Abstract
Spatial and temporal control of cell growth is central for the morphogenesis of multicellular organisms. For some cell types that undergo extensive post-mitotic cell growth, such as neurons and hair cells, orchestrating the extent of post-mitotic cell growth with development is vital for their physiology and function. Previous studies suggested that the extent of cell growth is linked with an increase in ploidy by endoreduplication but how developmental signals control endocycling and cell growth is not understood in both animals and plants. In this study we show that a trihelix transcription factor, GT2-LIKE 1 (GTL1), actively terminates ploidy-dependent cell growth and its developmentally regulated expression is one of the key determinants of cell size in Arabidopsis leaf hair cells (trichomes). Through genome-wide chromatin-binding studies (ChIP-chip) coupled with transcriptional profiling, we further demonstrate that GTL1 directly represses the transcription of CDH1/FZR/CCS52, an activator of the anaphase-promoting complex/cyclosome (APC/C), to stop the endocycle progression and ploidy-dependent cell growth. Thus, our findings uncover a previously uncharacterised key molecular link between developmental programming and cell-size control, highlighting the central role of APC/C in post-mitotic cell growth.
Keywords: APC/C, cell growth, endocycle, trichome, trihelix transcription factor
Introduction
Organ morphogenesis in multicellular organisms relies on the coordinated progression of cell proliferation and cell growth. Compared with animals, plants often undergo far more extensive post-mitotic cell growth, sometimes up to 1000-fold of original size, and an increase in cell size generally contributes to a larger extent to organ growth. It is also known that some cell types, such as neurons and various hair cells in insects, animals and plants, undergo massive cell growth during differentiation and this is vital for their specialised physiology and function (Sugimoto-Shirasu and Roberts, 2003). Since the final size of cells is often fairly constant under given conditions, the duration of post-mitotic cell growth is likely to be developmentally programmed. We still know surprisingly little about how cell size is determined and how developmental signals link to this control. Most of loss-of-function mutants with developmental defects display reduced cell-size phenotypes, pointing to the existence of robust mechanisms to promote cell growth (Breuer et al, 2010). Several mutations, on the contrary, cause larger cells (Perazza et al, 1999; Horiguchi et al, 2005; Imai et al, 2006; Lasorella et al, 2006; Breuer et al, 2009) but whether there are some mechanisms to actively restrain or terminate cell growth has not been extensively studied.
In various eukaryotes, cell size often positively correlates with the nuclear DNA content or ploidy (Nagl, 1976; Melaragno et al, 1993). This correlation was already proposed early last century, referred to as the ‘nuclear–cytoplasmic ratio’ theory (Wilson, 1925), but there have been also reports in which the ploidy level does not match with cell size (Beemster et al, 2002; Schnittger et al, 2003). The situation is also more complex in plants where the vacuoles constitute most of cell volume and the cytoplasm accounts very little for overall cell size (Sugimoto-Shirasu and Roberts, 2003). Although it is generally accepted that some proportion of cell growth is ploidy-dependent, how far this contributes to total growth and importantly, how cells grow in a ploidy-dependent and/or -independent manner still remains elusive.
One strategy to increase the ploidy level is to initiate an aberrant version of the mitotic cell cycle called endoreduplication cycle or endocycle. Endocycling cells skip the mitotic phase, thus they re-enter into the S phase without cytokinesis, resulting in the duplication of the nuclear DNA content (Nagl, 1976; Edgar and Orr-Weaver, 2001). Endoreduplication is widespread in plants and many mitotically dividing cells switch into the endocycle as they start to differentiate. Endocycling also occurs in specialised cell types of insects and mammals, potentially accounting for up to half of the biomass in the world (Zielke et al, 2011). Studies in Arabidopsis and Drosophila have revealed that some of the regulatory mechanisms governing the onset and progression of endocycles are shared between plants and animals, and an important step to promote the endocycle is to inactivate the mitotic cyclin (CYC)–cyclin-dependent kinase (CDK) complexes (Lee et al, 2009; Breuer et al, 2010; Komaki and Sugimoto, 2012). In Arabidopsis, for example, CDKB1;1, in complex with CYCA2;3, represses the mitotic-to-endocycle transition and its down-regulation results in an early endocycle onset (Boudolf et al, 2004, 2009). The abundance of active CYC–CDK complexes is controlled by several regulatory mechanisms, but one key event tightly connected to the endocycle is the proteolytic degradation of mitotic CYCs by the 26S proteasome (Hershko, 1997; Peters, 2002; De Veylder et al, 2011; Komaki and Sugimoto, 2012). The anaphase-promoting complex/cyclosome (APC/C) is the multisubunit E3 ubiquitin ligase complex that plays central roles in this control and its activation strongly depends on the presence of evolutionary conserved proteins, such as CELL DIVISION CYCLE 20 (CDC20)/Fizzy and CDC20 HOMOLOG 1 (CDH1)/CELL CYCLE SWITCH 52 (CCS52)/Fizzy-related (FZR) (Sigrist and Lehner, 1997; Visintin et al, 1997; Cebolla et al, 1999). The CDC20 and CDH1 activators interact with mitotic CYCs and target them to the core complex of the APC/C to facilitate their ubiquitination for prompt degradation. The Arabidopsis genome encodes five CDC20 (CDC20.1–CDC20.5), and 3 CDH1 (CCS52A1, CCS52A2 and CCS52B) homologues. Previous loss- and gain-of-function studies have uncovered strong association of CCS52A1 and CCS52A2 with the control of endocycle onset and progression (Lammens et al, 2008; Larson-Rabin et al, 2009; Kasili et al, 2010; Mathieu-Rivet et al, 2010). The function of CCS52B and the CDC20 homologues is less characterised but recent studies suggest that CCS52B also participates in this control (Iwata et al, 2011; de Almeida Engler et al, 2012).
With the tight connection between endocycling and cell growth or cell differentiation, its onset and progression are likely to be developmentally regulated. Accumulating evidence suggests that transcriptional regulation of the CDH1/CCS52/FZR activators is one common mechanism mediating this control in various organisms. For instance, endocycle establishment and progression during follicle cell development in the Drosophila ovary is controlled through the Notch signalling pathway (Deng et al, 2001). Several transcriptional regulators have been identified to modulate the fzr expression although whether they directly target the fzr locus is not known (Sun and Deng, 2005, 2007). The Notch pathway is not conserved in plants (Wigge and Weigel, 2001) but in contrast to flies, both CCS52A1 and CCS52A2 are under the direct transcriptional control of the E2FA-RETINOBLASTOMA-RELATED (RBR) complex and an atypical E2F transcription factor DEL1 (Lammens et al, 2008; Magyar et al, 2012). The E2FA–RBR complex transcriptionally represses expression of both CCS52A1 and CCS52A2, whereas DEL1 represses only CCS52A2 in mitotically dividing cells to suppress early developmental transition from the mitotic cycle into the endocycle.
Arabidopsis trichomes are unicellular and they serve as an excellent model system to study post-mitotic cell growth since they increase their cell volume by >100-fold in a developmentally specific window of time. Trichome cells also undergo several rounds of endocycle to increase their ploidy level on average up to 32C. Earlier molecular genetic studies have uncovered many regulators that participate in trichome differentiation and have shown that trichome cell growth is promoted by complex transcriptional networks (Ishida et al, 2008; Morohashi and Grotewold, 2009). Previously, we found that a mutation in GTL1, a GT-2 family trihelix transcription factor, results in extended endocycles and cell growth (Breuer et al, 2009) but the molecular mechanism underlying this control remains unknown. Here, we show that overexpression of GTL1 is sufficient to arrest the endocycle and cell growth in both trichome and other leaf epidermal cells. Furthermore, a combination of ChIP-chip and trichome-specific gene expression analysis revealed that GTL1 directly binds to the promoter of CCS52A1 and represses its expression to terminate the endocycle. We also provide genetic evidence that trichome cells grow through both ploidy-dependent and -independent pathway and that transcriptional repression of CCS52A1 by GTL1 accounts only for ploidy-dependent cell growth. Our data thus highlight that terminating cell growth is an active process and that plants accomplish this by programming the developmental timing to down-regulate the APCCDH1/FZR/CCS52 activity.
Results
GTL1 is sufficient to terminate ploidy-dependent cell growth
GTL1 is expressed only at late stages of trichome development and is required to terminate its cell growth (Breuer et al, 2009). To investigate whether the expression of GTL1 is sufficient to block cell growth, we fused GTL1 to the green fluorescence protein (GFP) and expressed the GFP–GTL1 protein at an early stage of trichome differentiation using the GLABRA2 (GL2) promoter (Szymanski et al, 1998; Schnittger et al, 2002). These GFP–GTL1 proteins are functional and complement the gtl1-1 mutant phenotypes when expressed under its own promoter (Breuer et al, 2009). We confirmed the expression of GFP–GTL1 proteins in developing trichomes of pGL2:GFP:GTL1 plants (Supplementary Figure S1A). Our scanning electron microscopy revealed that fully mature trichomes from 17-day-old wild-type (WT) plants have characteristic size and morphology with three branches (Figure 1A and B). In contrast, the pGL2:GFP:GTL1 plants display severe defects in trichome development, ranging from an early arrest to complete loss of cellular outgrowth and branching (Figure 1A and B).
Figure 1.
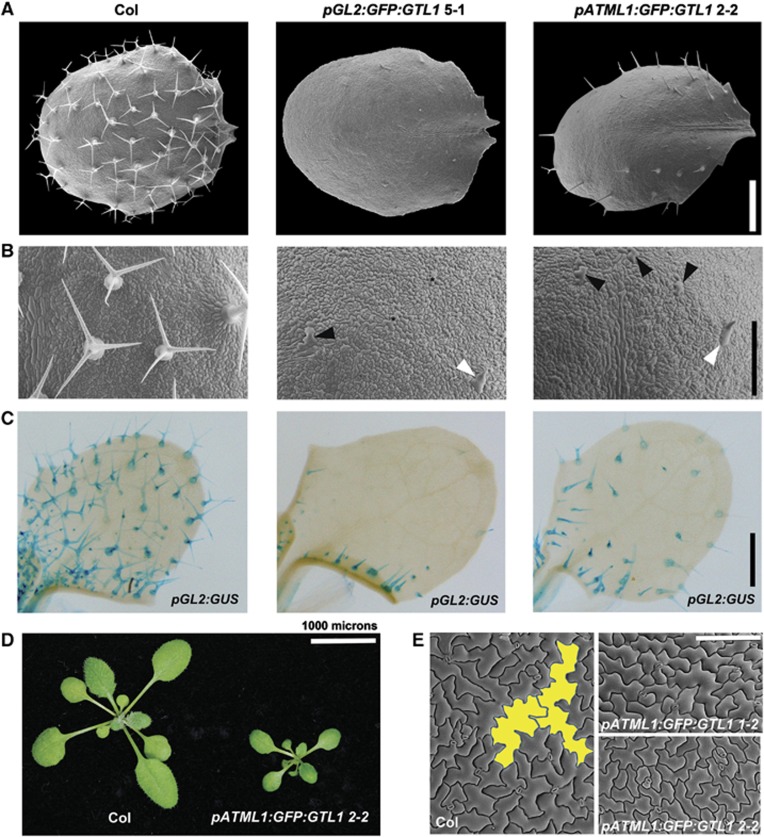
Overexpression of GTL1 dramatically represses cell growth and cell differentiation. (A, B) Scanning electron micrographs of the fifth true leaves of 17-day-old WT (Col), pGL2:GFP:GTL1 and pATML1:GFP:GTL1 plants (A) and corresponding close-up views of their adaxial epidermis (B). White arrows mark out-grown, unbranched trichomes. Black arrows point to abnormally enlarged and pointed epidermal cells that may possess trichome identities. Another type of abnormally enlarged cells, marked with black asterisks, exhibit cellular morphology characteristic to epidermal pavement cells. (C) Expression of the trichome identity marker pGL2:GUS in the third true leaves of 10-day-old Col, pGL2:GFP:GTL1 and pATML1:GFP:GTL1 plants. (D, E) A light micrograph of 21-day-old Col and pATML1:GFP:GTL1 plants (D) and scanning electron micrographs of their adaxial epidermis on fully expanded first true leaves (E). Two large pavement cells are highlighted in yellow in Col leaves. Epidermal cells of similar size are not found in pATML1:GFP:GTL1 leaves. Scale bars: 300 μm in (A), 250 μm in (B), 500 μm in (C), 1 cm in (D), and 150 μm in (E).
Previous studies have shown that GTL1 is also expressed in non-trichome cells (Breuer et al, 2009; Yoo et al, 2010). To examine whether GTL1 regulates cell growth in a more general context, we expressed GFP–GTL1 proteins using the ATML1 promoter that drives gene expression in cells at the outermost layer of shoot apical meristem and leaf epidermis (Sessions et al, 1999). The GFP–GTL1 proteins are expressed in both trichomes and other epidermal cells in pATML1:GFP:GTL1 plants (Supplementary Figure S1A) and as expected, they repress cell growth in trichomes (Figure 1A and B). We occasionally saw some outgrowth of trichomes at the leaf margin but they hardly underwent branching and full growth. In addition, we found that epidermal pavement cells in pATML1:GFP:GTL1 plants are small compared with WT cells that have undergone extensive cell growth (Figure 1E; Supplementary Figure S1E). Consequently, the final size of pATML1:GFP:GTL1 leaves is also strongly reduced (Figure 1D; Supplementary Figure S1D) although overall number of adaxial epidermal cells is similar to that of WT (Supplementary Figure S1E and F).
We have previously shown that gtl1 mutant trichomes undergo on average an additional round of endocycle, resulting in an increase of trichome ploidy (Breuer et al, 2009). To test whether overexpression of GTL1 also interferes with the progression of endocycles, we first quantified the nuclear DNA content of individual trichomes isolated from 25-day-old WT and pATML1:GFP:GTL1 plants. Compared with the average nuclear DNA content of WT trichomes, the average DNA content of pATML1:GFP:GTL1 trichomes is strongly reduced (Supplementary Figure S1B), suggesting that excess levels of GTL1 delays the endocycle progression in trichomes. Because of the strong repression of trichome differentiation in pGL2:GFP:GTL1 plants, we could not isolate trichomes for the quantification of their ploidy. Furthermore, flow cytometric analysis using the first pair of leaves from 21-day-old pATML1:GFP:GTL1 plants shows an increase in 8C nuclei and reduction in 16C and 32C nuclei (Supplementary Figure S1C). These results establish that GTL1 is not only necessary but is also sufficient to repress ploidy-dependent cell growth and that temporally regulated transcription of GTL1 is one of the key determinants of final cell size in Arabidopsis trichomes.
The GTL1 expression interferes with trichome differentiation
We noticed that some of the leaf epidermal cells in the pGL2:GFP:GTL1 plants are abnormally enlarged, yet they retain a relatively normal morphology similar to other epidermal pavement cells (Figure 1B). To examine whether these cells possess trichome cell fate, we introduced a trichome identity marker, pGL2:GUS, into pGL2:GFP:GTL1 plants. As shown in Figure 1C, early expression of GTL1–GFP proteins by the GL2 promoter leads to a profound reduction of GUS-positive cells in pGL2:GFP:GTL1 plants. Most of out-grown and unbranched cells are GUS positive while those that are abnormally enlarged but not out-grown are GUS negative although they must have had the GL2 promoter active to express GTL1 earlier in development. The impact of GTL1 expression under the ATML1 promoter is less severe probably because of its weaker promoter activity during trichome development but compared with WT leaves, we also detected less pGL2:GUS positive cells in pATML1:GFP:GTL1 leaves (Figure 1C). These results suggest that overexpression of GTL1 also interferes with full acquisition and/or maintenance of trichome cell fate.
Genome-wide identification of chromatin regions bound by GTL1
To understand how GTL1 regulates cell growth, we first identified its potential direct targets by the chromatin immunoprecipitation followed by the hybridisation on an Affymetrix Arabidopsis Tiling 1.0R array (ChIP-chip). To enrich the genomic region bound by GTL1 in vivo, we harvested whole aerial parts of 12-day-old gtl1-1 plants complemented with the pGTL:GTL1:GFP constructs (Breuer et al, 2009) and immunoprecipitated the chromatin fragments associated with GTL1–GFP proteins using antibodies against GFP. After applying a cutoff P-values of 0.001, we identified a total number of about 3900 putative immediate target genes that showed consistent binding by GTL1 (Supplementary data S1). To assess the false discovery rate of these ChIP-chip experiments, we randomly chose 17 putative targets and subjected to validation by ChIP-qPCR. Out of 17 genes, 13 showed robust and reproducible binding of GTL1 within the predicted binding region (Supplementary Figure S2), suggesting a false discovery rate of <0.25. This is similar to previous studies that reported false discovery rates of 0.2 and 0.3 using the same experimental design (Morohashi and Grotewold, 2009). By examining the GTL1 binding within the 3-kb chromatin region of annotated transcriptional start sites (TSSs), we found that GTL1 preferentially interacts with the target promoter region within the first 0.5 kb upstream of TSSs (Figure 2A). Further gene structure analysis also supported that the binding of GTL1 is statistically enriched for the upstream region of predicted genes compared to 5′UTR, exons, introns and 3′UTR (P<10−5; χ2 test) (Figure 2B). GT transcription factors generally bind to GT elements (GT1 box, 5′-GGTTAA-3′; GT2 box, 5′-GGTAAT-3′; GT3 box, 5′-GGTAAA-3′) (Ni et al, 1996) and GTL1 was recently shown to bind to a GT3 box in the promoter of STOMATA DENSITY AND DISTRIBUTION 1 (SDD1) (Yoo et al, 2010). Using a promoter element analysis tool MEME (Bailey et al, 2009), we found that a highly conserved GT3 box and an inverted partial GT3 box (5′-TTTAC-3′) are over-represented among the sequences identified by ChIP-chip (Figure 2C). Together, these results strongly suggest that the ChIP-chip experiments have been successful to identify potential targets of GTL1.
Figure 2.
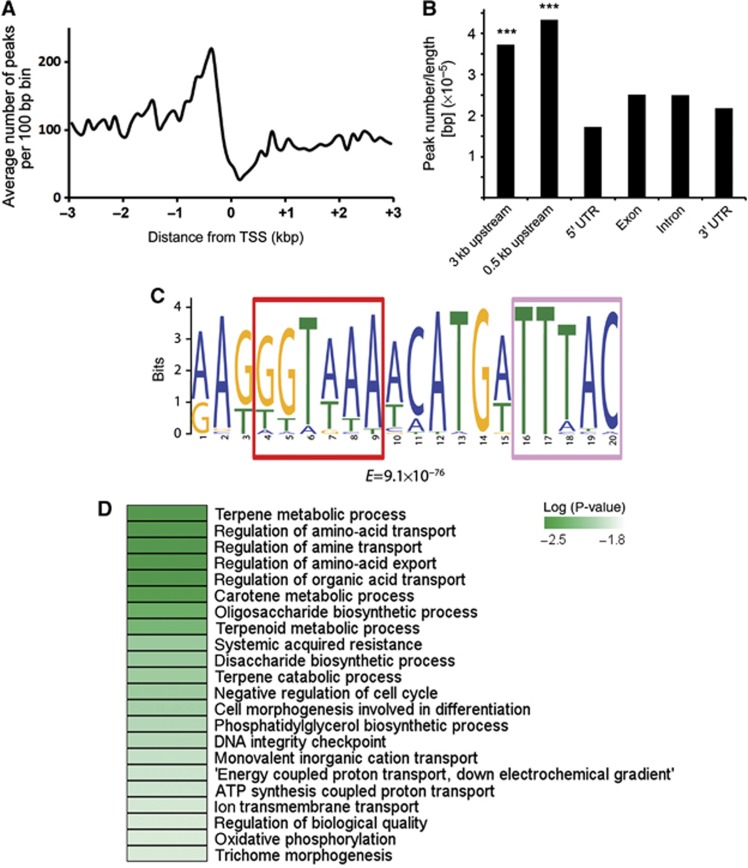
Genome-wide identification of chromatin regions bound by GTL1. (A) Distribution of GTL1 binding within the ±3 kb region of the TSS. (B) Peak distribution of GTL1 binding to the structural elements of genes. Peak centres of individual GTL1-binding sites were used to score the distribution. The peak number was normalised by the length of the entire genome for the category of gene structure. ***P<10−5. The P-value was calculated based on the empirical null distribution with the numbers of 1000 permutations of randomly chosen from GTL1-binding peaks. (C) Putative GTL1 targets share a common DNA-binding motif that contains a GT3-box (5′-GGTAAA-3′) marked with red frame and a palindromic partial GT3 element (5′-TTTAC-3′) marked with light red frame. (D) Representation of the most enriched gene ontology (GO) categories among the putative GTL1 targets identified by ChIP-chip.
To investigate the biological function of these putative GTL1 target genes, we used the BiNGO analysis tool (Maere et al, 2005) and identified significantly enriched Gene Ontology (GO) categories. Among the enriched GO categories, we found ‘negative regulation of cell cycle’ (P<0.01) and ‘DNA integrity checkpoint’ are over-represented (P<0.01) (Figure 2D), suggesting that GTL1 may directly affect transcription of cell-cycle-related genes. Consistent with the role of GTL1 in trichome development, the enriched GO categories also included ‘cell morphogenesis involved in differentiation’ (P<0.01) and ‘trichome morphogenesis’ (P<0.1). In addition, we noticed that ‘terpene metabolic process’ (P<0.01), ‘regulation of amino-acid transport’ (P<0.01), ‘oligosaccharide biosynthetic process’ (P<0.01) are highly ranked, implying that GTL1 may target diverse biological processes linked to plant development.
Genome-wide identification of GTL1 response genes
Previous studies suggested that among putative direct target genes identified by ChIP-chip, only <10% respond transcriptionally to the misregulation of bound transcription factors and they include functional relevant targets (Busch et al, 2010). We therefore identified GTL1-responsive genes by microarray analysis. The gtl1-1 mutants exhibit the extended cell growth phenotype at a late stage of trichome development (Figure 3A), thus we reasoned that changes in the expression of direct GTL1 target genes should be most prominent in trichome cells at the same developmental stage. To this end, we extracted total RNA from trichomes of fifth and sixth rosette leaves of 3-week-old WT and gtl1-1 mutants (Figure 3B). With the significance threshold of P<0.075 (Hochberg and Benjamini, 1990), we found a total number of 1694 probes, corresponding to 1759 genes on the ATH1 chip that show differential expression of at least 1.3-fold. Out of these 1759 genes, 47.2% are positively regulated and 52.8% are negatively regulated by GTL1 (Figure 3C; Supplementary data S2). To our surprise, the significantly enriched GO category among positively regulated genes included ‘photosynthesis’ (P<10−19), ‘response to red light’ (P<10−7) and ‘response to blue light’ (P<10−6) (Figure 3D). None of these processes has been previously linked to trichomes but it is worth mentioning that GT2, a close homologue of GTL1, was previously shown to bind to the promoter of PHYTOCHROME A (PHYA) gene (Kuhn et al, 1993; Ni et al, 1996). Our ChIP-chip experiments, however, suggest that none of the PHYA genes is directly bound by GTL1 (Supplementary data S1 and S2), implying that these transcriptional changes are indirect. Many of the highly ranked GO categories for genes negatively regulated by GTL1 were related to metabolic processes (Figure 3D), probably reflecting the transcriptional response accompanied with extended cell growth of gtl1 trichomes.
Figure 3.
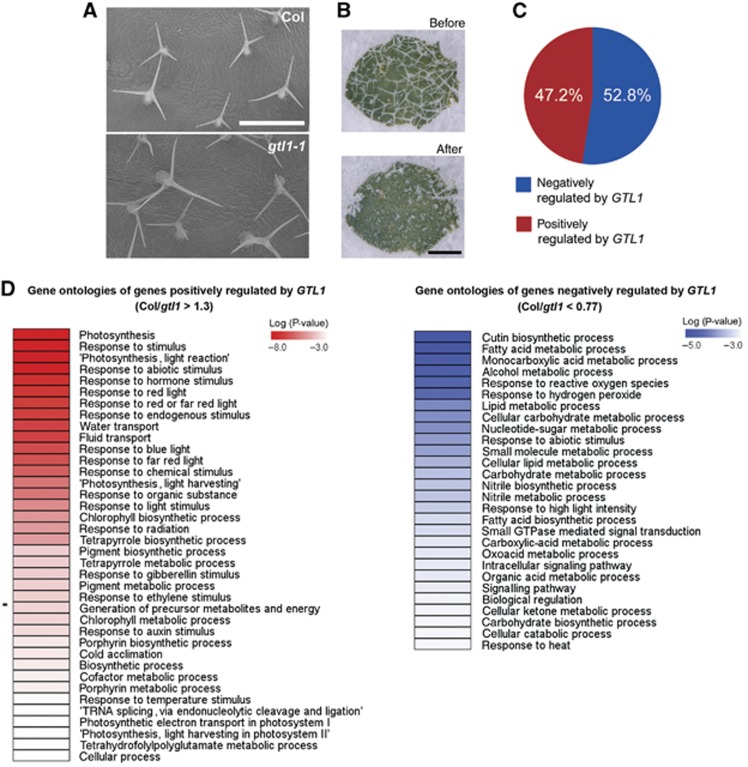
Genome-wide identification of GTL1-responsive genes. (A) Scanning electron micrographs of the adaxial epidermis of Col and gtl1-1 leaves showing enlarged trichomes in gtl1-1. (B) Light micrographs of gtl1-1 leaves before and after trichome removal. (C) A pie-chart showing the percentage of genes positively or negatively regulated by GTl1. (D) GO categories enriched for genes positively or negatively regulated by GTL1. Scale bars: 250 μm in (A), 500 μm in (B).
GTL1 binds to the GT3 box within the CCS52A1 promoter
Among the 3900 genes identified by ChIP-chip, 2398 genes were represented on the ATH1 microarray chip used for the transcriptional analysis. By combining the list of these genes and 1759 GTL1-responsive genes, we narrowed down putative direct targets of GTL1 to 182 genes (Figure 4A) that represent about 7.6% of all putative targets. Considering that we used the whole aerial tissue for the ChIP-chip analysis and trichomes only for the microarray analysis, this is a reasonable overlap between these experiments. Out of these 182 genes, 79 genes are positively regulated by GTL1 while 103 genes are negatively regulated, suggesting that GTL1 may act as both a transcriptional activator and repressor. Given that GTL1 impacts on the endocycle, we first asked whether this gene list includes cell-cycle-related genes and found that two genes, APC10 and an APC/C activator CCS52A1, associate with GTL1 and are down- and up-regulated, respectively, in gtl1-1 (Supplementary data S3). Accumulating evidence suggests that modulation of CCS52A1 levels affects both endoreduplication and cell growth in various cell types (Lammens et al, 2008; Larson-Rabin et al, 2009; Vanstraelen et al, 2009; Kasili et al, 2010). In contrast, APC10 has not been directly linked to the endocycle and importantly, down-regulation of APC10 does not affect trichome development in Arabidopsis (Roodbarkelari et al, 2010), suggesting that CCS52A1 is a more likely functional target of GTL1 during trichome development. Using the Integrated Genome Browser (IGB), we indeed detected distinct peaks of GTL1 binding in the upstream region of CCS52A1, −1,389 and −765 base pairs upstream of the TSS. We also examined the upstream sequence of two other CCS52 homologues, CCS52A2 and CCS52B, but GTL1 seems to interact specifically with the CCS52A1 promoter because we did not identify any significant signals around the loci of CCS52A2 and CCS52B (Figure 4B).
Figure 4.
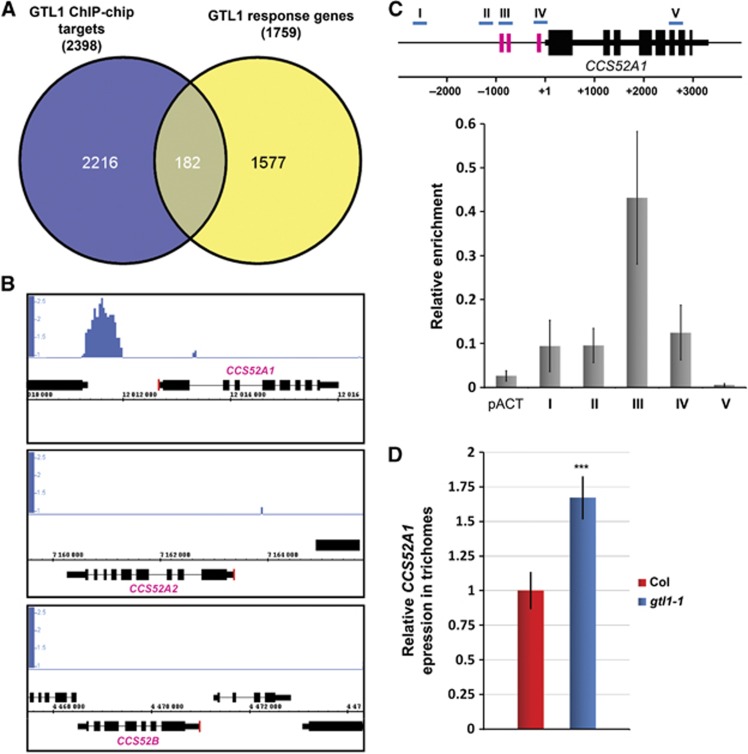
GTL1 directly binds to the promoter of the CCS52A1 gene and represses its transcription. (A) Venn diagram analysis identifies 182 genes as a potential target of GTL1. (B) The IGB analysis of the CCS52A1, CCS52A2 and CCS52B loci shows a clear signal enrichment in the 5′-region upstream of the CCS52A1 open reading frame. Red lines mark the TSS. (C) (Upper panel) Schematic representation of the CCS52A1 locus highlighting the position of GT3 boxes (pink) and primer sets used for ChIP-qPCR (I–V, blue). (Lower panel) ChIP-qPCR analysis reveals the strongest association of GTL1–GFP fusion proteins in the −850 bp region of the 5′UTR. The IP experiments were repeated three times with independent plant samples and the average enrichment of qPCR products from immunoprecipitated DNA is normalised against the corresponding input DNA. pACT shows signal enrichment in the promoter of actin (At3g18780) and error bars represent ±s.d. of the means (n=3). (D) Relative expression of CCS52A1 in trichomes of fifth and sixth leaves from 17-day-old Col and gtl1-1 plants. Errors represent ±s.d. of the means (n=3). Asterisks indicate a significant difference between Col and gtl1-1 (Student’s t-test, P<0.009).
To validate that GTL1 binds to the promoter sequence of CCS52A1, we performed ChIP-qPCR using several gene-specific primer sets. The promoter of CCS52A1 contains three GT elements, a complete GT3 box at −200 and −800 bp, and a partial GT3 box at −980 bp (Figures 4C and 5A). Using at least three independent biological replicates, we found strongest enrichment of GTL1 binding at the genomic region between −800 and −1000, bp (Figure 4C), suggesting that GTL1 preferentially binds to a GT3 box at −800 bp and/or −980 bp. To further confirm GTL1 binding to the CCS52A1 promoter, we also carried out the yeast-one-hybrid assay and found that GTL1, but not GT-1 and GT-3, other trihelix family proteins carrying s single trihelix domain, interacts with a 1.5-kb fragment of the CCS52A1 promoter in yeast cells (Figure 5B). To test whether any of the GT3 boxes is responsible for the GTL1 binding, we subsequently substituted nucleotides in the −800 bp box (5′-GGTAAAT-3′ to 5′-GATTCAG-3′) or the −980 bp box (5′-TTTAC-3′ to 5′-GCACG-3′) (Figure 5A). As shown in Figure 5B, nucleotide substitutions in the −980 bp box completely abolishes the GTL1 binding whereas mutations in the −800 box do not interfere with the binding. These results establish that GTL1 binds to the promoter sequence of the CCS52A1 gene through preferential interaction with the GT3 box at −980 bp.
Figure 5.
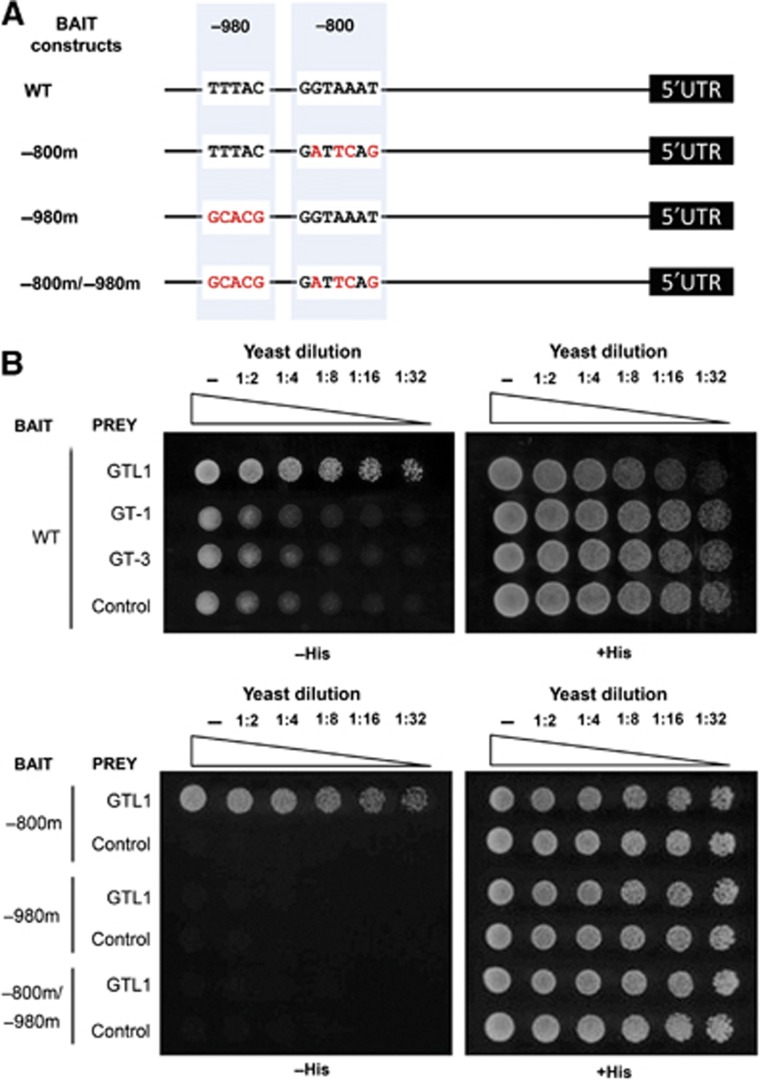
GTL1 binds to the GT3 box within the CCS52A1 promoter. (A) Schematic representation of the bait constructs illustrating the two putative GT3 boxes, one at −980 bp and other at −800 bp upstream of 5′UTR, in the WT CCS52A1 promoter. Substituted nucleotides are highlighted in red. (B) (Upper panel) Yeast cells transformed with the GAL4-fusions of trihelix transcription factor constructs (GTL1, GT-1, GT-3) and empty vector control were diluted and spotted on plates without (−His, +5 mM 3-AT, left) or with histidine (+His, right). GTL1, but neither GT-1 nor GT-3, binds to the CCS52A1 promoter. (Lower panel) The same assay was carried out with the bait constructs harbouring the mutated CCS52A1 promoter. Only the mutations in the GT3 box at −980 bp abolish the binding of GTL1 to the CCS52A1 promoter.
GTL1 represses the CCS52A1 expression at a late stage of trichome development
Our trichome-specific microarray data suggest that GTL1 negatively regulates the expression of CCS52A1 (Supplementary data S2). To confirm this, we isolated trichomes from fourth and fifth leaves of WT and gtl1-1 plants, and performed quantitative RT–PCR experiments using their total RNA. As shown in Figure 4D, the level of CCS52A1 transcripts is significantly up-regulated in gtl1-1 trichomes, supporting the view that GTL1 acts as a transcriptional repressor of CCS52A1. In our previous study we also recognised the elevated expression of CCS52A2 in gtl1-1 trichomes (Breuer et al, 2009) but CCS52A2 was neither identified by ChIP-chip nor detected by the microarray experiment in this study.
The GTL1 gene is not expressed at an early stage of trichome development when cells are still undergoing branching and its transcription starts while cells reach their maximum size (Figure 6A). We therefore hypothesised that this temporal transcription of GTL1 may result in the down-regulation of CCS52A1, and hence the cessation of endocycling, at a late stage of trichome development. To resolve the temporal activity of the CCS52A1 promoter during trichome development, we generated a construct that drives the expression of nuclear targeted histone 2B (H2B) fused with GFP under the 1.5-kb CCS52A1 promoter and introduced them into plants heterozygous for gtl1-1. We subsequently identified T2 plants homozygous for the pCCS52A1:H2B:GFP construct and heterozygous for gtl1-1, and assessed the expression of H2B:GFP fusion proteins in WT and gtl1-1 mutants segregating at T3 generation. Our fluorescence microscopy analysis reveals that WT plants carrying pCCS52A1:H2B:GFP display clear nuclear accumulation of H2B–GFP proteins in young trichomes that have not completed the second branching while these signals are no longer detectable in older trichomes (Figure 6B). We detected background fluorescence signals in old trichomes but the nuclei we identified by bright-field microscopy never emitted the GFP fluorescence. In sharp contrast to this, we found that the nuclear expression of H2B:GFP proteins is extended into the late stage of development in gtl1-1 (Figure 6B), indicating that loss of GTL1 allows prolonged activity of the CCS52A1 promoter in vivo. These results confirm that GTL1 directly represses the CCS52A1 expression in trichome cells.
Figure 6.
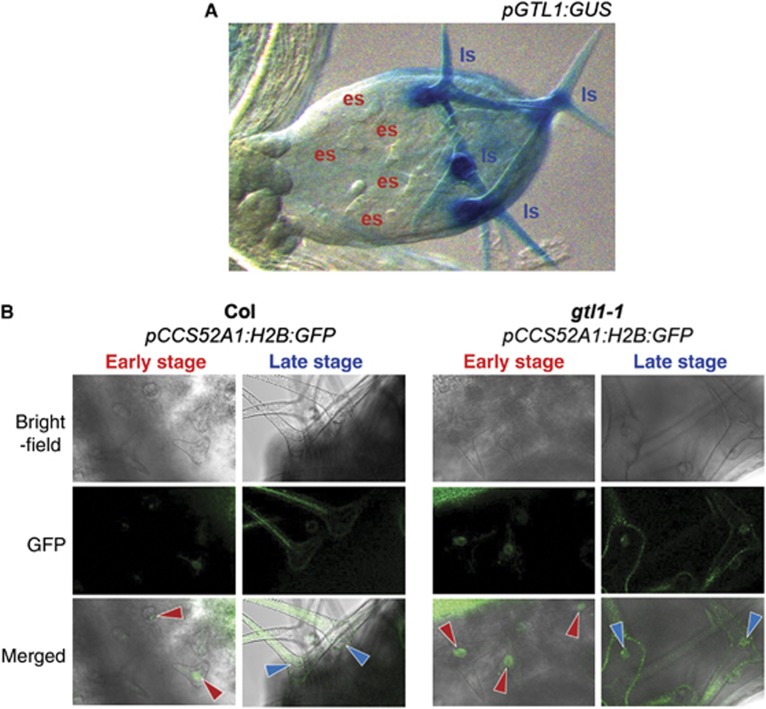
GTL1 represses the activity of the CCS52A1 promoter in maturing trichomes. (A) The pGTL1:GUS reporter shows that the GTL1 promoter activity is not detected at an early stage (es) of trichome development and it is only found during the late stage (ls) when trichome cells reach their maximum size. (B) The representative expression of pCCS52A1:H2B:GFP markers in trichomes from sixth leaves of 15-day-old Col and gtl1-1 plants. Red and blue arrows mark trichome nuclei during early and late stage of development, respectively. Nuclear accumulation of H2B:GFP proteins is detected only at an early stage in Col but is extended to maturing trichomes in gtl1-1. Broad green signals found in the cytoplasm derive from autofluorecence in trichomes. Scale bars: 150 μm in (A), 50 μm in (B).
CCS52A1 mediates ploidy-dependent cell growth governed by GTL1 but not ploidy-independent cell growth
The function of CCS52A1 as a positive regulator of the endocycle progression during early trichome development has been recently described (Larson-Rabin et al, 2009; Kasili et al, 2010; Heyman et al, 2011). Compared with WT, ccs52a1 mutants develop smaller, under-branched and under-endoreduplicated trichomes while overexpression of CCS52A1 under the GL2 promoter leads to larger, overbranched and over-endoreduplicated trichomes. To test whether the prolonged expression of CCS52A1 is responsible for the extended ploidy-dependent cell growth of gtl1-1, we generated gtl1-1 ccs52a1-2 double mutants. As shown in Figure 7A, trichome cells in gtl1-1 ccs52a1-2 plants are clearly smaller than those of WT or gtl1-1 mutants and most of them are under-branched, similar to those in ccs52a1-2 mutants. We quantified the average number of branching per trichomes and found that the ccs52a1-2 mutation is indeed epistatic to gtl1-1 for the trichome branching phenotype (Supplementary Figure S3). Consistently, the average ploidy level of gtl1-1 ccs52a1-2 trichome nuclei is identical to that of ccs52a1-2 (Figure 7B), indicating that the ccs52a1-2 mutation completely suppresses the trichome ploidy phenotype of gtl1-1. Interestingly, however, a thorough analysis of trichome cell size, as represented by the length of individual branches, revealed that trichome cells in gtl1-1 ccs52a1-2 are significantly larger than those in ccs52a1-2 single mutants (Figure 7A and C), suggesting that the ccs52a1-2 mutation does not completely suppress the cell growth phenotype of gtl1-1. These observations provide strong genetic evidence that CCS52A1 is a central regulator of the endocycle and cell growth acting directly downstream of GTL1. Our data also uncouple ploidy-dependent cell growth from ploidy-independent growth and predict the existence of additional targets acting downstream of GTL1 to control ploidy-independent cell growth.
Figure 7.
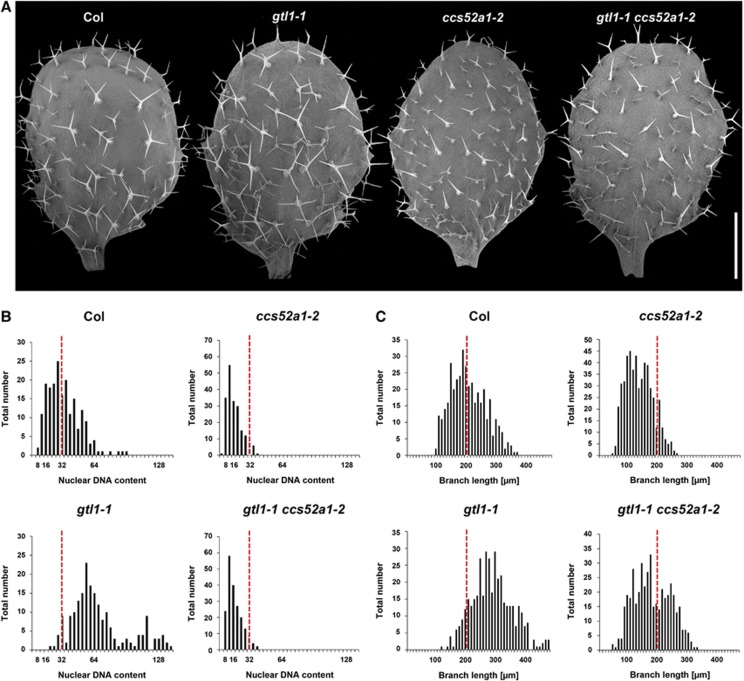
CCS52A1 functions downstream of GTL1 to mediate ploidy-dependent cell growth. (A) Scanning electron micrographs of adaxial epidermis from third leaves of 12-day-old Col, gtl1-1, ccs52a1-2 and gtl1-1 ccs52a1-2 double mutants. (B) Quantification of trichome nuclear DNA contents isolated from third and fourth leaves of 22-day-old plants. Values on the y-axis represent the number of trichomes counted for a given DNA content level. All ploidy measurements were normalised to the mean of 32C in WT trichome. (C) Quantification of the trichome branch lengths of the corresponding trichomes. Red lines in (B, C) represent the mean values of WT. Scale bar: 750 μm in (A).
Discussion
How cell size is controlled in multicellular organisms is an important question in biology but molecular mechanisms underlying this control remain poorly understood. Previous studies uncovered many regulators that promote cell growth but whether an active regulatory mechanism operates to cease cell growth was not known before. In this study we demonstrate that plants possess a genetic programme to actively terminate cell growth and that this is largely mediated through the transcriptional repression of CDC20/FZR/CCS52, an evolutionally conserved regulator of the endocycle. An involvement of the endocycle in cell-size control has been long debated but this study establishes that an increase in ploidy by the endocycle has major impacts on cell growth.
Developmental control of cell growth and cell differentiation
This study shows that a trihelix transcription factor GTL1 directly represses the endocycle and its developmentally programmed expression is one of the key determinants of final cell size in plants. Our model predicts that developmentally controlled expression of GTL1 shuts down the transcription of CCS52A1, leading to the reduced activity of APC/C and thus cessation of the endocycle (Figure 8). While this CCS52A1-dependent pathway fully accounts for ploidy-dependent cell growth controlled by GTL1, our data hint that GTL1 also represses other pathways mediating ploidy-independent cell growth. An involvement of endoreduplication in the control of cell size has long been debated (Sugimoto-Shirasu and Roberts, 2003; Breuer et al, 2010) and the existence of ploidy-independent growth was previously proposed for trichome cells (Schnittger et al, 2003). Our results are consistent with this idea and strongly suggest that both ploidy-dependent and -independent cell growth contribute to the overall increase in cell size. We do not know the nature of other GTL1 targets acting in the ploidy-independent pathway but the remaining 181 genes identified by ChIP-chip and microarray analyses are the best candidates. Further validation of ChIP-chip results coupled with functional analyses of candidate genes should uncover how GTL1 coordinates ploidy-dependent and ploidy-independent cell growth.
Figure 8.
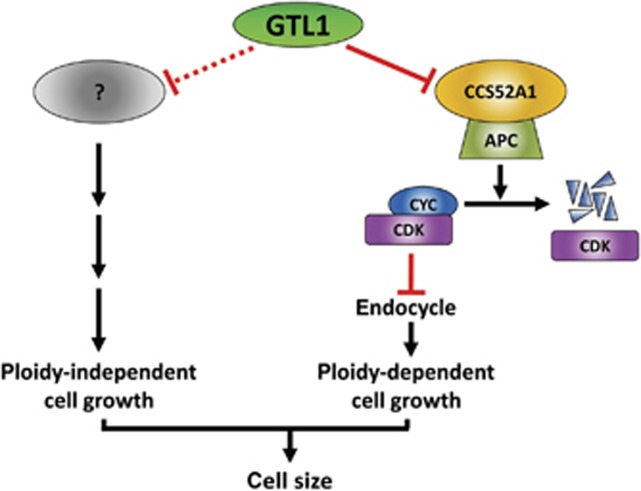
A schematic model illustrating how GTL1 regulates ploidy-dependent and -independent cell growth. During late trichome development, GTL1 represses the CCS52A1 expression, resulting in a decline of the APC/C activity. Reduced APC/C activity then leads to stabilisation of active CYC/CDK complexes, which blocks further progression of the endocycle. This active repression of ploidy-dependent cell growth is the major pathway that determines final size of trichome hair cells. In addition, GTL1 is likely to act as a negative regulator of ploidy-independent cell growth by repressing the expression of other unknown genes.
The next important question is whether the same mechanism also governs general plant cell growth. Given that GTL1 is expressed in various cell types, for example, in leaves and roots, undergoing rapid cell expansion (Breuer et al, 2009; Yoo et al, 2010), we think this is very likely. Indeed our data show that overexpression of GTL1 represses growth of non-trichome leaf epidermal cells. Expressing a repressor of cell growth in expanding cells sounds counterintuitive but we speculate that such a mechanism may allow adjusting the pace and duration of cell growth in the context of development and/or in response to the environmental change. Although developmental defects of gtl1 mutants are restricted to trichomes under our standard growth conditions, lack of other phenotypes might be due to the functional redundancies among other closely related GT-2 homologues. We should note that gtl1 mutant trichomes are larger than those in WT but they do stop growing, hence additional mechanisms may repress further cell growth. Systematic identification of new mutations that permit extended growth should help uncovering other pathways involved in the repression of plant cell growth.
Our overexpression studies imply that GTL1 may also interfere with trichome cell differentiation either directly or indirectly. Our ChIP-chip experiment identified 10 putative GTL1 targets implicated in the control of trichome morphogenesis but none of them is directly linked to the acquisition and/or maintenance of trichome identities. Although GTL1 may target some uncharacterised genes functioning in these processes, an alternative hypothesis is that GTL1 indirectly influences trichome differentiation through the modulation of the endocycle. It is indeed shown that misregulation of endocycles has immediate consequences on trichome differentiation (Bramsiepe et al, 2010), thus it might be possible that the reduced level of endocycles in pGL2:GFP:GTL1 trichomes is inhibitory to trichome cell differentiation.
Transcriptional control of the endocycle
Orchestrating the progression of the mitotic cycle and endocycle in space and time is vital for the optimum growth and development of multicellular organisms. In many plant species, cells proliferate through the mitotic cycle in the meristem and as soon as cells leave the meristem, they switch from the mitotic cycle to the endocycle. Temporal transcription of cell-cycle regulators is one core mechanism underpinning the timely mitotic-to-endocycle transition but how this is regulated is not well understood. Previous studies suggest that FZR/CDH1/CCS52 is a key regulator of this transition and that its expression is under the control of both typical and atypical E2F transcription factors (Lammens et al, 2008; Heyman et al, 2011; Magyar et al, 2012). In the Arabidopsis shoot meristem, an atypical E2F protein DEL1 prevents the premature transition into the endocycle by repressing the expression of CCS52A2 (Lammens et al, 2008). In addition, a recent study shows that the E2FA–RBR complex represses the expression of both CCS52A1 and CCS52A2 in proliferating cells to inhibit an entry into the endocycle (Magyar et al, 2012). How the progression of consecutive endocycles is controlled is even less clear and an important discovery of this study is that at least in Arabidopsis the endocycle needs to be actively terminated through the transcriptional repression of CCS52A1. Trihelix transcriptional regulators are specific to plants and whether similar mechanisms operate in non-plant species is not currently known. In Drosophila, FZR/CDH1/CCS52 proteins control the mitotic-to-endocycle transition and the APC/C activity is necessary to sustain the endocycle progression (Sigrist and Lehner, 1997). Thus, it would not be surprising if transcriptional down-regulation of core APC/C submits or FZR/CDH1/CCS52 also triggers the endocycle cessation in Drosophila.
This study also highlights an involvement of transcription factors other than typical and atypical E2Fs in controlling the plant endocycle. Several developmental regulators are implicated in the transcriptional control of mitotic cycle genes (Sozzani et al, 2010; Xie et al, 2010; Vanneste et al, 2011) and our data extend this to the endocycle. Non-E2F transcription factors also regulate the endocycle initiation and maintenance in other organisms. For example, a Drosophila zinc-finger transcription factor escargot and its mouse homologue mSna prevent an entry into the endocycle and they are required to maintain the Cdk1 activity (Hayashi, 1996; Nakayama et al, 1998). A basic helix-loop-helix transcription factor Hand1 promotes endoreduplication although its exact molecular function is not yet known (Martindill and Riley, 2008). Based on our overexpression studies, we speculate that GTL1 may also control earlier progression of endocycles in non-trichome cells potentially by modulating the CCS52A1 expression. How E2F and non-E2F transcription factors cooperate during development for the spatio-temporal control of endocycles is an interesting question and should be further investigated in future studies.
Functional roles of the APC/C in post-mitotic cells
Recent studies are beginning to uncover previously uncharacterised roles of the APC/C in post-mitotic cells (Eguren et al, 2011). Several lines of evidence suggest an involvement of the APC/C in plant cell differentiation (Komaki and Sugimoto, 2012) and our data substantiate that sustained APC/C activity is the prerequisite for post-mitotic cell growth. Interestingly, previously reported APC10 or APC11 RNAi lines do not cause any defects in trichome cell growth and endocycle, proposing that at least those APC/C components are not required for the endocycle regulation in trichomes (Roodbarkelari et al, 2010). Loss- and gain-of-function lines of CDH1/FZR/CCS52, however, affect the endocycle progression in both animals and plants (Narbonne-Reveau et al, 2008; Larson-Rabin et al, 2009; Kasili et al, 2010; Heyman et al, 2011). Our results agree with these observations and support an involvement of the APC/C in the endocycle progression.
The targets of the APCCCS52A1 in trichome cells is not established but one good candidate is CYCA2;3 which together with CDKA;1, is required to terminate the endocycle in trichomes (Imai et al, 2006). In Arabidopsis roots, APCCCS52A1 promotes the degradation of CYCA2;3, leading to the reduced activity of CYCA2;3–CDKB1;1 and thus an endocycle onset (Boudolf et al, 2009). If a similar mechanism operates in trichomes, the repression of APCCCS52A1 towards the end of cell growth may increase the CYCA2;3–CDKA;1 activity which in turn triggers the endocycle cessation. According to this scenario, the repression of APCCCS52A1 is lifted in gtl1 trichomes, thus delaying the accumulation of CYCA2;3–CDKA;1 to terminate the endocycle.
Materials and methods
Plant material and growth conditions
The gtl1-1, ccs52a1-2 and pGL2:GUS lines were previously described (Masucci et al, 1996; Lammens et al, 2008; Breuer et al, 2009). Plants were germinated and grown either on plates containing Murashige and Skoog salts, pH 5.7, 1% sucrose (w/v), and 0.5% phytogel (w/v) or on a 1:1 mixture of soil Supermix A (SAKATA, Japan) and vermiculite (VS Kako, Japan) in continuous light at 22°C.
Construction of binary plasmids for Arabidopsis transformation
The generation of gtl1-1 plants carrying the pGTL1:GTL1:GFP constructs was described previously (Breuer et al, 2009). For the construction of the pCCS52A1:H2B:GFP plasmid, a 1977-bp fragment of the CCS52A1 promoter was amplified with the oligonucleotides attB4-proCCS52A1-Fw 5′-GGGGACAACTTTGTATAGAAAAGTTGAAGAACTCGTCAGTCTTGTG-3′ and attB1r-proCCS52A1-Rv 5′-GGGGACTGCTTTTTTGTACAAACTTGCATTGGTTTTTTTTTTTTGTTGACT-3′, and cloned into the vector pDONR-P4P1R by Gateway BP clonase II enzyme reaction (Invitrogen, Japan). Histone 2B cDNA was amplified from Arabidopsis cDNA derived from whole seedlings with the oligonucleotides H2B-TOPO-F 5′-CACCATGGCGAAGGCAGATAAGAAACC-3′ and H2B-ΔSTOP-R 5′-AGAACTCGTAAACTTCGTAACCGCC-3′ and cloned into pENTR-D-TOPO (Invitrogen, Japan). The CCS52A1 promoter and the H2B cDNA fragments were transferred into the R4pGWB504 destination vector by LR clonase II enzyme (Invitrogen, Japan) in a one-step tripartite cloning reaction (Nakagawa et al, 2008). Both pATML1:GFP:GTL1 and pGL2:GFP:GTL1 were generated by infusion into a previously generated pENTR-based pGTL1:GFP:GTL1 construct (Breuer et al, 2009). First, the pGTL1:GFP:GTL1 plasmid was amplified using the oligonucleotides GFP–GTL1-INF-R1 5′-AAGGGTGGGCGCGCCGACCCAG-3′ and GFP–GTL1-INF-F1 5′-ATGCCCGGGGGTGGCATGG-3′ and digested with DpnI (Toyobo, Japan). For the infusion reaction a 3384-bp genomic fragment of the ATML1 promoter and a 1748-bp genomic fragment of the GL2 promoter were amplified from the BAC clones F17L22 and F19K16 using the oligonucleotide pair ATML1p-F1 5′-CGGCGCGCCCACCCTTAAGCTTATCAAAGAA-3′ and ATML1p-R1 5′-CCATGCCACCCCCGGGCATAACCGGTGGATTCAGGG-3′ and the pair GL2p-F1 5′-CGGCGCGCCCACCCTTACAATTCCCTAGGCCGTACGACG-3′ and GL2p-R1 5′-CCATGCCACCCCCGGGCATACAAATCCTGTCCCTAG-3′, respectively. The promoter fragments were then cloned into the amplified GFP:GTL1 backbone using the In-fusion Advantage PCR Cloning Kit (TAKARA, Japan). The fused fragments were then cloned by LR reaction into the binary vector pGWB1 (Nakagawa et al, 2007). All stable plant transformations were performed via floral dip (Clough and Bent, 1998).
RNA extraction and microarray analysis
Trichomes were removed with forceps from the fifth and sixth rosette leaves of 3-week-old WT and gtl1-1 mutant plants according to a previously established protocol (Jakoby et al, 2008). Total RNA was extracted from three biological replicates by a Qiagen plant RNeasy kit, and treated with DNaseI (Takara Bio, Japan) following the manufacturer’s instructions. RNA probes were labeled using an Affymetrix 3′ IVT Express Kit (Affymetrix, Japan) and hybridised on ATH1 chips. Microarray data analysis was performed using the R software with AffylmGUI of the Bioconductor package (Wettenhall et al, 2006). The expression ratio of WT versus gtl1-1 mutants were subjected to the Benjamini Hochberg method (Hochberg and Benjamini, 1990), and statistical significance was considered with P<0.075, which is equivalent to an FDR <0.075.
Quantitative PCR
Total RNA from dissected trichomes was treated with DNaseI (Takara Bio, Japan) and reverse transcribed using the Invitrogen Superscript III kit. Transcript levels were determined by quantitative real-time PCR using the Toyobo SYBR Green Supermix kit and an Agilent Mx3000P QPCR system. The oligonucleotide pairs of qCCS52A1-F2 5′-CACGCTGCAAGAGAACAAGA-3′, qCCS52A1-R1 5′-ACCACTTGAGTCCGCATACC-3′ and of ACT2F 5′-CTGGATCGGTGGTTCCATTC-3′, ACT2R 5′-CCTGGACCTGCCTCATCATAC-3′ were used for PCR reaction to quantify the CCS52A1 and ACT2 expression, respectively.
ChIP-chip and ChIP-qPCR experiments
Whole aerial tissues of 2-week-old gtl1-1 plants carrying pGTL1:GTL1:GFP were used for ChIP experiments as previously described (Morohashi et al, 2007; Morohashi and Grotewold, 2009). For amplification of precipitated and input DNA, the GenomePlex Whole Genome Amplification Kit (Sigma) was used. DNA fragmentation, labeling, hybridisation, washes and detection were performed following the Affymetrix 100K protocol ( http://www.affymetrix.com/products/arrays/specific/100k.affx). CEL files were further analysed by MAT (Model-based Analysis of Tiling array; http://chip.dfci.harvard.edu/wli/MAT/) using the following parameters: BandWith=300, MaxGap=300, MinProbe=10 and P-value=0.001. Peaks consisting of continuous probes with significant MAT scores were evaluated using IGB (IGB, Affymetrix) with the additional criteria that the minimum gap should be <100 bp. We defined target genes as those for which 3 kb upstream regions contained at least one peak showing significant MAT scores (Morohashi and Grotewold, 2009). Sequences corresponding to peaks (FDR<0.1) were subjected to MEME (meme-maxsize 120000-nmotifs 5-minw 6-maxw 20). Computational analyses were performed using R ( http://www.r-project.org/), BedTool ( http://code.google.com/p/bedtools/) and custom made Perl scripts, which will be provided upon request.
ChIP-qPCR experiments were performed using immunoprecipitated and input DNA of WT and gtl1-1 plants carrying pGTL1:GTL1:GFP as templates. Three independent ChIP replicates were used as material for the validation of targets by ChIP-chip. The qPCR reactions were run using an iQ SYBR Green Supermix (Bio-Rad) in an iCycler thermocycler. Oligonucleotides used for individual genes are listed in Supplementary Table S1.
Yeast-one-hybrid assay
The CCS52A1 promoter, containing 1490, bp fragment upstream of the start codon, was amplified with the oligonucleotides CCS52A1-1490F1 5′-ATGGCGGCCGCTTATCATTTGTTTTCTGATT-3′ and CCS52A1-1490R1 5′-ATGGCGGCCGCTGGTTTTTTTTTTTTGTTGACT-3′, and cloned into the pINT1-HIS3NB vector (Lopato et al, 2006) resulting in the bait vector pINT1-HIS3-CCS52A1 that harbours a transcriptional fusion between the CCS52A1 promoter and the HIS3 gene. For making the yeast reporter strain, the reporter vector was introduced into the yeast strain AH109 (MATa; trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, MEL1, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ; Clontech) and incubated at 30°C for 4 days. The open reading frame of GTL1, GT-1, and GT-3 was fused to the GAL4 activation domain in the vector pGADT7 (Clontech) and transformed into the reporter yeast strain. Transformed yeast strains were gown to an ODE600=0.1 and spotted on plates in a dilution series of 1:2, 1:4, 1:8, 1:16 and 1:32 with or without histidine (+5 mM 3-AT). Transformation and selection of yeast were carried out according to the Yeast Protocol Handbook (Clontech).
To generate nucleotide substitutions in a GT3 box at the −800 or −980 bp regions of the CCS52A1 promoter, the following oligonucleotide pairs were used to amplify the pINT1-HIS3-CCS52A1: -800mF1 5′-TTTTGATTCAGTTTCTTTTATATCTTTGATATT-3′, -800mR1 5′-AAGAAACTGAATCAAAAAAATATTCAAAAAATAATCC-3′ and -980mF1 5′-TCAATTGCACGTCATTTTAGTAAGGTTTTGT-3′, -980mR1 5′-AAAATGACGTGCAATTGATTGCAAGTTGCAATG-3′, respectively. The resulting PCR products were digested with DpnI and propagated in Escherichia coli DH10B to produce plasmids we named pINT1-HIS3-CCS52A1-800m and pINT1-HIS3-CCS52A1-980m. For the construction of the bait construct pINT1-HIS3-CCS52A1-800m/-980m harbouring substitutions in both GT3 boxes, the pINT1-HIS3-CCS52A1-800m plasmid was amplified with the oligonucleotide pair -980mF1 and -980mR1, digested with DpnI and propagated in E. coli DH10B.
Microscopy techniques
Confocal microscopy of young true leaves was carried out with a Zeiss 510 Meta confocal microscope. Recorded pictures were converted to JPEG format using the Zeiss LSM Image Browser version 4.0.0.157. For scanning electron microscopy of the adaxial epidermis of true leaves, a tabletop Hitachi T-1000 scanning electron microscope was used. Expression of the translational GFP-fusions in pATML1:GFP:GTL1 and pGL2:GFP:GTL1 lines was examined using a Leica MZ 16 FA dissection microscope equipped with a digital Leica DFC 500 camera. Contrast and brightness were adjusted with Adobe Photoshop CS3 version 10.0.
Ploidy measurements, histological tissue clearing and GUS staining
Nuclear ploidy level of whole leaves was measured with a ploidy analyser PA-I (Partec) as previously described (Sugimoto-Shirasu et al, 2002). To quantify ploidy levels of trichome nuclei, trichome cells were isolated from leaves (Marks et al, 2008) and their nuclei were stained with DAPI. DAPI-stained nuclei were visualised using an Olympus BX51 fluorescence microscope equipped with a digital Olympus DP70 camera and Olympus DP Manager software version 1.2.1.107 (Zhang and Oppenheimer, 2004), and fluorescent signals were measured with ImageJ version 1.37 as previously described (Schnittger et al, 1998). The average ploidy level of WT trichomes was set to 32C to normalise the ploidy level of mutant trichomes.
To quantify the area of epidermal pavement cells, fully expanded first true leaves were fixed overnight in glacial acetic acid: ethanol (1:1) at room temperature. The fixed plant material was de-hydrated by an ascending ethanol series of 30, 50, 75, 85 and 95% ethanol for 20 min in each concentration, and cleared in chloral hydrate: H2O: glycerol (8:2:1) overnight at 4°C. The cleared tissue was unfurled on microscopy slides in clearing solution and used for differential interference contrast (DIC) microscopy using an Olympus BX51. ImageJ version 1.37 was used to quantify the cell area of individual epidermal pavement cells. GUS staining in young seedlings was performed as previously described (Breuer et al, 2009).
Accession numbers
Sequence data of genes from this publication can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GTL1, At1g33240; ATML1, At4g21750; GL2, At1g79840; CCS52A1/FZR2, At4g22910; CCS52A2/FZR1, At4g11920; CCS52B/FZR3, At5g13840. The microarray data from this publication have been submitted to the NCBI GEO database [ http://www.ncbi.nlm.nih.gov/geo/] and assigned the accession number GSE40520.
Supplementary Material
Acknowledgments
We thank all the members of KS’s laboratory for discussions and comments on the manuscript. We are grateful to Tsuyoshi Nakagawa (Shimane University, Japan) for sharing the R4 gateway binary plasmid library, Liam Dolan (University of Oxford, UK) for the pGL2:GUS reporter, and Lieven DeVeylder (VIB Gent, Belgium) for the ccs52a1-2 mutant. This work was supported by a grant from MEXT to KS (No. 22119010) and a grant from National Science Foundation to EG (MCB0418891). CB and TI were recipients of the RIKEN postdoctoral fellowship.
Author contributions: CB and KS conceived and designed the experiments. CB and AK performed most of the experiments at RIKEN. KM and EG carried out the ChIP-chip, ChIP-qPCR experiments and the computational analyses at OSU. NT and MU performed the yeast-one-hybrid assay at NAIST. TI contributed the pCCS52A1:H2B:GFP line. CB and KS wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, De Vusser K, De Tavernier E, De Bock K, Inze D (2002) Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol 129: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E, De Jaeger G, Inze D, De Veylder L (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hulskamp M, Schnittger A (2010) Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Ishida T, Sugimoto K (2010) Developmental control of endocycles and cell growth in plants. Curr Opin Plant Biol 13: 654–660 [DOI] [PubMed] [Google Scholar]
- Breuer C, Kawamura A, Ichikawa T, Tominaga-Wada R, Wada T, Kondou Y, Muto S, Matsui M, Sugimoto K (2009) The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21: 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, Haubeiss S, Ha N, Chan RL, Lohmann JU (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 841–853 [DOI] [PubMed] [Google Scholar]
- Cebolla A, Vinardell J, Kiss E, Oláh B, Roudier F, Kondorosi A, Kondorosi E (1999) The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J 18: 4476–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Almeida Engler J, Kyndt T, Vieira P, Van Cappelle E, Boudolf V, Sanchez V, Escobar C, De Veylder L, Engler G, Abad P, Gheysen G (2012) CCS52 and DEL1 genes are key components of the endocycle in nematode induced feeding sites. Plant J 72: 185–198 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Deng W-M, Althauser C, Ruohola-Baker H (2001) Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128: 4737–4746 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL (2001) Endoreplication cell cycles: More for less. Cell 105: 297–306 [DOI] [PubMed] [Google Scholar]
- Eguren M, Manchado E, Malumbres M (2011) Non-mitotic functions of the anaphase-promoting complex. Semin Cell Dev Biol 22: 572–578 [DOI] [PubMed] [Google Scholar]
- Hayashi S (1996) A Cdc2 dependent checkpoint maintains diploidy in Drosophila. Development 122: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Hershko A (1997) Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol 9: 788–799 [DOI] [PubMed] [Google Scholar]
- Heyman J, Van den Daele H, De Wit K, VR Boudolf, Berckmans B, Verkest A, Kamei CLA, De Jaeger G, Koncz C, De Veylder L (2011) Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23: 4394–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y (1990) More powerful procedures for multiple significance testing. Stat Med 9: 811–818 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim G-T, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T (2006) The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Iwata E, Ikeda S, Matsunaga S, Kurata M, Yoshioka Y, Criqui MC, Genschik P, Ito M (2011) GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23: 4382–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hulskamp M, Larkin J, Schnittger A (2008) Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol 148: 1583–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasili R, Walker JD, Simmons LA, Zhou J, De Veylder L, Larkin JC (2010) SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S, Sugimoto K (2012) Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol 53: 953–964 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Caspar T, Dehesh K, Quail PH (1993) DNA binding factor GT-2 from Arabidopsis. Plant Mol Biol 23: 337–348 [DOI] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, Inze D, De Veylder L (2008) Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA 105: 14721–14726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Rabin Z, Li Z, Masson PH, Day CD (2009) FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol 149: 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A (2006) Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 442: 471–474 [DOI] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ (2009) Endoreplication: polyploidy with purpose. Genes Dev 23: 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Bazanova N, Morran S, Milligan A, Shirley N, Langridge P (2006) Isolation of plant transcription factors using a modified yeast one-hybrid system. Plant Methods 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Horvath B, Khan S, Mohammed B, Henriques R, De Veylder L, Bako L, Scheres B, Bogre L (2012) Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J 31: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD, Betancur L, Gilding E, Chen F, Bauer S, Wenger JP, Dixon RA, Haigler CH (2008) A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J 56: 483–492 [DOI] [PubMed] [Google Scholar]
- Martindill DMJ, Riley PR (2008) Cell cycle switch to endocycle: the nucleolus lends a hand. Cell Cycle 7: 17–23 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Mathieu-Rivet E, Gévaudant F, Sicard A, Salar S, Do PT, Mouras A, Fernie AR, Gibon Y, Rothan C, Chevalier C, Hernould M (2010) Functional analysis of the anaphase promoting complex activator CCS52A highlights the crucial role of endoreduplication for fruit growth in tomato. Plant J 62: 727–741 [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E (2009) A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Zhao M, Yang M, Read B, Lloyd A, Lamb R, Grotewold E (2007) Participation of the Arabidopsis bHLH Factor GL3 in trichome initiation regulatory events. Plant Physiol 145: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl W (1976) DNA endoreduplication and polyteny understood as evolutionary strategies. Nature 261: 614–615 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nakamura S, Tanaka K, Kawamukai M, Suzuki T, Nakamura K, Kimura T, Ishiguro S (2008) Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci Biotechnol Biochem 72: 624–629 [DOI] [PubMed] [Google Scholar]
- Nakayama H, Scott IC, Cross JC (1998) The transition to endoreduplication in trophoblast giant cells is regulated by the mSNA zinc finger transcription factor. Dev Biol 199: 150–163 [DOI] [PubMed] [Google Scholar]
- Narbonne-Reveau K, Senger S, Pal M, Herr A, Richardson HE, Asano M, Deak P, Lilly MA (2008) APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 135: 1451–1461 [DOI] [PubMed] [Google Scholar]
- Ni M, Dehesh K, Tepperman JM, Quail PH (1996) GT-2: in vivo transcriptional activation activity and definition of novel twin DNA binding domains with reciprocal target sequence selectivity. Plant Cell 8: 1041–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hulskamp M, Brown S, Dorne AM, Bonneville JM (1999) Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell 9: 931–943 [DOI] [PubMed] [Google Scholar]
- Roodbarkelari F, Bramsiepe J, Weinl C, Marquardt S, Novak B, Jakoby MJ, Lechner E, Genschik P, Schnittger A (2010) CULLIN 4-RING FINGER-LIGASE plays a key role in the control of endoreplication cycles in Arabidopsis trichomes. Proc Natl Acad Sci USA 107: 15275–15280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Jurgens G, Hulskamp M (1998) Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125: 2283–2289 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Schobinger U, Stierhof Y-D, Hulskamp M (2002) Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol 12: 415–420 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schobinger U, Hulskamp M (2003) Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky MF (1999) The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF (1997) Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90: 671–681 [DOI] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JAH, Benfey PN (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) ‘Big it up’: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC (2002) DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr Biol 12: 1782–1786 [DOI] [PubMed] [Google Scholar]
- Sun J, Deng W-M (2005) Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development 132: 4299–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Deng W-M (2007) Hindsight mediates the role of Notch in suppressing hedgehog signaling and cell proliferation. Dev Cell 12: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Coppens F, Lee E, Donner TJ, Xie Z, Van Isterdael G, Dhondt S, De Winter F, De Rybel B, Vuylsteke M, De Veylder L, Friml J, Inze D, Grotewold E, Scarpella E, Sack F, Beemster GTS, Beeckman T (2011) Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J 30: 3430–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Vr Boudolf, Brown SC, De Veylder L, Mergaert P, Kondorosi E (2009) APC/CCCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA 106: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463 [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK (2006) affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22: 897–899 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Weigel D (2001) Arabidopsis genome: life without Notch. Curr Biol 11: R112–R114 [DOI] [PubMed] [Google Scholar]
- Wilson EB (1925) The Cell in Development and Heredity. 3rd edn, New York, USA: The Macmillan Company, [Google Scholar]
- Xie Z, Li D, Wang L, Sack FD, Grotewold E (2010) Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J 64: 731–739 [DOI] [PubMed] [Google Scholar]
- Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV (2010) The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Oppenheimer DG (2004) A simple and efficient method for isolating trichomes for downstream Analyses. Plant Cell Physiol 45: 221–224 [DOI] [PubMed] [Google Scholar]
- Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo MJ, Nagarajan S, van Straaten M, Woods B, von Dassow G, Rottig C, Lehner CF, Grewal SS, Duronio RJ, Edgar BA (2011) Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


