Abstract
EMBO J (2013) 32 1, 45–59 doi:; DOI: 10.1038/emboj.2012.306; published online November 23 2012
Sequence-specific DNA-binding transcription factors (TFs) bind throughout the genome to orchestrate the process of adipocyte differentiation in an exquisitely timed and precise manner. TF binding recruits histone-modifying enzymes, resulting in dynamic epigenomic modifications that correlate with extensive gene expression changes. In this issue of The EMBO Journal, Wang et al (2012) demonstrate a critical role for the SET-containing histone methyltransferase G9a in generating repressive chromatin marks that are lost in the process of adipogenesis.
Adipocytes are central to energy homeostasis and their dysfunction is associated with metabolic diseases such as obesity. Understanding the mechanisms driving adipocyte differentiation, also known as adipogenesis, has become increasingly important as these diseases reach epidemic levels. Extensive work has revealed that this process is directed during both white and brown adipocyte differentiation by a highly regulated transcriptional cascade, culminating in the robust expression of the master adipocyte regulator PPARγ during the terminal differentiation stage of adipogenesis (Cristancho and Lazar, 2011). PPARγ combines with C/EBPα and C/EBPβ to localize to enhancer regions neighbouring genes induced during differentiation (Lefterova et al, 2008). These enhancer regions are characterized by increased DNAse I hypersensitivity or are enriched for histone modifications associated with increased transcription, such as Histone 3 Lysine 9 acetylation (H3K9ac) or H3K27ac (Mikkelsen et al, 2010; Steger et al, 2010; Siersbæk et al, 2011).
The current study from Ge and colleagues provides evidence of transcriptional repression of PPARγ in the preadipocyte (Wang et al, 2012). In contrast to activating histone modifications that co-localize to common enhancer regions, they show that in precursor cells the repressive histone modifications H3K9me2 and H3K27me3 do not overlap and H3K9me2 is specifically located throughout the entire PPARg gene body and its enhancer regions. H3K9me2 levels decrease globally during differentiation and are undetectable at the PPARg locus of mature adipocytes, suggesting an epigenomic mechanism for silencing the PPARg locus in preadipocytes that is alleviated as transcriptional activators work in concert to promote PPARγ expression during adipogenesis (Figure 1).
Figure 1.
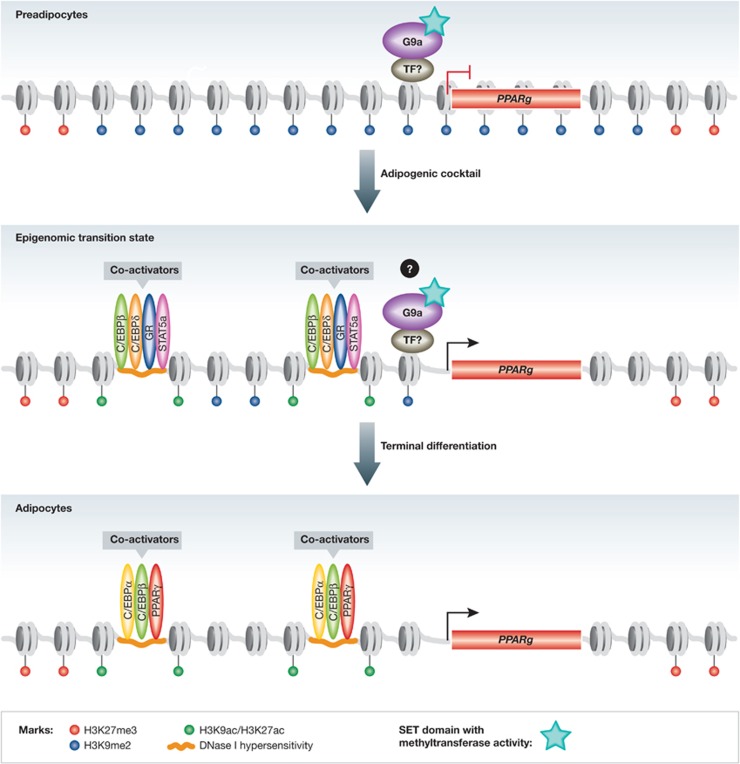
G9a represses PPARγ levels in preadipocytes. In preadipocytes, the repressive H3K27me3 mark surrounds the PPARg locus, but Wang et al have shown that H3K9me2 is a repressive mark found throughout this region. G9a, an SET domain containing histone methyltransferase, is required for the deposition of H3K9me2 at this locus. However, the sequence-specific TF(s) that localize it to the genome are unknown. During early differentiation, as G9a levels begin to decrease and H3K9me2 levels also diminish at the PPARg locus, transcriptional activators of adipogenesis (C/EBPβ, C/EBPδ, GR and STAT5a) are recruited to enhancers at hotspots of open chromatin and epigenomic change, driving the increase in PPARγ levels. The exact temporal relation of changes in H3K9me2 levels to the epigenomic transition state remains to be determined. In mature adipocytes, the major sequence-specific positive regulators of adipogenesis (PPARγ, C/EBPα and C/EBPβ) bind enhancers characterized as regions with DNAse I hypersensitivity and histone modifications of active transcription, like H3K9ac and H3K27ac, H3K9me2 is no longer present and robust PPARγ expression is sustained.
This novel layer of regulation is significant because, while histones throughout the genome are extensively modified during adipocyte differentiation, perhaps no other locus is more extensively regulated than PPARg (Mikkelsen et al, 2010; Steger et al, 2010; Siersbæk et al, 2011). Indeed, Wang et al report that H3K9me2 is mostly absent from loci of other positive regulators of adipogenesis. Previous studies have revealed that an epigenomic transition state is initiated within hours of adding an adipogenic cocktail to preadipocytes. During this period early adipogenic transcription factors (TFs), such as glucocorticoid receptor (GR), C/EBPβ, C/EBPδ and STAT5a, bind multiple functional enhancers upstream of the PPARg transcription start site, opening chromatin, recruiting co-activators, and generating activating histone modifications (Mikkelsen et al, 2010; Steger et al, 2010; Siersbæk et al, 2011). The time course of H3K9me2 disappearance at the PPARg locus in the context of the previously described epigenomic transition state could provide insight into signals governing the switch between gene repression and activation.
Key to understanding the role of H3K9me2 in preadipocytes is determining how this mark is deposited in a loci-specific manner. Ge and colleagues demonstrate that G9a is likely the histone methyltransferase that catalyses the formation of H3K9me2 in adipogenic cells. It is present at the PPARg locus in preadipocytes, maintains low PPARγ levels in both white and brown precursor cells and decreases during differentiation. Furthermore, knockdown, deletion or chemical inhibition of this enzyme promotes adipogenesis. G9a, however, does not itself bind DNA in a sequence-specific manner, and thus it will be critical to determine the sequence-specific TFs that recruit it to the PPARg locus in preadipocytes (Shinkai and Tachibana, 2011). Candidates would include multiple TFs that have recently been identified as necessary for maintaining adipogenic competency in precursor cells (Seale et al, 2007; Gupta et al, 2010; Cristancho et al, 2011).
Intriguingly, Wang et al (2012) also demonstrate that an intact SET domain is not required for G9a to promote expression of Wnt10a, a canonical Wnt ligand that has been shown to inhibit differentiation. Many chromatin-modifying enzymes have been implicated in adipogenesis, but in some cases conflicting studies have mired our understanding of their role (Cristancho and Lazar, 2011). Potentially, one explanation for the contradictory roles of these enzymes is that histone modifiers have both enzyme-dependent and -independent roles. Future studies that tease out these distinct mechanisms could mark a new set of targeted therapeutics for metabolic disease.
Footnotes
The authors declare that they have no conflict of interest.
References
- Cristancho AG, Lazar MA (2011) Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 12: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Schupp M, Lefterova MI, Cao S, Cohen DM, Chen CS, Steger DJ, Lazar MA (2011) Repressor transcription factor 7-like 1 promotes adipogenic competency in precursor cells. Proc Natl Acad Sci USA 108: 16271–16276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM (2010) Transcriptional control of preadipocyte determination by Zfp423. Nature 464: 619–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Liu XS, Lazar MA (2008) PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 22: 2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143: 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM (2007) Transcriptional control of brown fat determination by PRDM16. Cell Metab 6: 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M (2011) H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 25: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R, Nielsen R, John S, Sung M-H, Baek S, Loft A, Hager GL, Mandrup S (2011) Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J 30: 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Lazar MA (2010) Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 24: 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu S, Lee J-E, Baldridge A, Grullon S, Peng W, Ge K (2012) Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J 32: 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]


