of Th17 cells after viral infections, little is known about their regulation and function. In contrast to IFN-γ, Th17 cells have been shown to play an important role in host defense against fungal infection as Candida (25). Mice with targeted defects in IL-17 receptors or IL-23p19 have showed increased tissue fungal burdens and reduced survival (29). In most cases, inflammation subsides after infection is cleared; however, an overexuberant proinflammatory Th1 or Th17 response can lead to tissue damage that results in an inflammatory disease and autoimmunity, such as RA and multiple sclerosis (MS).
We hypothesize that during evolution, MHC molecules might have been positively selected on the basis of their ability to activate the Th1 (IFN-γ), Th2 (IL-4), or Th17 (IL-17) subset of T cells, required for protection from extracellular or intracellular, or both, pathogens and parasitic infections (Figure 1). Since protection from intracellular pathogens requires IFN-γ (Th1), the MHC class II molecule HLA-DQ6 (*0601) or comparable class II molecules with ability to induce strong IFN-γ response were positively selected. Similarly, MHC class II molecules, such as DQ8 (*0302), were selected for their ability to induce the Th17 subset of helper CD4 T cells, which are required for clearance of extracellular pathogens. Yet, the HLA-DR molecule seems to be more promiscuous because it activates both Th1 and Th17 subsets, though at a lower intensity than DQ molecules. Interestingly, recent studies have showed that IL-17– secreting Th17 subsets play an important role in control of infection at mucosal surfaces, such as lungs and gut (30).
Figure 1.
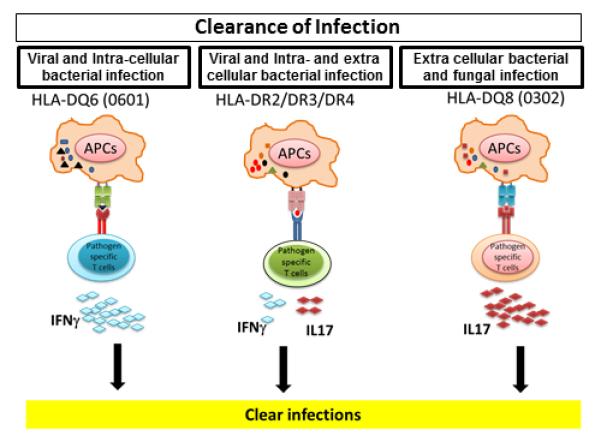
Evolution of MHC class II molecules based on their ability to clear different kind of infections. During evolution, MHC class II molecules might have been selected on the basis of their ability to clear particular types of pathogens through a cytokine network. Because IFN-γ has an important role in clearance of viral and intracellular infections, class II molecules such as HLA-DQ6 (DQB1*0601) and DRβ1*0402 might have been selected on the basis of their ability to induce IFN-γ or TGFβ and IL-10 from CD4 T cells. Since clearance of extracellular bacterial and fungal infection requires Th17 cytokine IL-17, class II molecules such as DQ8 (DQB1*0302) were selected for their ability to induce production of IL-17 from CD4 T cells. At the same time, HLA-DR molecules were able to take care of both kinds of infection; however, they did so at lower levels because of their ability to produce moderate amounts of both IFN-γ and IL-17 compared with HLA-DQ molecules.
MHC and Autoimmunity
Affinity binding of a self-peptide to MHC leads to positive selection of T cells in the thymus (2). CD4 T cells with moderate affinity for the MHC self-peptide complex are selected to fight infection; those with high or low affinity are deleted. However, this selection is not absolute because some CD4 T cells of low affinity escape deletion and join the T cell pool in the periphery. Activation of these CD4 T cells that recognize self-protein leads to development of autoimmunity later in life. A promiscuous MHC class II molecule is good for host defense against infection but might lead to activation of autoreactive T cells.
Three main steps occur in the development of autoimmune diseases. First, predisposition to disease susceptibility is due to the presence of certain HLA class II allele/haplotypes. However, not all individuals who carry a susceptible haplotype have disease, indicating involvement of other factors, such as environment, that might act as a second hit leading to precipitation of disease in certain individuals. Progression of the disease depends on the cytokine and chemokines secreted by the various CD4 T cell subsets.
Autoimmunity-Prone HLA Haplotypes
HLA-DQ8/DR4 Haplotype
HLA-DQ8/DR4 is the most autoimmune-prone haplotype in humans with a high relative risk for RA, type 1 diabetes, celiac disease, MS, and other diseases (31, 32). Yet, it occurs with the highest frequency among all human HLA-DR and DQ haplotypes. During evolution, HLA-DQ8 (DQβ1*0302) might have been selected on the basis of its ability to induce the Th17 subset of T cells to secrete IL17 and protect from infections due to extracellular pathogens. Since most extracellular pathogens gain entry through mucosal surfaces, the DQ8 allele might have played an important role in host survival because IL-17 and Th17 subsets are vital.
How did the DQ8 molecule acquire this ability? A crystal structure of the DQ8 molecule shows that the peptide-binding groove has a deep P4 pocket and a shallow P9 pocket capable of binding multiple peptides with moderate affinity (33). Certain peptides such as gliadin can bind in multiple orientations (34). Thus, the DQ8-peptide complex can activate a large number of CD4 T cell subsets (Th17) to clear infection. Still, DQ8 can also bind multiple self-peptides during thymic education, to positively select autoreactive CD4 T cells.
A person with DQ8 expression is born with a large pool of autoreactive CD4 T cells waiting to be activated to cause autoimmunity. Naïve HLA-DQ8 transgenic mice contain multiple autoreactive T cells (12). Thus, the present DQ8 gene resulted from thousands of years of evolution and gene conversion to produce a promiscuous molecule that is efficient in presenting peptides from infectious agents to activate Th17 cells to clear infection, but which also can cause autoimmunity. The DQ8 allele occurs in linkage with 1 of the more than 50 different HLA-DR4 subtypes in making the HLA-DQ8/DR4 haplotype. The DQ8/*0401 is linked to RA, but DQ8/*0402 is not (35).
HLA-DQ6/DR2 Haplotype
HLA-DQ6.1 (DQB1*0601) allele is found at increased frequency in Asia but is mostly absent in whites, who carry the DQ6.2 (DQB1*0602) allele. Data from our transgenic mice showed that HLA-DQ6.1 induced high levels of IFN-γ (36) and thus might have been positively selected in Asia because of the ability to control infection caused by intracellular pathogens, such as M tuberculosis. HLA-DQ6.1 is linked with HLA-DRB1*1502; HLA-DQ6.2 is linked mostly with HLA-DRB1*1501. All HLA-DR2 subtypes (HLA-DRB1*1501, 1502, and 1503) produce moderate levels of Th1 and Th17 cytokines. In MS, the strongest genetic association is observed in the presence of HLA-DQβ1*0602/DRβ1*1501 (DQ6.2/DR2) haplotype in the white population and HLA-DQβ1*0602/DRβ1*1503 in the African American population (37). At the same time, HLA-DQ6.1 (DQB1*0601) allele is associated with decreased risk of MS.
To better understand the role of DQ6 vs. DR2 in susceptibility to MS, we generated HLA class II transgenic mice expressing HLA-DQ6.1 (DQB1*0601), HLA-DQ6.2 (DQB1*0602), and the HLA-DRB1*1501, 1502, and 1503 alleles and investigated their susceptibility to EAE, an animal model to study human MS. We observed that in contrast to RA, where HLA-DR4 and HLA-DQ8 mice were susceptible to disease, only HLA-DR2 transgenic mice were susceptible to MOG-induced EAE. All 3 DR2 subtypes (DRB1*1501, *1502, and *1503 alleles) were susceptible to MOG-induced EAE and produced moderate levels of both Th1 and Th17 cytokines, indicating a role of both pathways in disease development (31). At the same time, HLA-DQ6.1 (DQB1*0601) transgenic mice and HLA-DQ6.2 (DQB1*0602), were resistant to disease development. Double transgenic DR2/DQ0601 mice had a much lower incidence and severity of disease, confirming human data that DQ6.1 is protective. Theiler’s Murine Encephalomyelitis Virus (TMEV) infection of susceptible mouse strains leads to acute inflammation and subsequent demyelination. Introduction of human class II transgene such as HLA-DR2 and DQ6 protects these mice from demyelination compared to wild-type mice. The clearance of TMEV is associated with increased levels of IFN-γ in the CNS of the protected mice (38).
HLA- DQ6 (0601) Downregulates Autoimmunity
To understand the mechanism of the protection induced by HLA-DQ6, we compared cytokine response between DQ6 and DR3 in mice immunized with PLP91-110 peptide. The EAE-susceptible HLA-DR3 mice immunized with PLP91-110 produced moderate levels of IFN-γ, IL-17, IL-22, and IL-23 while the EAE-resistant DQ6 and DR3/DQ6 mice produced high levels of IFN-γ (36). To determine whether high levels of IFN-γ produced by DQ6.1 restricted CD4 T cells were downregulating EAE, we did neutralization studies. Neutralization of IFN-γ in DQ6/DR3 mice led to increased disease incidence, confirming a protective role of IFN-γ. IFN-γ can suppress inflammation through multiple pathways, such as induction of CD4+CD25+ FoxP3+ Tregs, induction of NO, and direct apoptosis of activated CD4+ T cells. The protective role of IFN-γ in autoimmune disease had been shown earlier because IFN-γ–deficient mice had severe EAE, as well as CIA, on immunization with their respective autoantigens (39).
To further confirm the role of IFN-γ in protection to EAE, we crossed HLA-DR3 transgenic mice with IFN-γ–deficient mice. These HLA-DR3.IFN-γ−/− mice have severe EAE characterized with severe brain plaque, a hallmark of human MS pathology. Antigen-specific CD4 T cells produce high levels of IL-17, indicating that IFN-γ might regulate propagation of Th17 subsets. High IL-17 levels have been shown previously to result in more brain-specific disease and pathologic characteristics (40). Thus, presence of DQ6.1 is associated with induction of high IFN-γ–producing CD4 T cells, an advantage in fighting infection and in downregulating autoimmunity. In whites, MS is linked with HLA-DQ6.2 (0602) allele; DQ6.2 transgenic mice are susceptible to EAE induced by PLP178-197 and MOBP (41, 42). These studies suggest some interesting evolutionary development of DR2, DR3, and DQ6 genes. DR3 transgenic mice produce moderate levels of Th1 and Th17 cytokine, suggesting they had to survive both intracellular and extracellular infection (Figure 2). DQ6.1 induces high levels of IFN-γ to combat intracellular infections (Figure 3). The high levels of IFN-γ also downregulate IL-17 produced by HLA-DR–restricted Th17 cells, to prevent autoimmunity. As early humans moved into Europe, the DQ6.1 gene mutated to DQ6.2 to produce both IFN-γ and IL-17 to combat new pathogens. This conversion resulted in increased autoimmunity linked to this haplotype.
Figure 2.
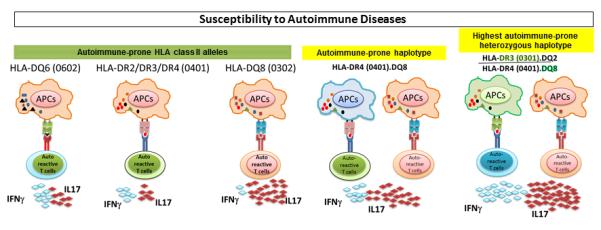
HLA class II alleles predispose to autoimmune diseases through secretion of proinflammatory cytokines. HLA-DR2, DR3, DR4, DQ6 (0602), and DQ8 (0302) alleles are associated with development of autoimmune diseases, such as multiple sclerosis, rheumatoid arthritis, type 1 diabetes, and systemic lupus erythematosus. These molecules activate autoreactive CD4 T cells, secreting proinflammatory cytokine such as IL-17 and IFN-γ. In a disease-prone haplotype, both class II molecules synergize to induce more severe disease through production of increased levels of proinflammatory cytokines, such as IFN-γ and IL-17. Heterozygosity for 2 disease-prone haplotypes results in a cascade of proinflammatory cytokines.
Figure 3.
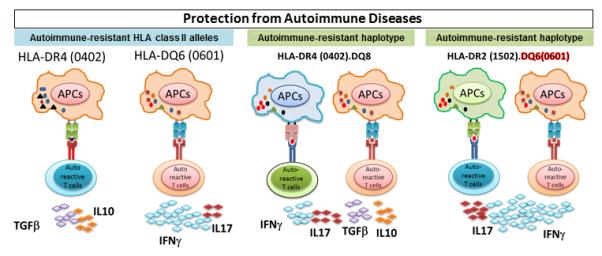
Protective class II haplotypes can modulate the cytokine network. HLA-DQ6 (0601) and DR4 (0402) alleles are associated with protection from multiple sclerosis and rheumatoid arthritis, respectively. The studies done with use of single- and double-HLA transgenic mice have suggested that in protective alleles, protection is mediated through production of anti-inflammatory cytokines. DQ6 (0601) molecules protect from multiple sclerosis through anti-inflammatory IFN-γ, while HLA-DR4 (0402) modulate rheumatoid arthritis through production of anti-inflammatory IL-10 and TGF-β.
HLA-DQ8 Exacerbates Autoimmunity
The presence of DQ8 gene in either cis or trans results in high incidence and severity of autoimmune diseases. For example, individuals heterozygous for DR3.DQ2/DR4.DQ8 have the highest disease incidence and severity for type 1 diabetes and celiac disease (43). We tested the role of DQ8 in disease exacerbation in our EAE model. Presence of DQ8 with either DR2 (DQ8/DR2), DR3 (DQ8/DR3) or DR4 (DQ8/DR4) results in severe disease and CNS pathology (31, 44). DQ8/DR3 transgenic mice with EAE showed widespread brain pathologic characteristics, with severe inflammation and demyelination in all parts of the brain, including cerebellum, brain stem, cortex, corpus callosum, striatum, and meninges. Pathologic analysis showed typical loss of parenchymal white matter, the classic pathologic sign observed in MS. DQ8/DR3 double-transgenic mice also showed increased inflammation and demyelination in the spinal cord.
Using DQ8/DR3 double-transgenic mice, we had observed that the increased disease severity in DQ8/DR3 transgenic mice was due to induction of IL-17 and GM-CSF–producing CD4 T cells by the DQ8 molecule (44, 45) (Figure 2). To confirm the proinflammatory role of IL-17, we neutralized IL-17 in DQ8/DR3 mice immunized for EAE induction. Blocking of IL-17 suppressed the disease incidence, as well as severity, indicating that IL-17 has an important role in induction and progression of disease in EAE. IL-17 binds to IL-17 receptor (IL17R) expressed on numerous cells, including endothelial cells, epithelial cells, and cells of the immune system.
Activation of IL17R leads to a cascade of cytokine and chemokines, culminating in the generation of a strong inflammatory response. IL-17 can induce production of other inflammatory cytokines, such as IL-1β, IL-6, TNF-α, and GM-CSF, besides several chemokines such as CCL20, CXCL1, and CXCL2 (21). The chemokine helps in recruitment of neutrophils to inflamed tissue. IL-17 also causes increased production of reactive oxygen species from brain endothelial cells, leading to disorganization of tight junction and increased permeability of the blood brain barrier. These results cause increased infiltration of inflammatory cells inside the CNS. Once inside the CNS, IL-17 can induce microglia and astrocytes for increased production of IL-1β and IL-6, which help in induction of more Th17 cells. DQ8/DR3 mice with EAE show increased infiltration inside CNS besides production of high levels of IL-17 in brain-infiltrating cells (45). We further observed that DQ8/DR3 mice with EAE have increased levels of IgG and complement deposition in the brain, indicating that IL-17 might also help in induction of humoral immune response. Recently, it has been shown that IL-17 can induce formation of a germinal center inside CNS (46). Thus, IL-17, through modulation of multiple pathways, causes increased inflammation and demyelination in the CNS, leading to pathologic factors of disease. Similar destructive pathways might be activated in other autoimmune diseases such as RA. Observations in DQ8/*0401 mice support this hypothesis (47).
Summary and Conclusion
The main function of the MHC gene is clearing infection and thereby survival of species. HLA genes evolved during thousands of years as humans moved through different parts of the world. The major HLA class II haplotypes DR4/DQ8, DR3/DQ2, and DR2/DQ6 and class I molecules such as B27 are critical in generating efficient immune response to pathogens. They present multiple peptides to activate T cells, B cells, and NK cells and secrete cytokines to control pathogens. Unfortunately, these cells sometime target self-antigens and cause autoimmunity. Thus, autoimmunity is the price paid for clearance of infections and survival of the species.
Acknowledgments
We thank Julie A. Hanson and her staffs for mouse husbandry and Michele K. Smart for tissue typing of transgenic mice.
This work was supported by National Institutes of Health grants AI 75262, AR30752, AR60077, and NS52173.
Abbreviations
- CIA
collagen-induced arthritis
- CII
type II collagen
- EAE
experimental autoimmune encephalomyelitis
- MOG
myelin oligodendrocyte glycoprotein
- MS
multiple sclerosis
- RA
rheumatoid arthritis
References
- 1.Finch CE. Evolution in health and medicine Sackler colloquium: Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janeway CA, Travers P, Walport M, Shlomchik M. Immunobiology Garland Science. New York, NY: 2002. [Google Scholar]
- 3.Klein J. Natural History of the Major Histocompatibility Complex. John Wiley & Sons; New York, NY, USA: 1986. [Google Scholar]
- 4.Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity (Edinb) 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- 5.Trowsdale J. The MHC, disease and selection. Immunol Lett. 2011;137:1–8. doi: 10.1016/j.imlet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Yeager M, Hughes AL. Evolution of the mammalian MHC: natural selection, recombination, and convergent evolution. Immunol Rev. 1999;167:45–58. doi: 10.1111/j.1600-065x.1999.tb01381.x. [DOI] [PubMed] [Google Scholar]
- 7.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 8.Wills C, Green DR. A genetic herd-immunity model for the maintenance of MHC polymorphism. Immunol Rev. 1995;143:263–292. doi: 10.1111/j.1600-065x.1995.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 9.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 10.Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 11.Oliver MK, Piertney SB. Selection maintains MHC diversity through a natural population bottleneck. Mol Biol Evol. 2012;29:1713–1720. doi: 10.1093/molbev/mss063. [DOI] [PubMed] [Google Scholar]
- 12.Abraham RS, Wen L, Marietta EV, David CS. Type 1 diabetes-predisposing MHC alleles influence the selection of glutamic acid decarboxylase (GAD) 65-specific T cells in a transgenic model. J Immunol. 2001;166:1370–1379. doi: 10.4049/jimmunol.166.2.1370. [DOI] [PubMed] [Google Scholar]
- 13.Olerup O, Troye-Blomberg M, Schreuder GM, Riley EM. HLA-DR and -DQ gene polymorphism in West Africans is twice as extensive as in north European Caucasians: evolutionary implications. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:8480–8484. doi: 10.1073/pnas.88.19.8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence DN, Bodmer JG, Bodmer WF. Distribution of HLA antigens in Ticuna Indians of Brazil: results of typing a leprosy-affected family. Tissue antigens. 1980;16:152–160. doi: 10.1111/j.1399-0039.1980.tb00596.x. [DOI] [PubMed] [Google Scholar]
- 15.Yan ZH, Fan Y, Wang XH, Mao Q, Deng GH, Wang YM. Relationship between HLA-DR gene polymorphisms and outcomes of hepatitis B viral infections: A meta-analysis. World J Gastroenterol. 2012;18:3119–3128. doi: 10.3748/wjg.v18.i24.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chadwick D, Cardew G. Variation in the human genome. Wiley; 1996. [Google Scholar]
- 17.Tsuji KA,M, Sasazuki T. HLA 1991-Proceedings of the Eleventh International Histocompatbility Workshop and Conference; Oxford: Oxford University Press; 1992. [Google Scholar]
- 18.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 19.Mosmann TR, Sad S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 20.Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 21.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 22.Akdis M. The cellular orchestra in skin allergy; are differences to lung and nose relevant? Curr Opin Allergy Clin Immunol. 2010;10:443–451. doi: 10.1097/ACI.0b013e32833d7d48. [DOI] [PubMed] [Google Scholar]
- 23.Billiau A, Matthys P. Interferon-gamma: a historical perspective. Cytokine Growth Factor Rev. 2009;20:97–113. doi: 10.1016/j.cytogfr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Alexander J, Satoskar AR, Russell DG. Leishmania species: models of intracellular parasitism. J Cell Sci. 1999;112(Pt 18):2993–3002. doi: 10.1242/jcs.112.18.2993. [DOI] [PubMed] [Google Scholar]
- 25.Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pappu R, Ramirez-Carrozzi V, Sambandam A. The interleukin-17 cytokine family: critical players in host defence and inflammatory diseases. Immunology. 2011;134:8–16. doi: 10.1111/j.1365-2567.2011.03465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsdoerffer M, Lee Y, Jager A, Kim HJ, Korn T, Kolls JK, Cantor H, Bettelli E, Kuchroo VK. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc Natl Acad Sci U S A. 2010;107:14292–14297. doi: 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novelli F, Casanova JL. The role of IL-12, IL-23 and IFN-gamma in immunity to viruses. Cytokine Growth Factor Rev. 2004;15:367–377. doi: 10.1016/j.cytogfr.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis. 2004;190:624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 30.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Mangalam AK, Rajagopalan G, Taneja V, David CS. HLA class II transgenic mice mimic human inflammatory diseases. Adv Immunol. 2008;97:65–147. doi: 10.1016/S0065-2776(08)00002-3. [DOI] [PubMed] [Google Scholar]
- 32.Taneja V, David CS. HLA class II transgenic mice as models of human diseases. Immunol Rev. 1999;169:67–79. doi: 10.1111/j.1600-065x.1999.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol. 2001;2:501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 34.Hovhannisyan Z, Weiss A, Martin A, Wiesner M, Tollefsen S, Yoshida K, Ciszewski C, Curran SA, Murray JA, David CS, Sollid LM, Koning F, Teyton L, Jabri B. The role of HLA-DQ8 beta57 polymorphism in the anti-gluten T-cell response in coeliac disease. Nature. 2008;456:534–538. doi: 10.1038/nature07524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feitsma AL, van der Helm-van Mil AH, Huizinga TW, de Vries RR, Toes RE. Protection against rheumatoid arthritis by HLA: nature and nurture. Ann Rheum Dis. 2008;67(Suppl 3):iii61–63. doi: 10.1136/ard.2008.098509. [DOI] [PubMed] [Google Scholar]
- 36.Mangalam A, Luckey D, Basal E, Behrens M, Rodriguez M, David C. HLA-DQ6 (DQB1*0601)-restricted T cells protect against experimental autoimmune encephalomyelitis in HLA-DR3.DQ6 double-transgenic mice by generating anti-inflammatory IFN-gamma. J Immunol. 2008;180:7747–7756. doi: 10.4049/jimmunol.180.11.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gourraud PA, Harbo HF, Hauser SL, Baranzini SE. The genetics of multiple sclerosis: an up-to-date review. Immunol Rev. 2012;248:87–103. doi: 10.1111/j.1600-065X.2012.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez M, Zoecklein LJ, Howe CL, Pavelko KD, Gamez JD, Nakane S, Papke LM. Gamma interferon is critical for neuronal viral clearance and protection in a susceptible mouse strain following early intracranial Theiler’s murine encephalomyelitis virus infection. J Virol. 2003;77:12252–12265. doi: 10.1128/JVI.77.22.12252-12265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Pierson E, Simmons SB, Castelli L, Goverman JM. Mechanisms regulating regional localization of inflammation during CNS autoimmunity. Immunol Rev. 2012;248:205–215. doi: 10.1111/j.1600-065X.2012.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaushansky N, Altmann DM, Ascough S, David CS, Lassmann H, Ben-Nun A. HLA-DQB1*0602 determines disease susceptibility in a new “humanized” multiple sclerosis model in HLA-DR15 (DRB1*1501;DQB1*0602) transgenic mice. J Immunol. 2009;183:3531–3541. doi: 10.4049/jimmunol.0900784. [DOI] [PubMed] [Google Scholar]
- 42.Kaushansky N, Altmann DM, David CS, Lassmann H, Ben-Nun A. DQB1*0602 rather than DRB1*1501 confers susceptibility to multiple sclerosis-like disease induced by proteolipid protein (PLP) J Neuroinflammation. 2012;9:29. doi: 10.1186/1742-2094-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham J, Kockum I, Sanjeevi CB, Landin-Olsson M, Nystrom L, Sundkvist G, Arnqvist H, Blohme G, Lithner F, Littorin B, Schersten B, Wibell L, Ostman J, Lernmark A, Breslow N, Dahlquist G. Negative association between type 1 diabetes and HLA DQB1*0602-DQA1*0102 is attenuated with age at onset. Swedish Childhood Diabetes Study Group. Eur J Immunogenet. 1999;26:117–127. [PubMed] [Google Scholar]
- 44.Luckey D, Bastakoty D, Mangalam AK. Role of HLA class II genes in susceptibility and resistance to multiple sclerosis: studies using HLA transgenic mice. J Autoimmun. 2011;37:122–128. doi: 10.1016/j.jaut.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangalam A, Luckey D, Basal E, Jackson M, Smart M, Rodriguez M, David C. HLA-DQ8 (DQB1*0302)-restricted Th17 cells exacerbate experimental autoimmune encephalomyelitis in HLA-DR3-transgenic mice. J Immunol. 2009;182:5131–5139. doi: 10.4049/jimmunol.0803918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters A, Pitcher LA, Sullivan JM, Mitsdoerffer M, Acton SE, Franz B, Wucherpfennig K, Turley S, Carroll MC, Sobel RA, Bettelli E, Kuchroo VK. Th17 cells induce ectopic lymphoid follicles in central nervous system tissue inflammation. Immunity. 2011;35:986–996. doi: 10.1016/j.immuni.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behrens M, Trejo T, Luthra H, Griffiths M, David CS, Taneja V. Mechanism by which HLA-DR4 regulates sex-bias of arthritis in humanized mice. J Autoimmun. 2010;35:1–9. doi: 10.1016/j.jaut.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


