Summary
The abundance of Myc protein must be exquisitely controlled to avoid growth abnormalities caused by too much or too little Myc. An intriguing mode of regulation exists in which Myc protein itself leads to reduction in its abundance. We show here that dMyc binds to the miR-308 locus and increases its expression. Using our gain-of-function approach, we show that an increase in miR-308 causes a destabilization of dMyc mRNA and reduced dMyc protein levels. In vivo knockdown of miR-308 confirmed the regulation of dMyc levels in embryos. This regulatory loop is crucial for maintaining appropriate dMyc levels and normal development. Perturbation of the loop, either by elevated miR-308 or elevated dMyc, caused lethality. Combining elevated levels of both, therefore restoring balance between miR-308 and dMyc levels, resulted in lower apoptotic activity and suppression of lethality. These results reveal a sensitive feedback mechanism that is crucial to prevent the pathologies caused by abnormal levels of dMyc.
Key words: dMyc, miR-308, Drosophila melanogaster, Embryogenesis, Growth
Introduction
Myc is a transcription factor with a primary role in promoting cell proliferation and growth. Myc belongs to basic helix-loop-helix (bHLH) protein family together with Max. The Myc-Max heterodimer binds to E-boxes (CACGTG, CATGTG and alternative sequences) in the promoter region of target genes (Eilers and Eisenman, 2008; Blackwell et al., 1993). The biological effects of Myc are predominantly due to the up-regulation of genes that are involved in an increase of cell mass (Schmidt, 1999; Eilers and Eisenman, 2008), progression of cell division (Bouchard et al., 1998) and increased energy metabolism (Morrish et al., 2008). Myc's regulation of its targets occurs mainly at the transcriptional level. However, it has been shown that Myc's mechanisms of function also extend to regulation of translation and DNA replication (Cole and Cowling, 2008).
In Drosophila, Myc (dMyc) is required for oogenesis and the proper growth of Drosophila larvae (Maines et al., 2004; Pierce et al., 2004). Loss of dMyc has been shown to retard the growth of cells and cause lethality (Pierce et al., 2008). Conversely, overexpression of dMyc results in increased size of larvae and organs of adult flies (Johnston et al., 1999).
Certain types of human cancers lack proper regulation of Myc levels resulting in uncontrolled cell proliferation and tumor formation (Eilers and Eisenman, 2008; Meyer and Penn, 2008). Hence, tight control and maintenance of appropriate Myc protein levels is crucial.
We and others have demonstrated that Myc protein utilizes auto-regulation to repress its own transcription in Drosophila and rodents (Penn et al., 1990; Goodliffe et al., 2005). Moreover, additional post-transcriptional and post-translational regulatory pathways contribute to the critical maintenance of Myc protein levels in cells (Salghetti et al., 1999; Gregory and Hann, 2000). Growing evidence has shown that mammalian Myc uses microRNAs to partially regulate its targets (Wang et al., 2011; O'Donnell et al., 2005; Chang et al., 2008; Lin et al., 2009; Xiong et al., 2010) as well as its own transcript levels (Kress et al., 2011; Liao and Lu, 2011).
MicroRNAs are a group of short (21–25 nucleotides) non-coding RNAs involved in negative regulation of gene expression (He and Hannon, 2004). They block the translation of transcripts by binding to their complementary sequences in 3′ untranslated region (3′-UTR) regions of mRNAs (Berezikov et al., 2006). In mice and human cell lines, Myc abundance has been shown to be associated with changes in expression patterns of certain microRNAs, which in turn regulate the expression of other genes (Chang et al., 2008; Lin et al., 2009). In addition, Myc's mRNA itself has been shown to be a target of microRNAs in mice and human cell lines (Xiong et al., 2010) and it is also a predicted target of Drosophila “dme-miR-2a-1/6/11/13/308” microRNA family (Ruby et al., 2007).
In this study, we illustrate the regulatory relationship of Myc and microRNA 308 (miR-308) in Drosophila. We show that miR-308 levels increase in response to elevated levels of dMyc through the physical interaction of dMyc with the miR-308 locus, and dMyc's transcript itself is a target of miR-308. In a miR-308 knockdown study, we show that dMyc levels are under the control of miR-308 during embryogenesis. Furthermore, ectopic expression of miR-308 in embryos can rescue lethality and suppress apoptotic activity caused by dMyc overexpression. Finally, through analysis of dMyc's targets and comparing them to the predicted targets of miR-308, we propose models explaining a concerted gene regulation program by dMyc and miR-308.
Materials and Methods
microRNA microarray
Total RNA, including small RNAs, was isolated from 0–24 hour old embryos by miRNAeasy (Qiagene, California, USA). Small RNAs were subject to poly-adenylation and fluorescent labeling using a power labeling kit (Exiqon, Copenhagen, Denmark). Dual-color labeling with common reference method was used to hybridize the samples on Exiqon microRNA microarray version 11. The experiment was carried out in two biological replicates and four technical replicates on each slide. Spot intensities were acquired and normalized by GenePix Pro (Molecular Devices, California, USA). T-test statistical analysis was carried out by using Differential expression analysis module on Genepattern (Reich et al., 2006).
Affymetrix microarray
Total RNA, was isolated from 0–24 hour old embryos by Trizol (Invitrogen, California, USA), according to the manufacturer's protocol. Processing and analysis of microarrays were carried out by Expression Analysis (Durham, North Carolina, USA). Briefly, RNA samples form 3 biological replicates were labeled and hybridized on Affymetrix Drosophila Genome 2.0 Array. Data were analyzed using Two-Group Comparisons with Permutation Analysis for Differential Expression (PADE). Data have been deposited at the NCBI Gene Expression Omnibus (GEO) repository (Accession number: GSE38529).
Drosophila strains and genetics
Flies expressing Gal4 ubiquitously in embryos were used (w[*];P{w[+mW.hs] = GAL4-da.G32}UH1) for all the crosses involving yeast Gal4-UAS system (Fischer et al., 1988). UAS-dmyc flies (P{ry[+t7.2] = hsFLP}22, y[1] w[*]; P{w[+mC] = UAS-dm.Z}42) were used for ectopic expression of dMyc and generation of doubly homozygous flies. Both of the mentioned stocks were obtained from Bloomington Stock Center (Indiana University, Bloomington, IN, USA). For dMyc-knockdown study, we used a transgenic Drosophila expressing RNAi for dMyc, under the control of UAS (y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.JF01762}attP2). We crossed these flies to flies expressing Gal4 ubiquitously in embryos, as explained above. UAS-miR-308 flies were constructed by cloning 300 base pairs of the genomic region of microRNA-308 into the pUAST plasmid (Brand and Perrimon, 1993) and injecting the construct into Drosophila embryos (BestGene, California, USA). 10 independent lines were generated, four of which were homozygous for UAS-miR-308. One of the homozygous transgenic lines (transgenic line no. 10) that has UAS-miR-308 construct on the 3rd chromosome was used for all the experiments involving the ectopic expression of the miR-308. UAS-mutated-miR-308 flies were constructed by site-directed mutagenesis using Accuprime Pfx (Invitrogen, California, USA) on the same UAS-miR-308 construct. One of the homozygous transgenic (transgenic line no. 5) was used for all the experiments involving the ectopic expression of mutated-miR-308. All crosses were done in 25°C.
Quantification of miR-308 by qPCR
Total RNA including small RNAs was extracted by miRNeasy mini-kit (Qiagene, California, USA), according to the manufacturer's instruction. Universal cDNA synthesis kit (Exiqon, Denmark) was used to make cDNA form mature sequence of microRNAs. Briefly, mature microRNAs were poly-adenylated and cDNA were synthesized by using a poly-T primer with a 3′ degenerate anchor and a 5′ universal tag. Amplification was carried out using a microRNA-specific primer, a universal primer and SYBR green (all from Exiqon, Denmark) in an ABI 7500 Fast real-time PCR system (Applied Biosystems, California, USA). MicroRNA miR-1 was used as the reference gene as it showed no significant change in our microRNA microarray study. Comparative Ct method was used for analyzing the expression levels and relative fold changes (Schmittgen and Livak, 2008).
Quantitative RT-PCR of mRNAs
Total RNA was isolated using Trizol (Invitrogen, California). Power SYBR® Green RNA-to-CTTM 1-Step Kit and ABI 7500 Fast real-time PCR system (Applied Biosystems, California, USA) were used for quantification of mRNA transcripts. Comparative Ct method was used for quantification of all mRNA transcripts (Schmittgen and Livak, 2008). Ras was used as internal reference gene for quantification, as described previously (Khan et al., 2009). Q-RT-PCR Primers: dMyc (F:ATGCACATCACCGATCACAG, R:TGGGCCATCTGGAACTGTAG) dRas (F:ATATCGGCACCTACCGTGAG, R:GTCTTGGCGGATGTCTCAAT) RpS23 (F:CGTCCTGGAGAAGGTCGGCG, R:ACCTTGAAGCGCACACCGGG).
MiR-308 knockdown
For each independent experiment, approximately 200 wild-type (Canton-S) embryos were collected 30 minutes after egg deposition and were dechorionated in 50% sodium hypochlorite solution for 2 minutes and were rinsed several times with water. Embryos were desiccated for an appropriate, empirically determined time. Embryos were then injected with 5% egg volume as described (Kennerdell and Carthew, 1998). Injection solutions: 100 µM anti-miRNA-308 LNA inhibitor solution (Exiqon, Copenhagen, Denmark) (sequence of the inhibitor: CTCACAGTATAATCCTGTG) in PBS or PBS alone (control). The embryos were allowed to develop at 18°C for 15 hours before collecting them for either RNA or protein extraction.
Chromatin immuno-precipitation
Approximately 0.5 grams of 0–24 hours embryos were dechroniated and fixed in PBS/Hepatane/formaldehyde and sonicated in SDS-lysis buffer. The EZ-ChIP kit (Millipore, Massachusetts, USA) was used for precipitation and washes. Anti-MycN and anti-dMyc antibodies (SantaCruz Biotechnology, sc-28208 and sc-28207) were used for immuno-precipitation of dMyc protein. ChIP primers: ChIP region I: 5′-TGGCGAGATACGGCGGGACA-3′ and 5′-GGTTTGAGTCCAGGGTGGATGAACG-3′ ChIP region II: 5′- GCGAGGCGTCGAGACGTGTT-3′ and 5′-TCTACAACAGGCAAGCCAAGAGGT-3′ ChIP region III: 5′-TAGCAACTGCGGTAGGTACG-3′ and 5′-GCCTAAGCGAACTCAGGACT-3′. Fibrillarin promoter region: 5′-TTTTACGCACCTGGTTTGCCCA-3′ and 5′-CCTCTCCGCCTGGTGTTGAACTT-3′.
Immunoblots
8% poly-acrylamide gels were used for separation of whole cell protein extracts. Detection of dMyc was carried out using rabbit anti-dMyc (Santa Cruz biotechnology, California, USA, cat. no. 28207) and mouse anti-rabbit HRP. Detection of Fibrillarin was carried out using mouse anti-Fibrillarin (Abcam, Cambridge, USA, cat. no. ab4566) and rabbit anti-mouse HRP. Detection of Dronc was carried out using rabbit anti-cleaved caspase-3 (Cell Signaling Technology, Danvers, USA, cat. no. 9661) and mouse and rabbit-HRP. Anti-beta Actin antibody (Abcam, Massachusetts, USA, cat. no. ab8224) was used as loading control. Blots were quantified with NIH ImageJ software package (http://rsbweb.nih.gov/ij).
Growth and survival
Zero to 1 hour old embryos from each genotype were collected on grape agar plates and were counted and transferred to new plates. Survivals of the hatched larvae were assessed after 24, 48, 72 and 96 hours by observing their movement in response to mechanical stimulation. Fraction of the live animals was plotted in Microsoft Excel.
Functional analysis and graphical design
Functional analysis of gene lists was performed by Ontologizer 2.0 (Bauer et al., 2008). Heat map of created by ArcGIS (Esri, California, USA). Schematic structure of the genes was made using the CLC Genomic Workbench Utility (Aarhus, Denmark). Other graphics and diagrams were made using Microsoft PowerPoint. Images were cropped and adjusted using Adobe Photoshop (California, USA). Graphs were generated using GraphPad PRISM and Microsoft Excel.
Results
Elevated levels of dMyc results in an increase in miR-308 levels
Regulation of gene expression by dMyc occurs through the direct binding of dMyc to E-boxes in the promoter region of target genes (Gallant, 2009). However, many genes that are responsive to dMyc in Drosophila lack E-boxes or similar sequences (Goodliffe et al., 2007). We asked to what extent dMyc indirectly regulates its targets via microRNAs. Using a yeast Gal4-UAS system, we increased levels of dMyc in Drosophila embryos. We collected 0–24 hour embryos from a cross of daughterless-Gal4 females (w*; P{GAL4-da.G32}UH1) that express excessive Gal4 protein ubiquitously during embryogenesis, and UAS-dMyc males (P{hsFLP}22, y1 w*; P{UAS-dm.Z}42). A microRNA microarray was used to measure the genome-wide expression of microRNAs throughout embryogenesis. Out of 152 available probes for Drosophila microRNAs, 42 microRNAs were expressed in embryos. Ectopically increased levels of dMyc caused a significant, 1.8-fold (Cutoff: t-test p-value < 0.05, fold change > 1.5) increase in the expression of Drosophila microRNA-308 (miR-308), compared to that of control (Fig. 1A). Quantitative RT-PCR of miR-308 confirmed 2.9-fold±0.9 (mean ± s.d., n = 3) increase in miR-308 in response to elevated levels of dMyc (Fig. 1B). This result suggests a possible role for miR-308 in dMyc's regulation of gene expression.
Fig. 1. miR-308 responds to elevated levels of dMyc.

(A) microRNA microarray analysis of expression of microRNAs responding to elevated dMyc protein. X-axis indicates only those microRNAs that are expressed in embryos. Y-axis indicates relative fold change of each microRNA in embryos with overexpression of dMyc (da-Gal4 crossed to UAS-dMyc) compared to that of control (da-Gal4). Data are presented as means ± standard deviation of two independent biological replicates Asterisk represents the significant difference based on t-test (cutoff: P-value < 0.05, fold change > 1.5). (B) Quantitative RT-PCR analysis of miR-308 expression in control (da-Gal4) vs embryos with elevated amounts of dMyc (da-Gla4 crossed to UAS-dMyc), normalized to miR-1. Data are presented as means ± standard deviation of three independent biological replicates.
dMyc physically interacts with miR-308 and its host gene locus
miR-308 gene is located in the second intron of the ribosomal protein S23 (RpS23) gene. We identified two non-canonical E-boxes (CATGTG) upstream of the miR-308 gene (Fig. 2A). We asked whether dMyc directly binds to the locus of miR-308 gene. We carried out chromatin Immunoprecipitation (ChIP) using two different dMyc antibodies in wild-type (Canton-S) embryos, and results show a strong association (5.2-fold enrichment relative to IgG) of dMyc to the intronic region of the RpS23 gene. We also observed a lower association of dMyc (3.6-fold enrichment compared to IgG) to the upstream region of RpS23 transcription start site and no association to the downstream region of RpS23 gene (Fig. 2B,C). We used the locus of Fibrillarin gene as a positive control because it has multiple canonical E-boxes and is a known target of dMyc (Orian et al., 2003).
Fig. 2. dMyc physically binds to the miR-308 locus and regulates the transcript levels of miR-308 and its host gene, Ribosomal protein S23 (RpS23).

(A) miR-308 locus is shown in the second intron of the RpS23 gene. Arrows in the upstream region of the miR-308 gene indicate the positions of non-canonical E-boxes (CATGTG). PCR products for chromatin-IP are indicated as ChIP region-I and ChIP region-II. (B) Chromatin immuno-precipitation of dMyc by two different anti-dMyc antibodies and IgG, followed by electrophoresis of PCR product. (C) Quantification of ChIP by qPCR. Data are shown as fold enrichment compared to that of the IgG control. Data are presented as means ± standard deviation of three independent biological replicates. (D) Effect of elevated level of Myc on the transcript levels of RpS23, the gene that hosts miR-308. Data are presented as means ± standard deviation of three independent biological replicates.
It has been shown that intronic microRNAs are co-expressed with their host genes (Baskerville and Bartel, 2005; Truscott et al., 2011). We asked whether dMyc binding to the locus of miR-308 changes the expression of RpS23 as well. Quantitative RT-PCR analysis showed that the transcript levels of RpS23 increase in response to increased levels of dMyc (Fig. 2D). Overall, this result suggests that dMyc regulates the expression of miR-308 and its host gene, RpS23, by binding to their shared locus. However, we did not determine whether dMyc binding to the intronic region of RpS23 gene promotes transcription of miR-308, independent of RpS23 transcript.
dMyc's transcript is a predicted target of miR-308
miR-308 belongs to the Drosophila miR-2 conserved family of microRNAs. Members of miR-2 family (miR-2a, miR-6, miR-11, miR-13 and miR-308) are predominantly expressed during embryogenesis and share a vast number of predicted targets based on their common 6 nucleotide seed sequence (5′-AUCACA-3′) (Aravin et al., 2003). We identified the predicted targets of miR-308 by using two available algorithms, TargetScanFly (Ruby et al., 2007) and microRNA.org (Enright et al., 2003). Both indicated that a conserved sequence in the 3′-UTR region of dMyc's transcript is a putative target of miR-308 and other members of the miR-2 microRNA family (Fig. 3A).
Fig. 3. Ectopic miR-308 reduces the abundance of dMyc transcripts and protein.

(A) The conserved 6-nucleotide seed of miR-308 and its complementary sequence in the 3′-UTR region of the dMyc transcript. The number 292 denotes the position of the 5′ nucleotide relative to the first nucleotide in 3′-UTR of dMyc transcript. (B) Graphical presentation of the constructs used for in vivo overexpression of miR-308 and mutated-miR-308*. 5 out of 6 nucleotide of the seed sequence are randomly changed. (C) Expression of UAS-microRNA constructs in transgenic Drosophila lines. Top panel: Quantitative RT-PCR of miR-308 in control embryos (da-Gal4) and embryos with overexpression of miR-308 (da-Gal4 crossed to UAS-miR-308). Bottom panel: Quantitative RT-PCR of miR-308 in control embryos (da-Gal4) and embryos with overexpression of miR-308 (da-Gal4 crossed to UAS-mut-miR-308). Data are presented as means ± standard deviation of three independent replicates. P-value is calculated by two-tailed student t-test. (D) Quantitative RT-PCR analysis of dMyc expression in control vs embryos with elevated amounts of miR-308, normalized to Ras. Data from five independent biological replicates are plotted in box and whisker plots. Boxes represent 25%, 50% (median) and 75% quartiles. Bars indicate the maximum and minimum. Two-tailed student t-test is used for calculating p-value. (E) Western blot analysis of dMyc protein level in control embryos and embryos with overexpression of miR-308 and mutated-miR-308. Actin is used as loading control.
Based on the computational prediction, we hypothesized that dMyc's transcript is a target of miR-308 and that miR-308 is part of the negative feedback regulation of dMyc. To test the effect of miR-308 on the expression of dMyc in vivo, we generated transgenic Drosophila lines that express miR-308 under the control of the yeast Gal4 UAS enhancer. As a control, we generated flies expressing a mutated form of miR-308 with a change in the seed sequence from 5′-AUCACA-3′ to 5′-GGAUCC-3′ (Fig. 3B). We crossed these flies to flies expressing Gal4 and confirmed the expression of ectopic miR-308 and mutated-miR-308 in embryos by quantitative RT-PCR (Fig. 3C).
miR-308 overexpression results in reduced dMyc transcript and protein levels
microRNAs regulate the expression of their target genes primarily by inhibition of their translation. However, microRNAs can also destabilize their target mRNA (Baek et al., 2008). We tested whether the ectopic expression of miR-308 results in a decrease in dMyc transcript and protein levels. We collected 0–24 hour embryos from a cross of daughterless-Gal4 females and UAS-miR-308 males, and measured dMyc levels by qRT-PCR and Western blotting. We observed a modest 14%±5 (mean ± s.d., n = 5) reduction of dMyc transcripts in embryos expressing ectopic miR-308 compared to that of control (Fig. 3D). Despite the modest reduction in transcript levels, we observed a substantial 68%±10 (mean ± s.d., n = 3) decrease in dMyc protein levels in embryos expressing ectopic miR-308, compared to that of control (Fig. 3E). We asked whether these animals show reduced survival, potentially caused by the reduction in dMyc protein accumulation. Our survival assay showed that these animals have a reduced survival rate. 19%±9 (mean ± s.d., n = 97, P-value = 1.1E−15, Fisher exact test) of them survived past 96 hours after egg deposition, compared to 77%±6 (mean ± s.d., n = 81) of control. Together, these data suggest that miR-308 is sufficient to mediate a negative regulatory mechanism controlling dMyc in vivo. While our miR-308 and mutated-miR-308 overexpression models suggest that the 5′-seed sequence of miR-308 is crucial for this regulation, we were not able to conclude that this regulation is a result of direct interaction between miR-308 and dMyc mRNA. Further reporter gene assays, e.g. luciferase assays, on the 3′-UTR region of dMyc mRNA should be carried out to test the direct regulation.
miR-308 overexpression results in reduced Fibrillarin protein level
We asked whether the effect of miR-308 overexpression on the dMyc protein could alter the expression of dMyc targets. We selected fibrillarin as an indicator of dMyc activity. Fibrillarin is a marker for the nucleolus and nucleolar bodies (Daneshvar et al., 2011) and it is involved in biogenesis of ribosomal RNA (Tollervey et al., 1993). Both in Drosophila and vertebrates, fibrillarin has been shown to be a transcriptional target of the Myc protein (Orian et al., 2003; Coller et al., 2000). First, we asked whether a reduction in dMyc levels would alter the levels of fibrillarin in Drosophila embryos. We used transgenic Drosophila embryos with a dMyc-RNAi construct under the control UAS. We crossed these flies with flies expressing Gal4 and collected embryos that were 0–24 hours old for protein analysis. We tested the efficiency of the dMyc-RNAi model and observed reduced dMyc levels compared to the control embryos (Fig. 4A). In a western blot analysis using an anti-fibrillarin antibody, we observed that reduced dMyc levels caused a reduction in the abundance of fibrillarin. If a reduction in dMyc levels causes a decrease in fibrillarin levels, the overexpression of miR-308 should have the same effect on fibrillarin. Our western blot analysis showed a reduction in the abundance of fibrillarin in embryos in response to the overexpression of miR-308 (Fig. 4B).
Fig. 4. Reduction in abundance of Fibrillarin protein in response to a decrease in dMyc protein levels or an increase in miR-308 levels.

(A) Western blot analysis of dMyc protein level in control embryos (da-Gal4) and embryos with dMyc-RNAi (da-Gal4 crossed to UAS-dMyc-RNAi). (B) Western blot analysis of Fibrillarin protein in control embryos (da-Gal4), embryos with ectopic expression of miR-308 (da-Gal4 crossed to UAs-miR-308) and embryos with dMyc-RNAi (da-Gal4 crossed to UAS-dMyc-RNAi). Actin is used as loading control in both experiments.
In vivo knockdown of miR-308 leads to overexpression of dMyc
To determine whether dMyc transcripts are normally repressed by miR-308, we depleted miR-308 in early embryos by injecting LNA (Locked Nucleic Acids)-modified oligonucleotides complimentary to the sequence of miR-308. LNAs are modified DNAs with higher stability and specificity that can effectively inhibit microRNAs (Ørom et al., 2006). Wild-type embryos 30 minutes after egg deposition were injected with either a LNA inhibitor against miR-308 or with injection buffer. Quantitative RT-PCR showed that the inhibition of miR-308 causes an 18%±4 (mean ± s.d., n = 3) increase in dMyc mRNA levels (Fig. 5A). Immunoblotting showed an increase in dMyc protein accumulation after inhibition of miR-308, compared to control (Fig. 5B).
Fig. 5. Knockdown of miR-308 causes an increase in dMyc transcript and protein level.

(A) Quantitative RT-PCR with primers specific for dMyc in embryos injected with PBS (control) and embryos injected with inhibitor of miR-308. Data are presented as means ± standard deviation of three independent replicates. (B) Western blot with antibody specific to dMyc in embryos injected with PBS and embryos injected with inhibitor of miR-308. Actin is used as loading control. Bar graph shows the normalized quantification of the western blot bands, using ImageJ software.
These results show that miR-308, despite its low abundance (Aravin et al., 2003), can represses dMyc levels during embryogenesis. These data, taken with our chromatin-IP study demonstrating dMyc binding to the locus of miR-308 in wild-type embryos, suggest a feedback loop between dMyc and miR-308 that limits the accumulation of dMyc protein during embryogenesis. Since other members of miR-2 family share the same seed sequence, it would be interesting to determine whether dMyc is under repression by other members of miR-2 family.
miR-308 can rescue the dMyc overexpression phenotype
Overexpression of dMyc can cause lethality. It is believed that this lethality is due to the induction of apoptosis by dMyc (Montero et al., 2008). In our experiments, dMyc protein levels decrease upon overexpression of miR-308. As previously reported, we also observed that dMyc overexpressing embryos do not survive beyond 72 hours after egg deposition (Khan et al., 2009). We sought to determine whether miR-308 could rescue the dMyc overexpression lethal phenotype. By meiotic recombination of the two transgenes on third chromosome, we generated flies expressing both dMyc and miR-308 under the control of separate UAS enhancers that are responsive to the Gal4 transcription factor. Doubly transgenic flies were crossed to flies expressing Gal4 protein ubiquitously in embryos (daughterless-Gal4). Immunoblotting showed that dMyc protein accumulation was balanced by the addition of UAS-miR-308; it resembled wild type levels rather than ectopic levels (Fig. 6A). Consistent with these results, when crossed to Gal4 expressing flies, 40%±12 (mean ± s.d., n = 84, three independent crosses) of these double UAS transgenic animals survived beyond 72 hours, compared to 0% (n = 94, three independent crosses, p-value = 9.7E−14, Fisher exact test) UAS-dmyc alone and 19%±9 (mean ± s.d., n = 97, three independent crosses) p-value = 4E−4, Fisher exact test) UAS-miR-308 alone (Fig. 6B).
Fig. 6. Ectopic expression of miR-308 in embryos can rescue lethality and suppress apoptotic activity caused by dMyc overexpression.

(A) Western blot showing the dMyc protein level in control embryos, embryos with overabundance of dMyc and embryos with overabundance of both dMyc and miR-308. Actin is used as loading control. (B) Survival assay of the four genotypes: Blue, control (da-Gal4), Green, miR-308 overexpressing animals (da-Gal4 crossed to UAS-miR-308), Red, dMyc overexpressing animals (da-Gal4 crossed to UAS-dMyc) and Purple, animals having overexpression of dMyc and miR-308 (da-Gal4 crossed to UAS-dMyc; UAS-miR-308). Data are presented as means ± standard deviation of three independent replicates. P-values are calculated by Fisher exact test. (C) Western blot showing the levels of full length, cleaved large subunit and cleaved pro-domain of Dronc in control embryos (da-Gal4), embryos overexpressing dMyc (da-Gal4 crossed to UAS-dMyc) and embryos overexpressing miR-308 (da-Gal4 crossed to UAs-miR-308). Actin is used as loading control.
Although the Myc protein has been shown to promote proliferation and growth, the ectopic expression of Myc in normal cells does not cause overgrowth and cancer. It has been shown that overexpression of Myc in normal cells without background mutations triggers apoptosis (Pelengaris et al., 2002; Montero et al., 2008). We asked whether the overexpression of dMyc in Drosophila embryos causes an increase in apoptotic activity, and if this activity can be suppressed by the addition of ectopic miR-308. We used an anti-cleaved capase-3 antibody to measure apoptotic activities in the embryos. Anti-cleaved human caspase-3 recognizes the initiator caspase Dronc in Drosophila (Fan and Bergmann, 2010). Dronc is the ortholog of the human initiator caspase-9. After receiving upstream apoptotic signals, Dronc undergoes autocatalytic cleavage to become fully processed and activated. Processing of Dronc in the presence of Dark, the homolog of mammalian Apaf-1, results in the production of three major fragments: a large fragment, small fragment and pro-domain (Yan et al., 2006; Muro et al., 2004). Our western blot analysis shows that an increase in dMyc levels in Drosophila embryos causes an increase in the total level of the Dronc protein and its processed fragments. Consistent with our survival assay, dMyc-triggered apoptosis is balanced by additional ectopic expression of miR-308.
Overall, these results show a role for miR-308 to secure the balanced accumulation of dMyc during development. miR-308's response to elevated dMyc levels shows that this regulation is not passive and is precisely correlated to dMyc's levels.
Functional relationship between dMyc targets and miR-308 targets
In an effort to understand the broader significance of the feedback regulation between dMyc and miR-308, we examined the degree to which dMyc's transcriptional regulation of downstream targets may be antagonized by miR-308. We analyzed the targets of dMyc in embryogenesis using whole genome expression profiling. By crossing flies expressing Gal4 to flies carrying UAS-dmyc, we obtained embryos with elevated levels of Myc. Total RNA from this cross and the control wild-type embryos was obtained and analyzed using Affymetrix GeneChip Drosophila Genome 2.0 Array. Differentially expressed transcripts that were expressed in both groups were selected (P<0.05, FDR = 17%) for downstream functional analysis. Out of 624 affected genes, 607 transcripts were up-regulated and 17 transcripts were down-regulated. We considered the function of up-regulated targets of dMyc. We used a MGSA Byesian network model (Bauer et al., 2010) in Ontologizer 2.0 software to categorize those predicted targets according to their annotation of biological process (Bauer et al., 2008). This model categorizes a set of genes based on their functional annotation and assigns each category a score that corresponds with the significance of enrichment. A score closer to 1.0 shows a stronger support for the enrichment. Results show that dMyc's targets in embryogenesis are mainly involved in ribosome biogenesis, t-RNA metabolism and RNA modification (Cutoff = 0.5), consistent with previous reports (Grewal et al., 2005) (Fig. 7A).
Fig. 7. dMyc up-regulation targets have overlaps with predicted targets of miR-308.
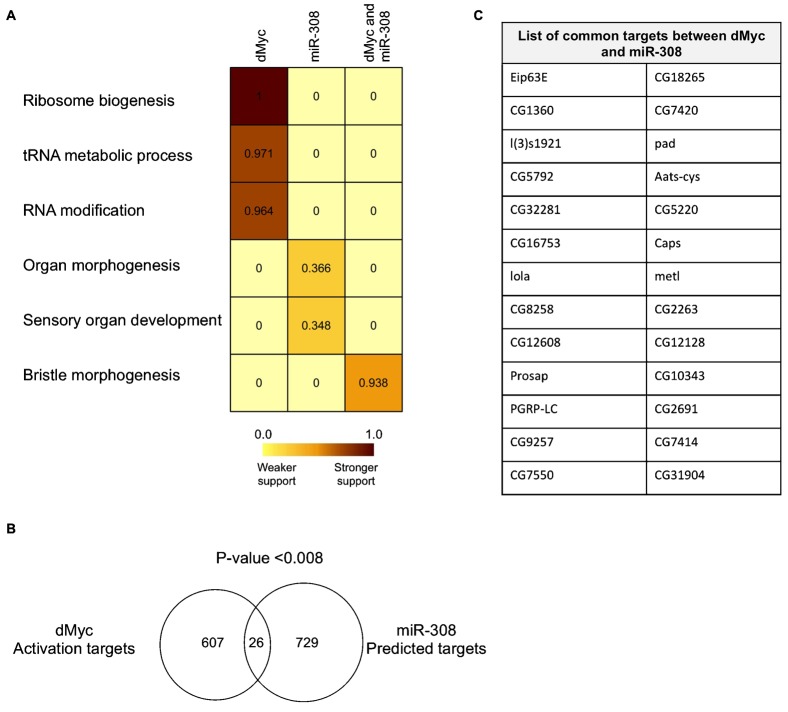
(A) Heat map of MGSA functional analysis for dMyc up-regulation targets, miR-308 predicted targets and their common targets. The MGSA model categorizes a set of genes based on their functional annotation and assigns each category a score (between zero and one) that corresponds with the significance of enrichment. A higher score means a stronger support for that enrichment. A cutoff score of 0.5 is considered to call a functional category significant, i.e. a level at which a gene set is estimated to be more likely to be related to certain functional category. In the heat map, a dark brown color corresponds with a higher score and shows stronger support. (B) Overlap of dMyc targets and miR-308 predicted targets. P-value shows the statistical analysis of significance of the overlap using hypergeometrical distribution. (C) List of common genes between dMyc over-expression targets and miR-308 predicted repression targets.
Given our observed inverse relationship between miR-308 and dMyc, we then determined whether predicted targets of miR-308 are functionally related to targets of dMyc in the context of its over-expression. We identified 729 conserved targets of miR-308 using microRNA.org tools (Enright et al., 2003), and classified these predicted targets using the method described above. Results show that miR-308 predicted targets are mainly involved in organ morphogenesis and development (Fig. 7A). However, the score of this enrichment was below the cutoff of 0.5, which probably reflects the difficulty in determining enrichment of a functional category in a large and broad list of target genes. This potential role of miR-308 in repression of sensory differentiation and morphogenesis is consistent with dMyc function in promoting a growth and proliferation program. Our analysis suggests that dMyc and miR-308 may have an overlapping role in coordinating the complex series of events that balance growth and differentiation during development.
Between the identified 624 dMyc up-regulated targets and 730 predicted targets of miR-308, we identified 26 common targets (Fig. 7B,C). This overlap was significantly different from any overlap that could occur randomly by this analysis (P-value<0.008, hypergeometric distribution) (Fig. 7B). Functional analysis of miR-308 and dMyc's common targets showed a significant enrichment in bristle morphogenesis (Fig. 7B). The observation that miR-308 may decrease expression of these genes while dMyc activates them suggests a specific role for miR-308 in fine-tuning dMyc's function as an inhibitor of morphogenesis.
Discussion
Transcription factors and microRNAs are both trans-acting regulators of gene expression, which can alter the expression of their many target genes at different levels, the former at the transcriptional level and the latter at the post-transactional level (Hobert, 2004). While transcription factors and microRNA can regulate their targets independently of each other, new findings suggest that they can cooperate in regulating each other and their target genes. In humans, transcription factors and microRNA can form various regulation motifs such as feedback and feed-forward loops that ultimately contribute to the gene regulation programs (Chen et al., 2011). In Caenorhabditis elegans, microRNAs are shown to participate in feedback circuits with key transcription factors, and these networks have a high capacity for the regulation of target genes (Martinez et al., 2008).
Here, we report the novel finding of a feedback circuit between dMyc, a key transcription factor in animal biology, and miR-308, a member of the conserved Drosophila miR-2 family. Our results suggest three models for dMyc and miR-308 interaction (Fig. 8). In the first model, supported by our loss-of-function and gain-of-function studies, we showed that dMyc and miR-308 are in cross-talk with each other and that miR-308 responds to dMyc levels and regulates dMyc's protein levels (Fig. 8, top panel).
Fig. 8.
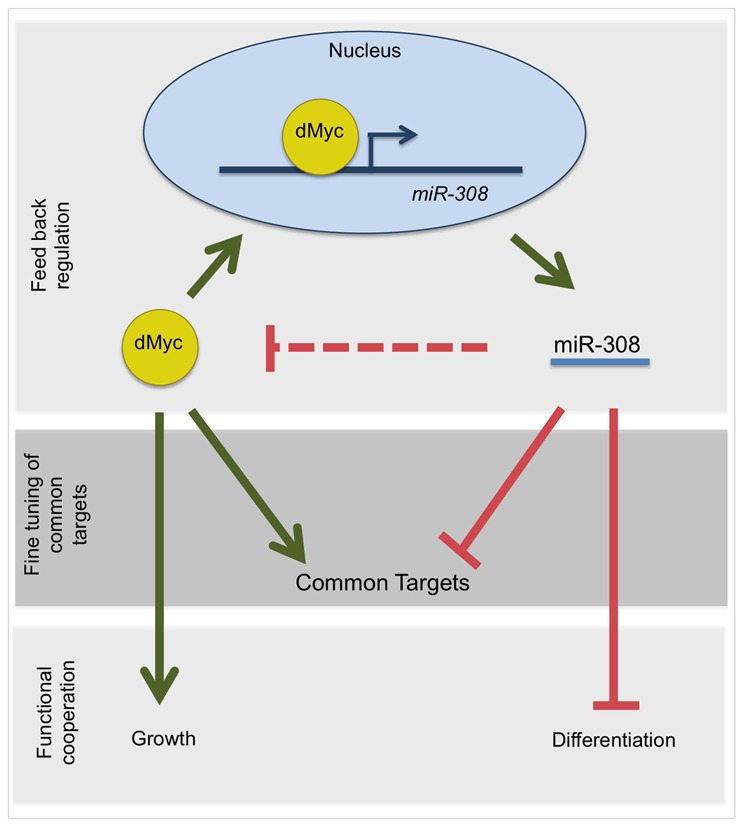
Three proposed models for the cross-talk of dMyc and miR-308 and its role in feedback regulation (top panel), target regulation (middle panel) and growth and differentiation programming (bottom panel).
In the second model, we identified common targets between dMyc and miR-308. This model shows a possible role for miR-308 in specific fine-tuning of regulation by dMyc (Fig. 8, middle panel). However, further experiments will be needed to confirm these common targets. It would be interesting to determine the extent to which levels of these common targets respond to different proportions of dMyc and miR-308. In the third model, we suggest a functional relationship between dMyc and miR-308 in the determination of a cell growth versus a differentiation program (Fig. 8, bottom panel). In this model, we suggest collaboration between dMyc and miR-308 in programming cells into rapid proliferation and the cessation of morphogenesis. Further experiments should be carried out to delineate the extent to which these functional programs are regulated by dMyc, miR-308 and the balance between the two.
All together, these results reveal a crucial role for miR-308 in feedback regulation of dMyc, and fine-tuning of target regulation by dMyc during Drosophila development.
Acknowledgments
We are thankful to Dr Cynthia Gibas and Dr Jennifer Weller of the UNC Charlotte Department of Bioinformatics and Genomics for training us and granting us access to the microarray core facility. We thank Dr Anthony Fodor and Dr Robert Reid for helping us with bioinformatics and microarray data analysis. We thank Mona Kashiha for helping us with graphical presentation of data. This work has been funded by the NIH (NCI, http://www.cancer.gov) to J.M.G., under grant number 1R15CA135481-01, and also by an NSF ADVANCE grant to UNC Charlotte.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Aravin A. A., Lagos–Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. (2003). The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350 10.1016/S1534-5807(03)00228-4 [DOI] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008). The impact of microRNAs on protein output. Nature 455, 64–71 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S., Bartel D. P. (2005). Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11, 241–247 10.1261/rna.7240905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S., Grossmann S., Vingron M., Robinson P. N. (2008). Ontologizer 2.0—a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24, 1650–1651 10.1093/bioinformatics/btn250 [DOI] [PubMed] [Google Scholar]
- Bauer S., Gagneur J., Robinson P. N. (2010). GOing Bayesian: model-based gene set analysis of genome-scale data. Nucleic Acids Res. 38, 3523–3532 10.1093/nar/gkq045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Cuppen E., Plasterk R. H. (2006). Approaches to microRNA discovery. Nat. Genet. 38 Suppl, S2–S7 10.1038/ng1794 [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Huang J., Ma A., Kretzner L., Alt F. W., Eisenman R. N., Weintraub H. (1993). Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 13, 5216–5224 10.1128/MCB.13.9.5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C., Staller P., Eilers M. (1998). Control of cell proliferation by Myc. Trends Cell Biol. 8, 202–206 10.1016/S0962-8924(98)01251-3 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Chang T.–C., Yu D., Lee Y.–S., Wentzel E. A., Arking D. E., West K. M., Dang C. V., Thomas–Tikhonenko A., Mendell J. T. (2008). Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40, 43–50 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.–Y., Chen S.–T., Fuh C.–S., Juan H.–F., Huang H.–C. (2011). Coregulation of transcription factors and microRNAs in human transcriptional regulatory network. BMC Bioinformatics 12 Suppl 1, S41 10.1186/1471-2105-12-S1-S41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D., Cowling V. H. (2008). Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat. Rev. Mol. Cell Biol. 9, 810–815 10.1038/nrm2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller H. A., Grandori C., Tamayo P., Colbert T., Lander E. S., Eisenman R. N., Golub T. R. (2000). Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97, 3260–3265 10.1073/pnas.97.7.3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K., Khan A., Goodliffe J. M. (2011). Myc localizes to histone locus bodies during replication in Drosophila. PLoS ONE 6, e23928 10.1371/journal.pone.0023928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Eisenman R. N. (2008). Myc's broad reach. Genes Dev. 22, 2755–2766 10.1101/gad.1712408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., John B., Gaul U., Tuschl T., Sander C., Marks D. S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5, R1 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Bergmann A. (2010). The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 17, 534–539 10.1038/cdd.2009.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J. A., Giniger E., Maniatis T., Ptashne M. (1988). GAL4 activates transcription in Drosophila. Nature 332, 853–856 10.1038/332853a0 [DOI] [PubMed] [Google Scholar]
- Gallant P. (2009). Drosophila Myc. Adv. Cancer Res. 103, 111–144 10.1016/S0065-230X(09)03005-X [DOI] [PubMed] [Google Scholar]
- Goodliffe J. M., Wieschaus E., Cole M. D. (2005). Polycomb mediates Myc autorepression and its transcriptional control of many loci in Drosophila. Genes Dev. 19, 2941–2946 10.1101/gad.1352305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodliffe J. M., Cole M. D., Wieschaus E. (2007). Coordinated regulation of Myc trans-activation targets by Polycomb and the Trithorax group protein Ash1. BMC Mol. Biol. 8, 40 10.1186/1471-2199-8-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory M. A., Hann S. R. (2000). c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol. Cell. Biol. 20, 2423–2435 10.1128/MCB.20.7.2423-2435.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S. S., Li L., Orian A., Eisenman R. N., Edgar B. A. (2005). Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7, 295–302 10.1038/ncb1223 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Hobert O. (2004). Common logic of transcription factor and microRNA action. Trends Biochem. Sci. 29, 462–468 10.1016/j.tibs.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., Gallant P. (1999). Drosophila myc regulates cellular growth during development. Cell 98, 779–790 10.1016/S0092-8674(00)81512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J. R., Carthew R. W. (1998). Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95, 1017–1026 10.1016/S0092-8674(00)81725-0 [DOI] [PubMed] [Google Scholar]
- Khan A., Shover W., Goodliffe J. M. (2009). Su(z)2 antagonizes auto-repression of Myc in Drosophila, increasing Myc levels and subsequent trans-activation. PLoS ONE 4, e5076 10.1371/journal.pone.0005076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress T. R., Cannell I. G., Brenkman A. B., Samans B., Gaestel M., Roepman P., Burgering B. M., Bushell M., Rosenwald A., Eilers M. (2011). The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol. Cell 41, 445–457 10.1016/j.molcel.2011.01.023 [DOI] [PubMed] [Google Scholar]
- Liao J.–M., Lu H. (2011). Autoregulatory suppression of c-Myc by miR-185-3p. J. Biol. Chem. 286, 33901–33909 10.1074/jbc.M111.262030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.–H., Jackson A. L., Guo J., Linsley P. S., Eisenman R. N. (2009). Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 28, 3157–3170 10.1038/emboj.2009.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines J. Z., Stevens L. M., Tong X., Stein D. (2004). Drosophila dMyc is required for ovary cell growth and endoreplication. Development 131, 775–786 10.1242/dev.00932 [DOI] [PubMed] [Google Scholar]
- Martinez N. J., Ow M. C., Barrasa M. I., Hammell M., Sequerra R., Doucette–Stamm L., Roth F. P., Ambros V. R., Walhout A. J. M. (2008). A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 22, 2535–2549 10.1101/gad.1678608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N., Penn L. Z. (2008). Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- Montero L., Müller N., Gallant P. (2008). Induction of apoptosis by Drosophila Myc. Genesis 46, 104–111 10.1002/dvg.20373 [DOI] [PubMed] [Google Scholar]
- Morrish F., Neretti N., Sedivy J. M., Hockenbery D. M. (2008). The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 7, 1054–1066 10.4161/cc.7.8.5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I., Monser K., Clem R. J. (2004). Mechanism of Dronc activation in Drosophila cells. J. Cell Sci. 117, 5035–5041 10.1242/jcs.01376 [DOI] [PubMed] [Google Scholar]
- O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H. J., Li L., Sawado T., Williams E., Loo L. W. M., Cowley S. M., Yost C.et al. (2003). Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17, 1101–1114 10.1101/gad.1066903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U. A., Kauppinen S., Lund A. H. (2006). LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 372, 137–141 10.1016/j.gene.2005.12.031 [DOI] [PubMed] [Google Scholar]
- Pelengaris S., Khan M., Evan G. I. (2002). Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109, 321–334 10.1016/S0092-8674(02)00738-9 [DOI] [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Land H. (1990). Negative autoregulation of c-myc transcription. EMBO J. 9, 1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. B., Yost C., Britton J. S., Loo L. W. M., Flynn E. M., Edgar B. A., Eisenman R. N. (2004). dMyc is required for larval growth and endoreplication in Drosophila. Development 131, 2317–2327 10.1242/dev.01108 [DOI] [PubMed] [Google Scholar]
- Pierce S. B., Yost C., Anderson S. A., Flynn E. M., Delrow J., Eisenman R. N. (2008). Drosophila growth and development in the absence of dMyc and dMnt. Dev. Biol. 315, 303–316 10.1016/j.ydbio.2007.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J. P. (2006). GenePattern 2.0. Nat. Genet. 38, 500–501 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., Lai E. C. (2007). Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17, 1850–1864 10.1101/gr.6597907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti S. E., Kim S. Y., Tansey W. P. (1999). Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18, 717–726 10.1093/emboj/18.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. V. (1999). The role of c-myc in cellular growth control. Oncogene 18, 2988–2996 10.1038/sj.onc.1202751 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C. (1993). Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72, 443–457 10.1016/0092-8674(93)90120-F [DOI] [PubMed] [Google Scholar]
- Truscott M., Islam A. B. M. M. K., López–Bigas N., Frolov M. V. (2011). mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes Dev. 25, 1820–1834 10.1101/gad.16947411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lin S., Li J. J., Xu Z., Yao H., Zhu X., Xie D., Shen Z., Sze J., Li K.et al. (2011). MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1∼let-7d via noncanonical E-box. J. Biol. Chem. 286, 39703–39714 10.1074/jbc.M111.293126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Du Q., Liang Z. (2010). Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene 29, 4980–4988 10.1038/onc.2010.241 [DOI] [PubMed] [Google Scholar]
- Yan N., Huh J. R., Schirf V., Demeler B., Hay B. A., Shi Y. (2006). Structure and activation mechanism of the Drosophila initiator caspase Dronc. J. Biol. Chem. 281, 8667–8674 10.1074/jbc.M513232200 [DOI] [PubMed] [Google Scholar]


