Abstract
High affinity antibodies are generated in mice and humans by means of somatic hypermutation (SHM) of variable (V) regions of Ig genes. Mutations with rates of 10−5–10−3 per base pair per generation, about 106-fold above normal, are targeted primarily at V-region hot spots by unknown mechanisms. We have measured mRNA expression of DNA polymerases ι, η, and ζ by using cultured Burkitt's lymphoma (BL)2 cells. These cells exhibit 5–10-fold increases in heavy-chain V-region mutations targeted only predominantly to RGYW (R = A or G, Y = C or T, W = T or A) hot spots if costimulated with T cells and IgM crosslinking, the presumed in vivo requirements for SHM. An ∼4-fold increase pol ι mRNA occurs within 12 h when cocultured with T cells and surface IgM crosslinking. Induction of pols η and ζ occur with T cells, IgM crosslinking, or both stimuli. The fidelity of pol ι was measured at RGYW hot- and non-hot-spot sequences situated at nicks, gaps, and double-strand breaks. Pol ι formed T⋅G mispairs at a frequency of 10−2, consistent with SHM-generated C to T transitions, with a 3-fold increased error rate in hot- vs. non-hot-spot sequences for the single-nucleotide overhang. The T cell and IgM crosslinking-dependent induction of pol ι at 12 h may indicate an SHM “triggering” event has occurred. However, pols ι, η, and ζ are present under all conditions, suggesting that their presence is not sufficient to generate mutations because both T cell and IgM stimuli are required for SHM induction.
The somatic hypermutation (SHM) of Ig variable (V) region genes (1) is required for the affinity maturation of antibodies that occurs during T-dependent responses to foreign and self-antigens. This process results in the introduction of point mutations and occasional deletions and insertions at rates of 10−5–10−3 per base pair per generation (/bp/gen) (2). This rate of mutation is about a million times higher than the rates observed in housekeeping genes. The hypermutation process is restricted to ∼1.5 kb downstream from the promoter in Ig heavy- and light-chain genes (3–5), requires cis-acting elements that regulate transcription (6–8), and occurs in B cell centroblasts in the dark zone of the germinal centers of the secondary lymphoid organs (9). There is a preference for transitions over transversions, and mutations are concentrated in hot-spot consensus motifs, especially RGYW (R = A or G, Y = C or T, W = T or A), or complementary sequence WRCY motifs on the other strand (10, 11).
Several models have been proposed to explain the high rate of somatic V-region mutation (7). One of the earliest suggestions was that an error-prone polymerase was involved (12), but attempts to identify the enzymes that are responsible have been so far unsuccessful. Recent studies suggesting that mutation is associated with breaks in the DNA proximal to SHM hot spots (13–15) provide further rationale for the possible involvement of error-prone polymerases. Such enzymes might copy short regions of DNA before double-strand end joining or copy small gaps in processing single-strand nicks (16). Another model proposes there are two distinct phases in the SHM process, the first phase targeting G and C in hot-spot motifs, and the second phase targeting A and T bases in surrounding sequences that depend on the Mut s homologue (MSH)2–MSH6 heterodimer and associated MMR proteins (17–19). In addition to mismatch repair proteins (reviewed in ref. 20), an activation-induced cytidine deaminase (AID) that is homologous to an RNA editing enzyme (21, 22) has been shown to play a role in the mutational process.
In searching for error-prone DNA polymerases that might introduce the V-region mutations, a number of authors have suggested recently that the mammalian homologues of the Escherichia coli and yeast UmuC/DinB/Rev1/Rad30 family of polymerases could be involved (reviewed in refs. 23 and 24). These enzymes participate in DNA synthesis when the processive replicative polymerase stalls because it cannot bypass a lesion in DNA. Then the replicative polymerase is replaced by a nonprocessive polymerase from the UmuC/DinB/Rev1/Rad30 family, which can bypass the block by introducing a nucleotide opposite the lesion. These enzymes lack editing functions and, depending on the activity of polymerase, the nature of the lesion, and the surrounding sequences, they may be error-prone and highly mutagenic even on undamaged DNA (24). Individually, or in combination, these polymerases introduce only one or a few bases, and then the newly synthesized DNA strand must be extended by other more processive polymerases.
A substantial number of mammalian homologues of the bacterial and yeast UmuC/DinB/Rev1/Rad30 family have been identified recently, four of which have been proposed as potential candidates for SHM (25–27): polymerase iota (pol ι), polymerase eta (pol η), polymerase kappa (pol κ), and deoxynucleotidyl transferase Rev1 (Rev1) (26). All of these enzymes have translesion bypass activity in cell-free systems, but most relevant for SHM, they also exhibit overall low fidelity on undamaged DNA. A role for pol η has been suggested recently based on the observation that patients with Xeroderma pigmentosum-variant (XP-V) with a mutant form of this enzyme have normal immune systems and undergo SHM, but most importantly, these patients have an altered mutation spectra, suggesting the absence of pol η has an effect on the targeting and type of mutations that occur (19). Polymerases mu (μ) and zeta (pol ζ), although not members of this family, have been proposed to play a role in V-region mutation (28, 29) also, perhaps in combination with one or more members of the UmuC/DinB/Rev1/Rad30 family (30, 31).
To investigate whether these enzymes play a role in V-region mutation, our first step was to perform expression analysis of pols ι, κ, ζ, η, and μ and Rev1 in the human Burkitt's lymphoma (BL)2 B cell line. BL2 cells have many of the surface markers of germinal center centroblasts that normally undergo somatic mutation. BL2 cells can be stimulated by crosslinking their surface IgMs and cocultivating them with T cells to undergo V-region mutation in tissue culture (32). On the basis of expression results and mutation data, we next took an in vitro biochemical approach to evaluate the fidelity of one of the candidate polymerases, pol ι, when replicating an RGYW hypermutation “hot spot” that is targeted in BL2 and a non-hot-spot motif at a template G site. The motifs were presented to the polymerase as model substrates designed to reflect several current “break and repair” models (14–16) that include putative single- and/or double-stranded break intermediates in the SHM process.
Materials and Methods
Cell Lines.
HuT 78 is a human T cell lymphoma cell line that was obtained from American Type Culture Collection. The human BL2 B cell lymphoma was provided kindly by Kay Hutchinson of the Fels Research Institute (Philadelphia). Cells were cultured in RPMI medium 1640 as described (32) with 10% heat inactivated FCS (HyClone).
Induction of BL2.
Plates were coated with anti-CD3 antibody (Santa Cruz Biotechnology) at 500 ng/ml in serum-free RPMI medium 1640 for 2.5 h at room temperature. HuT 78 cells that had been stimulated for 24 h with phytohemagglutinin (PHA; Sigma) at 2 μg/ml were gamma-irradiated with 4,000 rads from a 137Cs source. Washed HuT 78 cells were plated at 1 × 107 per well. BL2 cells were incubated with anti-human IgM antibody (Jackson ImmunoResearch) at 10 μg/ml for 30 min on ice, washed, and added to the appropriate wells at 1 × 106 per well. In most of the experiments, the BL2 cells were harvested after 72 h and the viable BL2 cells were separated from the killed T cells and any dead BL2 cells by using Ficoll/Paque PLUS (Amersham Pharmacia). In some experiments, treatment with anti-IgM and cocultivation with T cells was repeated 4 times (days 0, 4, 7, and 10) and the cells were analyzed on day 14. At the end of the experiment, the recovered cells were washed once in Hanks' balanced salt solution and once in staining buffer at 4°C. To further separate the BL2 cells from the T cells, the cells were labeled with FITC-conjugated anti-CD3 and phycoerythrin (PE)-conjugated anti-CD19 for 1 hour at 6°C with gentle swirling every 15 min. Then the cells were washed once with magnetic-activated cell sorting (MACS) buffer and incubated in anti-FITC microbeads (Miltenyi Biotec, Auburn, CA) for 15 min at 6°C. Cells were washed and resuspended in MACS buffer before sorting twice with the AUTOMACS apparatus (Miltenyi), using the depletion-sensitive program. The BL2 cell population was analyzed for purity by using the CD19 PE marker.
Semiquantitative Reverse Transcription (RT)-PCR.
Total RNA was isolated with TRIzol (GIBCO/BRL). After quantitation and DNaseI treatment (Roche, Indianapolis), cDNA was synthesized with Superscript preamplification system for first-strand cDNA synthesis with oligo(dT) primers according to the manufacturer's recommendations. cDNA created from 3.5 μg RNA was amplified in PCR reactions with Taq polymerase (Roche). First the cDNA was diluted 10-fold and then at 2-fold dilutions. Amplification was carried out with denaturation at 94°C for 5 min followed by 25 cycles: denaturation at 94°C 30 sec, annealing at 58°C for 30 sec, and strand extension for 2 min at 72°C. The parameters for pol η are 94°C for 45 sec, 60°C for 45 sec, and 72°C for 45 sec. The nucleotide sequences of the primers used are pol ι, forward 5′-AAGGGAAAGGAAGTGTGAGTTGTC-3′ and reverse 5′-TCTGGCTCTCTATTTTCTGTAAGT-3′; pol ζ, forward 5′-GCTCCAGTATGTGTACCATCTTGT-3′ and reverse 5′CATTTTGTGTTCAAGATGATGGC-3′; pol η, forward 5′-CGAAATGATAATGACAGGGTAGCC-3′ and reverse 5′-GGAGCAGTAAGAGATGAAAGCGAAG-3′; pol κ, forward 5′-TAGGAATGGGATAAGAAGGTGAT-3′ and reverse 5′-TGACAAGAAATGAAATACTGCCA-3′; Rev1, forward 5′-GGTATTTGCTGCCCTTCCTGCTGA-3′ and reverse 5′-GCACTTTGCAAATACCTCACAAGCAC-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward 5′-TCCACCACCCTGTTGCTGTAG-3′ and reverse 5′-GACCACAGTCCATGCCATCACT-3′. The products of reactions were separated on 0.8% agarose gels. The PCR products for each enzyme and for GAPDH were amplified for the same number of cycles at the same time and run on a single gel along with GAPDH. The gels were scanned, calibrated, and quantified by using National Institutes of Health image software. Each dilution at each time point was normalized by using the GAPDH amplified from the cDNA at that dilution.
Purification of Pol ι.
A glutathione S-transferase fusion of DNA pol ι was constructed by subcloning the original Rad30B clone (GenBank accession no. AF245438) into the BamHI and EcoRI sites of the pAcG2T Baculovirus expression vector kit from PharMingen. The fusion protein was expressed and purified as published (25) and stored in aliquots at −74°C.
RGYW Primer/Template Construction.
The primer/template oligonucleotides were synthesized on an Applied Biosystems model 392 DNA/RNA synthesizer. The hot-spot templates contained the RGYW motif 5′-AGCT-3′, and non-hot-spot control templates contained the same motif in the opposite polarity 5′-TCGA-3′, with the first templating base from the primer terminus being the central G of the RGYW motif for both hot-spot and non-hot-spot DNAs. The hot-spot primer sequence was 5′-ACTGACCCCGTTAAAACTTATTACCAGTAAG-3′ and the longest 65-mer hot-spot template was 5′-AAAGCGCAGTCTCTGAATTTACCGGTTCCATCAGCTTACTGGTAATAAGTTTTAACGGGGTCAGT-3′. The non-hot-spot primer sequence was 5′-ACTGACCCCGTTAAAACTTATTACCAGTACAGT-3′ and the longest 65-mer non-hot-spot template sequence was 5′-AAAGCGCAGTCTCTGAATTTACCGGTTCCTCGACTGTACTGGTAATAAGTTTTAACGGGGTCAGT-3′. Shorter templates were constructed by leaving off nucleotides on the 5′ end of the 65-mer templates during synthesis, resulting in primer/templates with 1-, 2-, 3-, and 4-nt overhangs on the 5′ end. Single nucleotide-gapped substrates were made by annealing a second 5′ chemically phosphorylated complementary oligonucleotide downstream of the G in the RGYW hot-spot and non-hot-spot 65-mer templates. Primers were 5′ end-labeled with 32P by using T4 polynucleotide kinase in enzyme reaction buffer at 37°C for 60min. Primer/template DNA was annealed in enzyme reaction buffer by using a ratio of 1 primer to 1.2 templates (and 1.44-fold 5′ phosphorylated downstream oligo for gap) by heating to 90°C and gradually cooling to room temperature, resulting in a final primer termini concentration of 100 nM.
Kinetic Analysis of Pol ι Fidelity in RGYW and WYGR Motifs.
A standing-start gel kinetic assay was used to determine the rate of incorporation of each of the four dNTPs opposite the target template G (33, 34). Reactions contained final concentrations of 10 nM primer/template DNA, 40 mM Hepes (pH 7.6), 250 μg/ml BSA, 60 mM KCl, 5 mM MgCl2, 2.5% glycerol, and 10 mM DTT. Reactions contained the following concentrations of glutathione S-transferase–pol ι: 10 nM for 34-nucleotide- and single-nucleotide-gapped DNA, 20 nM for 1–4-nt template non-hot-spot DNA, and 100 nM for 1–4-nt hot-spot templates. Reactions contained varying amounts of each dNTP in the following ranges and reaction times: dCTP (1–20 μM) for 5 min, dTTP (5–200 μM) for 10 min, dGTP (50–800 μM) for 15–30 min, and dATP (50–800 μM) for 10–30 min. The misincorporation specificity (finc) was obtained by measuring the ratio of apparent Vmax/Km values for incorporation of a mismatched (W) vs. a correctly matched (R) base pair opposite a template target base: finc = (Vmax/Km)W/(Vmax/Km)R. The reported values are averages calculated from at least three measurements, and the SE ± 30%.
Results
The Induction of V-Region Mutation in BL2 Cells.
The human BL2 cell line grows continuously in tissue culture and has many of the surface markers of germinal center B cells (32, 35). It produces an IgM λ antibody that is expressed on its surface and secreted into the medium. The V region that is part of the μ heavy-chain gene has been reported to have a low constitutive rate of mutation in BL2 cells (32). The rate of mutation of this endogenous μ V region can be increased 5–10 fold if the surface IgM is crosslinked with antibody and the cells are cocultivated with killed helper T cells (32, 35).
To confirm that our particular version of the BL2 cell line could be induced to undergo V-region hypermutation in culture by crosslinking its surface IgM and cocultivation with the HuT 78 T cell line, which had not been used previously for this purpose, we sequenced unselected μ V regions of stimulated and unstimulated cells. In BL2 cells that were treated only with anti-IgM for four cycles during the course of 14 days, 2 of 27 V regions sequenced had mutations and the overall frequency of mutation was 2.6 × 10−4 (Table 1 14-day coculture). Stimulation with anti-IgM and irradiated HuT 78 helper T cells for four cycles increased the frequency of V-region mutation so that 21/22 V regions sequenced had mutations and the overall frequency was 16.2 × 10−4 (Table 1 14-day coculture). These frequencies are similar to those reported by Denépoux et al. (32, 35). In the cells that were treated with anti-IgM and were cocultivated with T cells as well as the control, the majority of mutations were single-base changes in G or C targeted to an RGYW hot spot (Table 1 14-day coculture). In addition, the sequences revealed an extensive clonal genealogy in the BL2 cells treated with both anti-IgM and T cells, confirming that sequential cycles of mutation were occurring (data not shown). These results showed that the BL2 cells used in these studies are stimulated to hypermutate their endogenous heavy-chain V region by treatment with anti-IgM, mimicking the interaction of antigen with the B cell receptor, and activated HuT 78 CD4-positive T cells.
Table 1.
Sequence data for BL2 Ig V region
| % Sequences mutated | Mutation frequency | Mutation range | Hot spot incidence | |
|---|---|---|---|---|
| 14-day coculture | ||||
| +Anti-IgM | 7% (2/27) | 2.6 × 10−4 (3/11,340) | 0–2 | 2/3 |
| +Anti-IgM + HuT 78 | 95% (21/22) | 16.2 × 10−4 (15/9,240) | 0–4 | 29/40 |
| 3-day coculture | ||||
| Untouched | 12% (4/34) | 3.5 × 10−4 (5/14,280) | 0–2 | 0/5 |
| +HuT 78 | 8% (2/26) | 1.8 × 10−4 (2/10,920) | 0–1 | 1/2 |
| +Anti-IgM + HuT 78 | 66% (22/33) | 6.5 × 10−4 (9/13,860) | 0–2 | 21/26 |
Mutation frequency, number of independent mutations per total bp sequenced. Hot spot incidence, number of RGYW or WRGY mutations per total mutations.
Because the goal of these studies was to determine whether pols ζ, η, ι, and κ and Rev 1 were expressed in association with the onset of mutation in the BL2 cell line, we also examined whether V-region mutation could be detected only after 72 h of stimulation by sequencing unselected V regions. In uninduced BL2 cells that were not treated with either anti-IgM or T cells, 4/34 V regions had mutations but none were in hot spots (Table 1 3-day coculture). Cocultivation with T cells without anti-IgM resulted in mutations in 1/10 V regions. Stimulation with both anti-IgM and T cells for 72 h resulted in mutation in 22/33 V regions sequenced with 21 of the 26 base changes present in hot spots (Table 1 3-day coculture). Although the 5.5–6.6-fold increase in V regions mutated clearly indicates that mutation was underway 72 h after stimulation with anti-IgM and helper T cells, the 2–3-fold difference in the frequency of mutations (Table 1 3-day coculture, column 2) was lower than would be expected from the difference in the number of V regions mutated (Table 1 3-day coculture, column 1). Most of the early mutations were a G to A mutation in the RGYW hot spot at residue 30 in the V region. Because it was not possible to know whether each of these was caused by an independent mutational event, they were scored as only one mutation in Table 1 3-day coculture, artificially lowering the frequency of base changes. There were eight other base changes that also could be scored as independent mutations giving a minimal frequency of 6.5 × 10−4 (Table 1 3-day coculture), with the real frequency of mutation probably being higher. Similar results were obtained in seven independent experiments in which stimulation with anti-IgM and T cells resulted in 60% or more of the V regions having mutations (data not shown). Taken together, these studies show that crosslinking of surface IgM and cocultivation with T cells causes a rapid activation of the mutational process.
Because increases in mutation can be associated with increases in transcription (36), steady-state levels of μ heavy-chain mRNA were measured by using an RNase protection assay in which equal amounts of total cellular RNA were hybridized with a labeled 384-bp probe to the μ mRNA, digested with RNase, and the amounts of protected probe quantified on gels and compared with β-actin mRNA. No differences were detected between uninduced BL2 cells and those that were induced to undergo mutation by crosslinking their surface IgM and cocultivation with T cells (data not shown). This result suggests that in these BL2 cells, an increase in transcription of the heavy-chain mRNA is not required to induce mutation.
Expression of Pols ι, κ, ζ, and η and Rev1 mRNAs.
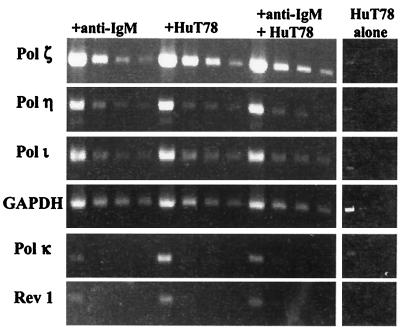
By using human immune tissue blots (CLONTECH), we were able to identify mRNAs of the expected size for pols ι and κ in tissues containing B cells as well as in the thymus (data not shown). These findings suggested that that the mammalian homologues of the UmuC/DinB/Rev1/Rad30 family might be expressed in human germinal center B cells that normally undergo V-region hypermutation. This tissue-expression result led us to use semiquantitative RT-PCR to study their expression in the human BL2 cell line at various times after the induction of somatic mutation. We also examined the expression of pol ζ, which has been suggested by others to play a role in V-region mutation (29, 37). As described in the previous section, BL2 cells were treated with anti-IgM alone, irradiated T cells alone, or with anti-IgM and irradiated T cells. When present, the T cells were removed by magnetic-activated cell sorting (MACS) separation and the B cells were examined by RT-PCR. Fig. 1 shows the semiquantitative RT-PCR products at 48 h. The first lane shows that mRNA for each of the polymerases was detectable in the BL2 cells. However, when the cDNA was diluted 10-fold and then amplified (second lane for each treatment), Rev1 and pol κ became undetectable, and there was no detectable increase in the levels of Rev1 and pol κ mRNA with the time or type of treatment.
Figure 1.
RT-PCR products of error-prone DNA polymerases at 48 h. cDNA from cells that were untreated or treated under various conditions (+ anti-IgM, + Hut 78, or + IgM + HuT 78) for 0, 12, 24, and 48 h were used in RT-PCR. Stock dilutions (1:10, 1:20, and 1:40) of the cDNA were made and used in the amplifications. For each time point and condition, undiluted cDNA and the aforementioned dilutions were used to detect pols ζ, η, and ι, and GAPDH. No PCR products were detected in the no-RT control cDNAs for each condition (data not shown).
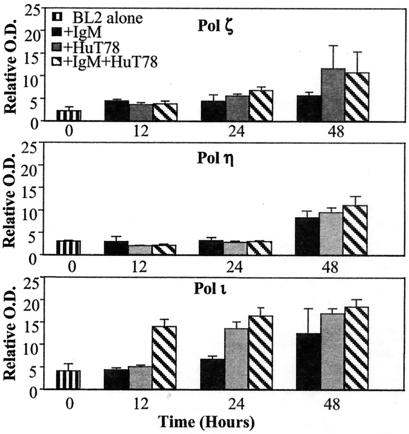
Fig. 2 summarizes the RT-PCR analysis expression of pols ζ, η, and ι at 12, 24, and 48 h after stimulation. Crosslinking surface IgM and cocultivating with T cells, which induces V-region mutation, is associated with a small but progressive increase in the levels of pol ζ from 12 to 48 h so that at 48 h there is 3–4 times as much pol ζ as there was at 0 time. Similar increases are seen with T cells alone, but there is little increase with anti-IgM alone (Fig. 2). Pol η shows no change until 48 h and there is no detectable difference between the different treatments (Fig. 2). Crosslinking surface IgM and cocultivating with T cells causes an approximate 3–4-fold increase in pol ι mRNA within 12 h, and the levels of pol ι remain elevated at 24 and 48 h. Treatment with T cells alone produces a slower increase in pol ι mRNA, whereas anti-IgM results in less of an increase. Reproducible results were obtained in independent experiments that included other time points. The only additional finding was that after 72 h of treatment with anti-IgM or with anti-IgM and T cells, pol η, which had increased at 48 h, returned to the 0 time levels by 72 h (data not shown). Thus, only pol ι mRNA was increased by stimulation with anti-IgM and T cells compared with treatment with either stimulus alone, and that occurred only at 12 h (Fig. 1).
Figure 2.
Semiquantitation of RT-PCR products of error-prone DNA polymerases in BL2. RT-PCR was performed for pols ζ, η, and ι at 0, 12, 24, and 48 h after various conditions. For each time point and condition, RT-PCR was performed for the housekeeping gene, GAPDH. The OD of each band for the polymerases was measured and normalized against the OD of GAPDH at the respective time point and condition. The PCR products from the 1:10 and 1:20 dilutions of the template cDNA were normalized individually to GAPDH. The normalized values of the PCR products of the 1:20 dilution were doubled and then averaged with the 1:10 PCR products. The average is shown in the bar graph, and the error between the two determinations is depicted by the error bars.
It was possible that mRNA for some of the polymerases could have come from T cells that might have contaminated the B cells in those samples treated with T cells alone, or with both T cells and anti-IgM. This possibility seems unlikely because FACS analysis of the BL2 cells that had been separated from the T cells showed no more than a 5–10% contamination with the irradiated killed T cells. In addition, we performed RT-PCR from the equivalent starting amount of activated and irradiated T cell RNA at various times after irradiation for each of the polymerases and found easily detectable levels in the 1/10 diluted cDNAs of only pol ζ (Fig. 1), making it very unlikely that a 5–10% contamination with T cell-derived pol ζ could explain the increases in mRNA for pol ζ or for any of the other polymerases seen in Fig. 2.
Biochemical Activities of Pol ι on RGYW Hot-Spot DNA.
Hypermutation events in our BL2 experiments occurred most frequently at G within the 5′ RGYW hot-spot motif at residue 30 of the heavy-chain V region. Because of its early induction by costimulation, we have analyzed the behavior of pol ι when copying this particular sequence and compared it to the reverse non-hot-spot sequence 5′ WYGR. Reported kinetic studies have measured the fidelity of pol ι on all four bases in a DNA template (30, 38–40), but none of these included an analysis of target sites within the context of a highly targeted SHM RGYW motif. It has been shown that sequence context (e.g., nearest-neighbor base-stacking interactions), can have a strong effect (∼5–50-fold) on misinsertion efficiencies by different DNA polymerases, exclusive of proofreading (41–43). In the RGYW hot spot, AGCT, the target template base G, is flanked by a 5′ A and 3′ C. The non-hot-spot motif TCGA has the opposite polarity 5′–3′ relative to the target G (see Materials and Methods).
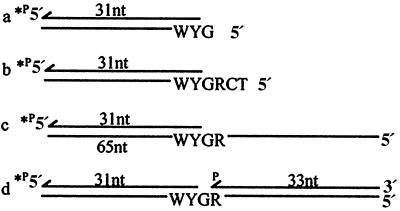
The possible effect of DNA sequence context was examined by placing the target hot-spot and non-hot-spot contexts into six different constructs, four of which are shown in Fig. 3. These constructs are intended to mimic several current break and repair models for hypermutation involving single- or double-stranded breaks occurring at or very near RGYW motifs (13–16). One construct is a 31-nt primer annealed to a 65-nt template strand having a single-stranded region of 34 nucleotides downstream from the RGYW motif (Fig. 3c). A second construct eliminates the downstream single-stranded region and “represents” a double-strand break two nucleotides from the template R followed by limited digestion by a 3′–5′ exonuclease, exposing G, R, and an adjacent C and T as template nucleotides (Fig. 3b). A third construct represents a double-strand break adjacent to template G, eliminating the R of the motif followed by an excision of just one nucleotide, exposing the “hot” nucleotide G at the 5′-template end (Fig. 3a). The fourth construct represents a single-strand nick on the strand opposite template G followed by the excision of one nucleotide, resulting in a single-nucleotide gap with G as the template target site (Fig. 3d).
Figure 3.
DNA primer/templates for fidelity studies. A schematic representation of DNAs used in primer-extension assays are illustrated showing the RGYW motif in the 5′–3′ template hot-spot orientation. Non-hot-spot DNAs were identical except for the reverse polarity of 5′–3′ WYGR, keeping G as the standing-start target site at the end of the primer 3′ terminus.
There is a large difference in primer/template utilization efficiency for the four constructs with dCMP incorporated opposite G, which is nearly 100-fold more efficient with the single nucleotide-gapped structure (kcat/Km = 2.4 × 10−2; Fig. 3d) compared with the single-nucleotide overhang in the double-strand break structure (kcat/Km = 3.0 × 10−4; Fig. 3a; Table 2). The favored misincorporation event is dTMP⋅G on each construct with the finc in a range of 1.8 × 10−1 [single-nucleotide overhang double-strand break structure (Fig. 3a; Table 2)] to 2.4 × 10−2 [34-nt template structure (Fig. 3c; Table 2)]. These are, of course, very high error rates consistent with those required for somatic mutation, but there is little difference in finc values at the hot-spot vs. non-hot-spot G for C and T on two of three break and repair model substrates (Fig. 3 b and d). We also investigated break and repair model substrates containing 5′ CRG and 5′ RG template overhangs, but these also showed no significant difference in finc values when comparing hot- and non-hot-spot sequence contexts (data not shown). The one exception is the double-strand break model template containing a 1-nt template G overhang (Fig. 3a), where the dTMP⋅G error rate is about 3.3-fold higher for the hot-spot G (finc = 1.8 × 10−1; Table 2) compared with the non-hot-spot G (finc = 5.4 × 10−2; Table 2).
Table 2.
Pol ι fidelity at template G in hot- and non-hot-spot DNAs
| P/T DNA | dNTP incorporated opposite G | 5-RGYW-3*
|
5 WYGR-3
|
||
|---|---|---|---|---|---|
| kcat†/Km | finc‡ | kcat†/Km | finc | ||
| 1-nt template | dCTP | 3.0 × 10−4 | 1 | 1.6 × 10−3 | 1 |
| dTTP | 5.5 × 10−5 | 1.8 × 10−1 | 8.7 × 10−5 | 5.4 × 10−2 | |
| dATP | ND | ND | ND | ND | |
| dGTP | ND | ND | ND | ND | |
| 4-nt template | dCTP | 9.8 × 10−4 | 1 | 1.8 × 10−2 | 1 |
| dTTP | 4.5 × 10−5 | 4.5 × 10−2 | 5.1 × 10−4 | 2.8 × 10−2 | |
| dATP | 2.5 × 10−5 | 2.5 × 10−2 | 8.4 × 10−4 | 4.6 × 10−2 | |
| dGTP | 9.2 × 10−6 | 9.3 × 10−3 | 3.6 × 10−5 | 2.0 × 10−3 | |
| 34-nt template | dCTP | 8.7 × 10−3 | 1 | 3.9 × 10−3 | 1 |
| dTTP | 2.1 × 10−4 | 2.4 × 10−2 | 1.4 × 10−3 | 3.5 × 10−1 | |
| dATP | 1.6 × 10−4 | 1.8 × 10−2 | 1.0 × 10−4 | 2.5 × 10−2 | |
| dGTP | 3.0 × 10−5 | 3.4 × 10−3 | 7.6 × 10−6 | 1.9 × 10−3 | |
| 1-nt gap template | dCTP | 2.4 × 10−2 | 1 | 4.7 × 10−2 | 1 |
| dTTP | 2.0 × 10−3 | 8.3 × 10−2 | 6.4 × 10−3 | 1.3 × 10−1 | |
| dATP | 4.4 × 10−4 | 1.8 × 10−2 | 5.7 × 10−3 | 1.2 × 10−1 | |
| dGTP | 7.0 × 10−6 | 2.9 × 10−4 | 2.7 × 10−3 | 5.7 × 10−2 | |
P/T, primer/templates. ND, not determined.
R = A, Y = C, W = T.
kcat/Km in units of min−1 μM−1.
finc = (Vmax/Km)W/(Vmax/Km)R.
We have verified that pol ι prefers to make dGMP⋅T mispairs rather than dAMP⋅T W-C pairs, by a factor of about 10-fold, which is in agreement with reported results (38, 39). A further reduction in fidelity for pol ι when copying single-nucleotide overhangs (e.g., Fig. 3a), was observed (44).
Discussion
The mutational process is thought to be activated in vivo by the combined crosslinking of surface Ig by antigen and the interaction of B cells with helper T cells. Mutation is most active during the centroblast stage of B cell differentiation, which occurs primarily in the germinal centers of secondary lymphoid organs (45). Denépoux et al. (32) have shown that the human BL2 B cell line can be induced to undergo V-region hypermutation in tissue culture by crosslinking its surface IgM with anti-IgM and cocultivating it with helper T cells. This cell line has surface markers that suggest it is a transformed centroblast (32) and thus represents the stage of B cell differentiation that normally undergoes mutation in vivo. The characteristics of the V-region mutation that are induced in BL2 are similar to those that occur in vivo, although, like other cell lines that undergo high rates of V-region mutation in culture, there is a greater predominance of mutations in G and C than is seen in vivo (13, 46–48).
In the studies reported here, we have shown that mRNAs of the human pols ι, κ, ζ, and η, and Rev1 homologues of the UmuC/DinB/Rev1/Rad30 family of polymerases are expressed in normal human B cells and in the BL2 cell line. The strongest relationship to the requirements for turning on SHM was an ∼4-fold increase in the expression of pol ι 12 h after costimulation with both anti-IgM and activated T cells; although by 48 h, this increase also is seen in the BL2 cells that have been treated with anti-IgM, T cells, or both. Pols η and ζ also exhibit modest inductions, but unlike pol ι, their expressions do not seem to be regulated significantly by the unique stimulus required to activate SHM in BL2. Modest changes in expression of pols ζ and η also have been observed by Zan et al. (37) in another Burkitt's lymphoma cell line, CL-01, that can be stimulated to undergo somatic mutation in tissue culture (48). Because pols ι, η, and ζ are present constitutively, regardless of T cells and/or IgM crosslinking, their presence per se is insufficient to generate V-region mutations.
The unique time course of induction for these polymerases may support the notion of two distinct phases targeting first G and C and next A and T in the somatic hypermutation process (17), where pol ι acts early in response to a “triggering” SHM stimuli, followed subsequently by the actions of pols η (19) and/or ζ, possibly in cooperation with MMR proteins. It has also been suggested that pol μ might be involved in V region mutation (28). Pol μ is present in induced and uninduced BL2 cells (data not shown). However, in parallel studies on the Ramos–Burkitt's lymphoma cell line that constitutively undergoes a high rate of mutation, we have identified highly mutating subclones that do not have detectable levels of pol μ mRNA by RT-PCR, whereas other Ramos subclones that do not have detectable V-region mutation express levels of pol μ mRNA comparable to GAPDH (A.M., C.J.W., and M.D.S, unpublished results). We have concluded on the basis of these studies that pol μ is probably not required for V-region mutation in the Burkitt's cell lines and we have not studied expression of its mRNA in further detail.
The two key findings in this study include the rapid induction of pol ι in response to SHM stimulus within 12 h, and the small but distinct preference of pol ι to make 3-fold more dTMP⋅G mismatches in the hot-spot single-nucleotide overhang. This event would lead to a G to A transition in the RGYW motif and a C to T in the complementary strand after the next round of DNA replication, the most commonly observed mutational events within the hot spot. These data support a role for pol ι in the first phase of the proposed two-step model for SHM. The lack of significant bias on the part of pol ι to make more mutations in the other RGYW constructs certainly does not exclude it as a viable candidate. These data may simply reflect that sequence context alone does not make the RGYW mutagenic. A targeting mechanism for SHM may involve a site-specific endonuclease that creates a particular broken DNA structure for subsequent low-fidelity synthesis. We and Woodgate and colleagues (44) find that pol ι is capable of using a variety of broken substrates, including a single-strand break (nick), a single-nucleotide gap, as well as a staggered or resected double-strand break, implying that pol ι has the ability to contribute to SHM at a number of unique DNA intermediates. Although our biochemical approach focused on pol ι, our data and that of others point to the fact that one polymerase alone is not responsible for SHM. Genetic data have implicated the requirement for enhancer-binding elements within the Ig gene as well as MMR proteins and activation-induced cytidine deaminase. The identification of polymerases that are involved is only one step in understanding this complicated and essential biochemical process.
Acknowledgments
We thank Drs. Casali, Gearhart, and Woodgate for allowing us to see their results before publication. This work was supported by grants from the National Institutes of Health (NIH) to V.P., who was supported by Training Grants 5T32CA09173 and CA73649; C.J.W. was supported by Training Grant T32GM 07491; B.T. was supported by NIH Grant T32 AG00093; A.M. was supported by a fellowship from the Cancer Research Institute; M.F.G. was supported by NIH Grants GM4554, GM21422, PO1 AG17179, and ADRC-AG05142; and M.D.S. was supported by NIH Grant CA73649.
Abbreviations
- SHM
somatic hypermutation
- V
variable region
- BL2
Burkitt's lymphoma 2
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- RT
reverse transcription
Footnotes
References
- 1.Weigert M G, Cesari I M, Yonkovich S J, Cohn M. Nature (London) 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 2.Harris R S, Kong Q, Maizels N. Mutat Res. 1999;436:157–178. doi: 10.1016/s1383-5742(99)00003-4. [DOI] [PubMed] [Google Scholar]
- 3.Winter D B, Sattar N, Gearhart P J. Curr Top Microbiol Immunol. 1998;229:1–10. doi: 10.1007/978-3-642-71984-4_1. [DOI] [PubMed] [Google Scholar]
- 4.Rothenfluh H S, Gibbs A J, Blanden R V, Steele E J. Proc Natl Acad Sci USA. 1994;91:12163–12167. doi: 10.1073/pnas.91.25.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rada C, Yelamos J, Dean W, Milstein C. Eur J Immunol. 1997;27:3115–3120. doi: 10.1002/eji.1830271206. [DOI] [PubMed] [Google Scholar]
- 6.Jolly C J, Wagner S D, Rada C, Klix N, Milstein C, Neuberger M S. Semin Immunol. 1996;8:159–168. doi: 10.1006/smim.1996.0020. [DOI] [PubMed] [Google Scholar]
- 7.Storb U. Immunol Rev. 1998;162:5–11. doi: 10.1111/j.1600-065x.1998.tb01424.x. [DOI] [PubMed] [Google Scholar]
- 8.Tumas-Brundage K, Vora K A, Giusti A M, Manser T. Semin Immunol. 1996;8:141–150. doi: 10.1006/smim.1996.0018. [DOI] [PubMed] [Google Scholar]
- 9.Kelsoe G. Semin Immunol. 1996;8:179–184. doi: 10.1006/smim.1996.0022. [DOI] [PubMed] [Google Scholar]
- 10.Dorner T, Foster S J, Brezinschek H-P, Lipsky P E. Immunol Rev. 1998;162:161–171. doi: 10.1111/j.1600-065x.1998.tb01439.x. [DOI] [PubMed] [Google Scholar]
- 11.Milstein C, Neuberger M S, Staden R. Proc Natl Acad Sci USA. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner S, Milstein C. Nature (London) 1966;211:242–243. [Google Scholar]
- 13.Sale J E, Neuberger M S. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- 14.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- 15.Papavasiliou F N, Schatz D G. Nature (London) 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 16.Kong Q, Maizels N. Genetics. 2001;158:369–378. doi: 10.1093/genetics/158.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rada C, Ehrenstein M R, Neuberger M S, Milstein C. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 18.Wiesendanger M, Kneitz B, Edelmann W, Scharff M D. J Exp Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng, X., Winter, D. B., Kasmer, C., Kraemer, K. H., Lehman, A. R. & Gearhart, P. J. (2001) Nat. Immunol.2, in press. [DOI] [PubMed]
- 20.Wiesendanger M, Scharff M D, Edelmann W. Cell. 1998;94:415–418. doi: 10.1016/s0092-8674(00)81581-0. [DOI] [PubMed] [Google Scholar]
- 21.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 22.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 23.Poltoratsky V, Goodman M F, Scharff M D. J Exp Med. 2000;192:F27–F30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman M F, Tippin B. Nat Rev Mol Cell Biol. 2000;1:101–109. doi: 10.1038/35040051. [DOI] [PubMed] [Google Scholar]
- 25.Tissier A, McDonald J P, Frank E G, Woodgate R. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson R E, Washington M T, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1999;96:12224–12226. doi: 10.1073/pnas.96.22.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tippin B, Goodman M F. Philos Trans R Soc London B Biol Sci. 2001;356:47–51. doi: 10.1098/rstb.2000.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominguez O, Ruiz J F, Lain de Lera T, Garcia-Diaz M, Gonzalez M A, Kirchhoff T, Martinez A C, Bernad A, Blanco L. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz M, Flajnik M F. Immunol Rev. 1998;162:13–24. doi: 10.1111/j.1600-065x.1998.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 30.Johnson R E, Washington M T, Haracska L, Prakash S, Prakash L. Nature (London) 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 31.Poltoratsky V, Goodman M F, Scharff M D. J Exp Med. 2000;192:F27–F30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denépoux S, Razanajaona D, Blanchard D, Meffre G, Capra J D, Banchereau J, Lebecque S. Immunity. 1997;6:35–46. doi: 10.1016/s1074-7613(00)80240-x. [DOI] [PubMed] [Google Scholar]
- 33.Boosalis M S, Petruska J, Goodman M F. J Biol Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 34.Creighton S, Bloom L B, Goodman M F. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 35.Denépoux S, Fournier N, Peronne C, Banchereau J, Lebecque S. J Immunol. 2000;164:1306–1313. doi: 10.4049/jimmunol.164.3.1306. [DOI] [PubMed] [Google Scholar]
- 36.Fukita Y, Jacobs H, Rajewsky K. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 37.Zan H, Komori A, Li Z, Cerrutti M, Flajnik M F, Diaz M, Casali P. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tissier A, Frank E G, McDonald J P, Iwai S, Hanaoka F, Woodgate R. EMBO J. 2000;19:5259–5266. doi: 10.1093/emboj/19.19.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Yuan F, Wu X, Wang Z. Mol Cell Biol. 2000;20:7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bebenek K, Tissier A, Frank E G, McDonald J P, Prasad R, Wilson S H, Woodgate R, Kunkel T A. Science. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 41.Petruska J, Goodman M F. J Biol Chem. 1985;260:7533–7539. [PubMed] [Google Scholar]
- 42.Mendelman L V, Boosalis M S, Petruska J, Goodman M F. J Biol Chem. 1989;264:14415–14423. [PubMed] [Google Scholar]
- 43.Bloom L B, Otto M R, Beechem J M, Goodman M F. Biochemistry. 1993;32:11247–11258. doi: 10.1021/bi00092a039. [DOI] [PubMed] [Google Scholar]
- 44.Frank E G, Tissier A, McDonald J P, Rapic-Otrin V, Zeng X, Gearhart P J, Woodgate R. EMBO J. 2001;20:2914–2922. doi: 10.1093/emboj/20.11.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Przylepa J, Himes C, Kelsoe G. Curr Top Microbiol Immunol. 1998;229:85–104. doi: 10.1007/978-3-642-71984-4_8. [DOI] [PubMed] [Google Scholar]
- 46.Green N S, Lin M M, Scharff M D. Immunol Rev. 1998;162:77–87. doi: 10.1111/j.1600-065x.1998.tb01431.x. [DOI] [PubMed] [Google Scholar]
- 47.Bachl J, Wabl M. Proc Natl Acad Sci USA. 1996;93:851–855. doi: 10.1073/pnas.93.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. J Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]