Abstract
Mesenchymal stem cells (MSCs) represent one of the most promising stem cells for a number of degenerative conditions due to their multipotency, immunoprivileged properties, and easy expansion in vitro. However, the limited life span of primary MSCs during in vitro expansion greatly hampers their use in clinical applications and basic research. Immortalization of MSCs will overcome this problem and may provide a very useful tool with which to study MSC biology. Here we showed that silencing p53 expression with lentivirus-mediated small interfering RNA delayed the senescence by extended passage number, but was not sufficient to immortalize primary MSCs. However, combination of p53 knockdown and human telomerase reverse transcriptase (hTERT) overexpression was sufficient to immortalize MSCs. The effects of p53 knockdown and hTERT overexpression on MSCs, including proliferation, colony formation, and differentiation, were determined. The resultant immortal MSCs displayed similar surface antigen profile to primary MSCs and retained MSC differentiation potential. Gene expression profile showed high similarity between immortalized MSCs and primary MSCs. In addition, immortalization-associated genes were also identified. Our data suggested immortalization of MSCs related to upregulation of cell cycle regulator and DNA repair genes enabling them to bypass cell crisis and complete mitosis. This study provides a new cellular model for basic studies of MSCs and understanding of the molecular basis of MSC immortalization.
Introduction
Mesenchymal stem cells (MSCs) are multipotent stem cells present in several tissues, including bone marrow, adipose fat tissue, and many other tissues of mesenchymal origin. So far, MSCs are characterized through a panel of cell surface antigens and their differentiation potential into mesenchymal lineages. Like other adult stem cells, each cell division of MSCs is associated with shortening of telomeres during in vitro expansion. MSCs divide only a finite number of times in culture before going to the replicative senescence. At passage 7 to 12, MSCs appear enlarged, with lower expression of certain cell surface markers, leading to arrest of proliferation [1]. Such extended culture also limits differentiation potential [2]. These greatly compromise the benefit of human MSCs for therapeutic application. Thus, identifying methods for immortalizing MSCs and understanding the molecular basis underlying MSC immortalization have attracted great research interest.
Although spontaneous transformation of adult human MSCs was reported [3], it is not a commonly described phenomenon. It was shown that ectopic expression of human telomerase reverse transcriptase (hTERT), the catalytic component of telomerase, led to telomere elongation and extended life span in a number of cell types [4–6]. Overexpression of hTERT enhanced stem-like properties and inhibited spontaneous differentiation of hMSCs [7]. However, whether overexpression of hTERT alone is sufficient to immortalize human MSCs is a matter of controversy [8–11]. Our data showed that overexpression of hTERT alone was not sufficient to immortalize MSCs, which was consistent with previous reports [8,9]. However, the use of viral genes to prolong life span of MSCs may pose safety issues. The use of viral vectors has potential problems depending on the pathogenicity, toxicity, and stability of the virus. Viruses used as vectors should be modified so that they are unable to replicate after gene transfection. Lentivirus has been found to be fairly safe and some clinical trials have been approved for its use. However, to avoid potential problems with viral vectors, nonviral transfection, if effective, would be desirable. In this study, we determined the conditions for immortalizing MSCs by modulating human genes without the use of viral genes.
p53 plays an important role in cell cycle control. Deficiency in p53 induced transformation of MSCs [12]. Although combination of p53 knockdown and hTERT overexpression was sufficient to immortalize primary human ovarian surface epithelial cells [13], little is known about this combination in MSCs. So far, the mechanisms of MSC transformation and immortalization remain poorly understood. Aerobic glycolysis was not an intrinsic component of the transformation of adult stem cells [14]. In this study, our data showed that combination of p53 knockdown and TERT was able to immortalize MSCs. Furthermore, immortalization-related genes were also determined. This study provides a model system without viral genes to immortalize MSCs and to understand the molecular basis underlying immortalization of MSCs.
Materials and Methods
MSC culture and osteoblast, chondrocyte, and adipocyte differentiation
Human bone marrow-derived MSCs were harvested from the iliac crest after informed written consent and approval from Institutional Review Board of the National University Hospital of Singapore (NUS-IRB) and cultured as described [15]. To prevent spontaneous differentiation, cells were maintained at subconfluent levels. MSCs were induced to differentiate toward adipocytes for 14 days in adipogenic medium and osteoblasts for 14 days in osteogenic medium as described [16]. Pellet culture system as described [16,17] was used for chondrocyte differentiation for 28 days. Adipogenic medium contained 0.5 mM isobutyl-methylxanthine, 1 μM dexamethasone (Sigma), 10 μM insulin, 200 μM indomethacin, and 1% antibiotic/antimycotic. Osteogenic medium contained 0.1 μM dexamethasone, 50 μM ascorbate-2-phosphate, 10 mM β-glycerophosphate, and 1% antibiotic/antimycotic. Chondrogenic medium contained 10 ng/mL transforming growth factor (TGF)-β3, 10−7 M dexamethasone, 50 μg/mL ascorbate-2-phosphate, 40 μg/mL proline, 100 μg/mL pyruvate, and 50 mg/mL ITS+Premix (Becton Dickinson; 6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 μg/mL selenious acid, 1.25 mg/mL bovine serum albumin (BSA), and 5.35 mg/mL linoleic acid). The medium was replaced every 3–4 days. Differentiation of MSCs was evaluated by real-time reverse-transcription (RT)-polymerase chain reaction (PCR) and staining. Oil red O stain for lipoid deposits in adipogenesis, Alizarin red S stain for calcium deposits and AP stain for alkaline phosphatase in osteogenesis, Type II collagen immunostain for major collagen of cartilage, and alcian blue stain for cartilage proteoglycans in chondrogenesis were used in this study.
Stable knockdown of p53 and overexpression of hTERT in MSCs
Inducible lentiviral plasmids pLVTH-siP53 and pLV-tTRKRAB for knockdown of p53 and retroviral plasmid pBabe-hygro-hTERT for overexpression of hTERT were from Addgene.
Lentiviral plasmids were cotransfected with packaging vectors (Invitrogen) into 293FT cells. Retrotiviral plasmids were cotransfected with the envelope glycoprotein expression vector pVSV-G into GP2-293 cells. pLVTH-siGFP and pLV-tTRKRAB or pBabe-hygro were used as control. The supernatant was harvested after 48 h. MSCs were infected with viral supernatant containing 8 μg/mL polybrene to achieve stable knockdown or overexpression.
Real-time RT-PCR and RT-PCR
To quantify effect of MSC differentiation, quantitative real-time PCR was performed with TaqMan expression assay using ABI PRISM 7900HT sequence Detection System (Applied Biosystems). Briefly, 0.3 μg of total RNA was converted to cDNA using high-capacity cDNA archive kit in 30 μL and then diluted to 300 μL. Quantitative real-time PCR was done as follows: initial denaturation for 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of PCR (95°C for 15 s, 60°C for 1 min) using 5 μL of 2× Master mix, 0.5 μL of TaqMan probe, and 4.5 μL of cDNA. All probes were designed with a 5′ fluoregenic 6-carboxylfluorescein, and a 3′ quencher, tetramethyl-6-carboxyrhodamine. The expression of human GAPDH was used to normalize gene expression levels.
To validate microarray data, RT-PCR was performed, 1 μg total RNA was reverse transcribed into cDNA using SuperScript II reverse transcriptase (Invitrogen) for subsequent amplification.
Western blot analysis
Cells were collected by centrifugation, and cell pellet was resuspended in lysis buffer (25 mM Tris, pH7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) containing proteinase inhibitors and incubated on ice for 30 min. After centrifugation at 16,000 g for 15 min at 4°C, the supernatant containing total cell extract was collected and kept at −80°C. Protein from cell extracts in the gel was electrophoretically transferred onto a Hybond-PVDF membrane (Amersham Biosciences). The membrane was incubated in blocking buffer (TBS-T containing 5% skim milk) for 1 h at room temperature to block nonspecific protein binding and then incubated with primary antibody against human p53 (Santa Cruz) diluted (1:200) in blocking buffer for 1h at room temperature. After 4 washes with TBS-T, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody diluted (1:3000) in blocking buffer for 1h. Antibody binding was visualized with an ECL western blotting detection system (Amersham Biosciences).
Cell proliferation analysis
To determine the effects of p53 knockdown or hTERT overexpression or combination on MSC proliferation rate, 1×104 cells were plated in a 6-well plate in duplicate, cell proliferation was determined by counting cells with a hematometer at day 6 compared with control MSCs.
Colony-forming unit-fibroblast assay
To determine the effects of p53 knockdown or hTERT overexpression or combination on colony formation of MSCs, 500 MSCs were seeded into 10-cm-diameter dishes in triplicate. The colonies were counted at day 14 after Giemsa stain.
Fluorescence-activated cell sorting
The cells were harvested in 0.25% trypsin/ethylenediaminetetraacetic acid and washed with phosphate-buffered saline (PBS), and then incubated for 30 min in dark in fluorescence-activated cell sorting staining buffer (PBS with 3% FBS and 0.05% sodium azide) containing phycoerythrin (PE)-conjugated antibodies against the following surface antigens: CD34, CD45, CD29, CD44, CD73, CD90, CD105, and CD151. Cells were washed and resuspended in sorting buffer (PBS with 0.1% BSA) for analysis. Cells were stained with PE-conjugated nonspecific IgG to assess background fluorescence.
Senescence-associated β-galactosidase activity assay
The assay is based on detection of β-galactosidase at pH6 with senescence β-galactosidase staining kit (Cell signaling technology). Cells were washed once with PBS and fixed in the fixative solution, and then incubated in stain solution overnight.
Tumorigenicity assay
Immunodeficient nude mice were maintained in pathogen-free conditions. Immortalized MSCs were harvested by trypsinization and washed twice with PBS, and viable cell number was determined by trypan blue exclusion. About 3×106 immortalized MSCs were transplanted subcutaneously into the flanks of 6-week-old nude mice; 6 mice were performed. Mice were observed for 12 weeks to monitor tumorigenic growth.
cDNA microarray analysis
To compare gene expression profile between p53 knockdown or immortalized MSCs and primary MSCs, microarray analyses were performed by Illumina. Total RNA was isolated using RNeasy mini-kit (Qiagen) per the manufacturer's protocol. In brief, 0.5 μg total RNA was used to synthesize cRNA (Illumina TotalPrep RNA amplification kit; Ambion). The data were analyzed using Software Genespring V11. A t-test 0.05 on normalized intensity followed by ratio change (ratio of normalized intensity ≥2 or ≤−2) was used to generate the gene list with significant change in gene expression profile. In this study, MSCs from 3 different patients were used. We have deposited the raw data at GEO under accession number GSE28546, and we can confirm all details are MIAME-compliant.
Statistical analysis
Values are expressed as average and standard deviations. The probability associated with a Student's test was performed for comparisons of the parameters between 2 groups. P-values less than 0.05 were considered statistically significant.
Results
Silencing p53 in MSCs by lentivirus-mediated siRNA
p53 is an important general regulator that regulates various gene networks, including cell cycle [18], which is expressed at low level in normal cells. To determine the effects of p53 knockdown on MSCs, MSCs were infected by lentivirus containing p53 shRNA to stably knockdown p53 expression. Compared with control MSCs infected with GFP shRNA, p53 expression was reduced by 95% in p53 shRNA-infected MSCs in RNA level detected by quantitative real-time PCR (Fig. 1A). In protein level, p53 was detected in control MSCs but was not detected in p53 knockdown MSCs (Fig. 1B), suggesting that p53 is efficiently knocked down by specific shRNA against p53.
FIG. 1.
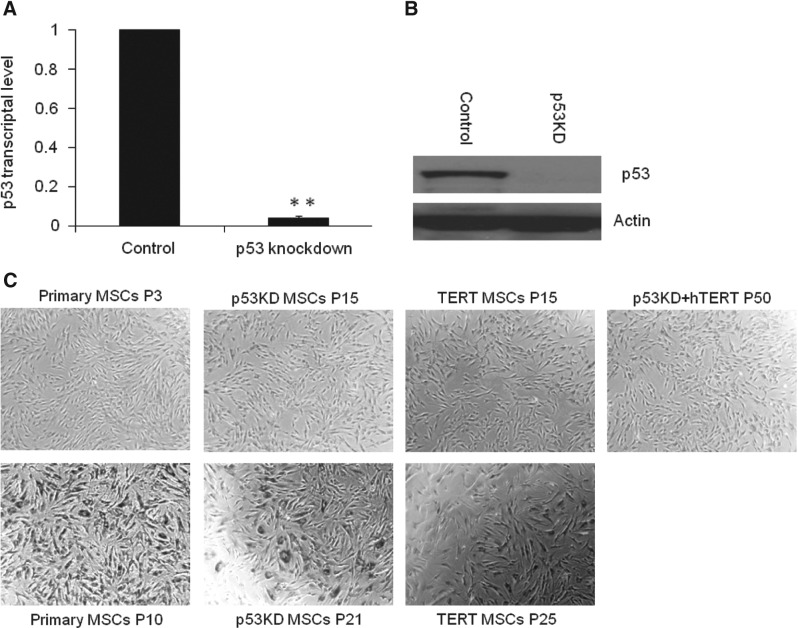
Combination of p53 knockdown and human telomerase reverse transcriptase (hTERT) overexpression is sufficient to immortalize mesenchymal stem cells (MSCs). (A) p53 knockdown in MSCs. Expression of p53 was efficiently knocked down by inducible lentiviral-mediated shRNA against p53 compared with siGFP control. Bars show the mean±SD fold change in transcript expression in p53 knockdown MSCs from 2 patients, tested twice, relative to siGFP control (arbitrarily set at 1, **P<0.01). (B) Western blotting showing p53 knockdown in p53 knockdown MSCs. (C) Combination of p53 knockdown and hTERT overexpression was sufficient to maintain MSC morphology. MSCs by p53 knockdown and hTERT overexpression retained MSC morphology and were negative for β-gal staining at passage 50, whereas p53 knockdown or hTERT overexpression MSCs lost MSC morphology and were positive for β-gal staining at passages 21 and 25, respectively, suggesting that MSCs were immortalized by p53 knockdown and hTERT overexpression.
Extension of life span of primary MSCs using lentivirus-mediated siRNA against p53 and hTERT overexpression
To examine the effect of p53 knockdown (p53 KD) on life span of MSCs, we generated p53 knockdown MSCs from patients. Control MSCs underwent senescence at passage 10, whereas p53 knockdown MSCs retained MSC morphology at passage 15, suggesting that p53 knockdown is able to delay the senescence of MSCs. However, p53 knockdown MSCs began to lose MSC morphology and were positive for β-gal staining, indicating senescence at passage 21 (Fig. 1C), suggesting that p53 knockdown alone is not sufficient to immortalize MSCs. Similarly to hTERT, overexpression of hTERT alone by retrovirus was not sufficient to immortalize MSCs. hTERT-MSCs began to lose MSC morphology and were positive for β-gal staining at passage 25 (Fig. 1C). These data show that p53 knockdown alone or hTERT overexpression alone is able to prolong life span of MSCs but is not sufficient to immortalize MSCs.
Effects of p53 knockdown and hTERT overexpression on proliferation, colony formation, and differentiation of MSCs
To determine the effects of p53 knockdown and hTERT overexpression on MSCs, we examined their effects on proliferation, colony formation, and differentiation of MSCs. Our data showed that p53 knockdown alone or combination of p53 knockdown and hTERT overexpression promoted proliferation of MSCs, whereas there was no significant difference in proliferation between hTERT overexpression alone and control MSCs (Fig. 2A). In colony formation, p53 knockdown alone or combination of p53 knockdown and hTERT overexpression enhanced colony formation of MSCs, whereas hTERT alone showed no significant difference from control MSCs (Fig. 2B).
FIG. 2.
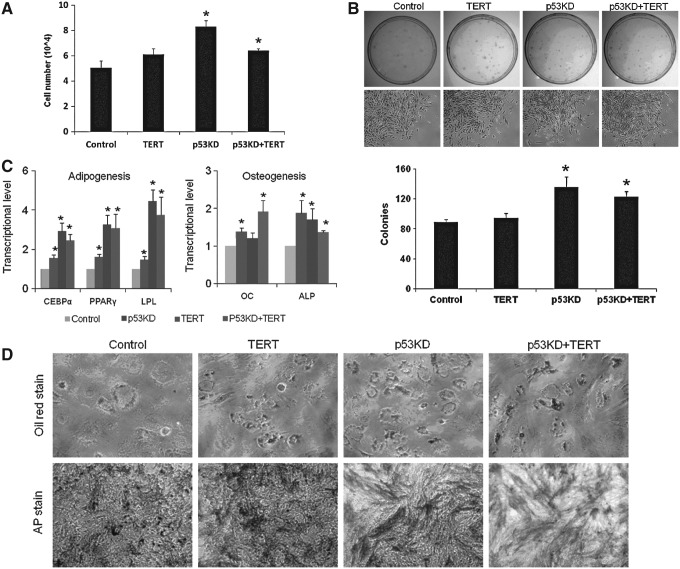
Effects of p53 knockdown and hTERT overexpression on proliferation, colony formation, and differentiation of MSCs. (A) Effects of p53 knockdown and hTERT overexpression on proliferation of MSCs. Bars show the mean±SD cells from 2 patients, tested twice. Asterisks indicate a significant difference compared with control MSCs, showing that p53 knockdown alone or combination of p53 knockdown and hTERT overexpression promoted MSC proliferation, whereas there was no significant difference in proliferation between hTERT-MSCs and control MSCs at passage 5 (*P<0.05). (B) Effects of p53 knockdown and hTERT overexpression on colony formation of MSCs. About 500 MSCs at passage 5 were seeded in 10-cm dishes; the colonies were counted after Giemsa stain 14 days after seeding. Asterisks indicate a significant difference compared with control MSCs, showing that p53 knockdown alone or combination of p53 knockdown and hTERT overexpression enhanced colony formation of MSCs (*P<0.05). (C) Effects of p53 knockdown and hTERT overexpression on adipogenic markers and osteogenic markers during differentiation of MSCs at passage 5 by quantitative real-time polymerase chain reaction (PCR). Bars show the mean±SD fold change in transcript expression in MSCs from 2 patients, tested twice, relative to control MSC (arbitrarily set at 1, *P<0.05). (D) p53 knockdown and hTERT overexpression enhanced oil red stain for lipoid deposits in adipogenesis and AP stain for alkaline phosphatase in osteogenesis of MSCs at passage 5.
Differentiation potential is the most important property of MSCs. Considering adipogenesis, compared with control, p53 knockdown alone or combination of p53 knockdown and hTERT overexpression enhanced adipogenic markers (Fig. 2C) and oil red stain for lipid deposits (Fig. 2D), suggesting that p53 knockdown and hTERT overexpression improves adipogenesis of MSCs. In osteogenesis, our findings showed that p53 knockdown alone or combination of p53 knockdown and hTERT or hTERT improved osteogenesis shown by enhanced osteogenic markers (Fig. 2C) and alkaline phosphatase activity (Fig. 2D) compared with control. Our studies in human were consistent with studies in mouse [19]. Similar to p53 knockdown, hTERT overexpression improved osteogenesis of human MSCs, which was consistent with previous studies [20], suggesting that p53 knockdown and hTERT overexpression improve osteogenesis of MSCs. The above data demonstrate that p53 knockdown or hTERT overexpression affects properties of MSCs.
Immortalization of hMSCs by combination of p53 knockdown and hTERT overexpression and characterization of immortalized MSCs
Telomerase activity is not detected or telomerase is expressed at low level in hMSCs. Similar to nonstem cells, MSCs' telomere shortens at similar rate and MSCs cease to divide when the telomere length is less than 10 kb [2]. Although ectopic expression of hTERT has been proven to extend life span [5,6], our data and previous studies [8,9] showed that overexpression of TERT alone is not sufficient to immortalize MSCs. When MSCs were infected with virus for p53 knockdown and TERT overexpression, infected MSCs maintained MSC morphology similar to primary MSCs after 1 year of continuous culture, suggesting that cells were immortalized by combination of p53 knockdown and TERT overexpression. To characterize immortalized cells, surface antigen profile analysis was performed. Immortalized cells displayed similar surface antigen profile to parental primary MSCs, which are negative for CD34 and CD45, but positive for CD29, CD44, CD73, CD90, CD105, and CD151 (Fig. 3A, Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd).
FIG. 3.
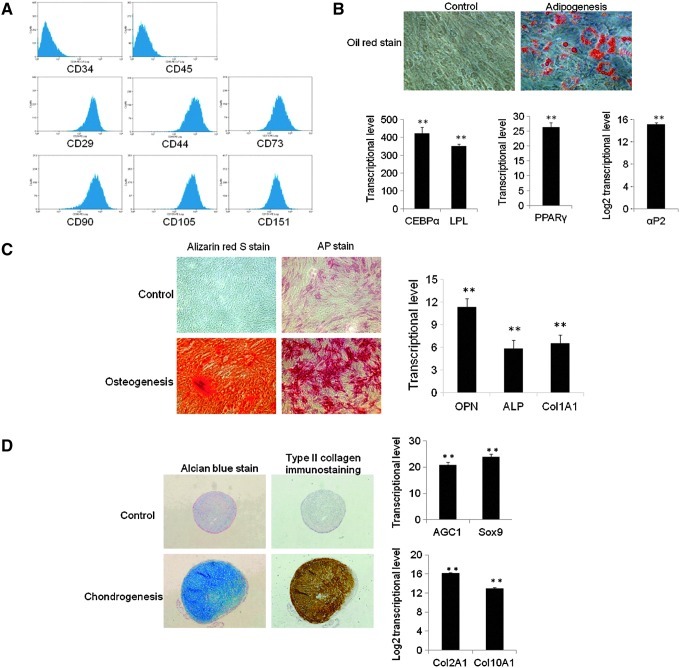
Characterization of immortalized MSCs. (A) Immortalized cells displayed surface antigen profile of MSCs. (B) Immortalized cells had adipogenic differentiation potential. Bars show the mean±SD fold change in transcript expression in MSCs from 2 patients, tested twice, relative to control MSCs in growth medium (arbitrarily set at 1). Immortalized MSCs were positive for oil red stain for lipoid deposits; adipogenic markers were uprgulated after 2 weeks of adipogenesis (**P<0.01). (C) Immortalized cells had osteogenic differentiation potential. Bars show the mean±SD fold change in transcript expression in MSCs from 2 patients, tested twice, relative to control MSCs in growth medium (arbitrarily set at 1). Immortalized MSCs were positive for alizarin red stain for calcium deposit and AP stain for alkaline phosphastase; osteogenic markers were upregulated after 2 weeks of osteogenesis (**P<0.01). (D) Immortalized cells had chondrogenic differentiation potential. Bars show the mean±SD fold change in transcript expression in MSCs from 2 patients, tested twice, relative to control MSCs in growth medium (arbitrarily set at 1). Immortalized MSCs were positive for alcian blue stain for cartilage proteoglycans and type II Collagen immunostaining for major collagen of cartilage; chondrogenic markers were upregulated after 4 weeks of chondrogenesis under pellet culture (**P<0.01).
The most important property of MSCs is their multilineage differentiation potential. To examine differentiation potential of immortalized cells after long-term culture, immortalized cells at passage 35 were induced into mesodermal lineage differentiation in differentiation medium. Considering adipogenesis, immortalized cells were positive for oil red stain after 2 weeks of induction in adipogenic medium. Consistently, adipogenic master regulators CCAAT/enhancer-binding protein alpha (CEBPα) and peroxisome proliferator-activated receptor gamma (PPARγ), and specific markers lipoprotein lipase (LPL) and αP2 were upregulated (Fig. 3B), showing that immortalized cells had adipogenic differentiation potential. In osteogenesis, immortalized cells were positive for alizarin red stain for calcium deposit and AP stain for alkaline phosphatase after 2 weeks of induction in osteogenic medium, osteogenic markers osteopontin (OPN), alkaline phosphatase (ALP), and Col1A1 were consistently upregulated (Fig. 3C), showing that immortalized cells had osteogenic differentiation potential. In chondrogenesis, immortalized cells were positive for type II collagen immunostain for major collagen of cartilage and alcian blue stain for cartilage proteoglycans after 4 weeks of induction in chondrogenic medium, and chondrogenic markers Col2A1, AGC1, Sox9, and Col10A1 were consistently upregulated (Fig. 3D), showing that immortalized cells had chondrogenic differentiation potential. Based on the above data, the immortalized cells were MSCs.
To test tumorigenesis of immortalized MSCs, 3×106 cells were transplanted subcutaneously into nude mice. No tumors were observed at 12 weeks. This result was consistent with previous reports on immortalized MSCs by combination of TERT and viral genes [8,9].
Global gene expression profile of p53 knockdown MSCs
Deficiency in p53 induced the transformation of MSCs [12]. So far, little is known about molecular basis of p53 knockdown in MSC. To decipher the molecular basis underlying transformation of p53 knockdown in MSCs, we compared gene expression profile of MSCs between p53 knockdown and control MSCs by microarray studies. Compared with control MSCs, 405 genes were changed by p53 knockdown, including 204 upregulated genes and 201 downregulated genes (Fig. 4A). The expression patterns of selected genes from parallel samples analyzed by microarray were subsequently compared by RT-PCR for validation. RT-PCR assays were consistent with microarray data (Fig. 4B). Upregulated genes by p53 knockdown included genes related to cellular growth and proliferation, cell cycle, and DNA replication, recombination, and repair (Fig. 4C). Compared with control MSCs, p53 was downregulated by 17.12-fold in p53 knockdown-MSCs. MDM2 as interacting protein of p53 for proteasomal degradation was downregulated by 4.95-fold. The changes of these genes may be responsible for extended life span of p53 knockdown MSCs.
FIG. 4.
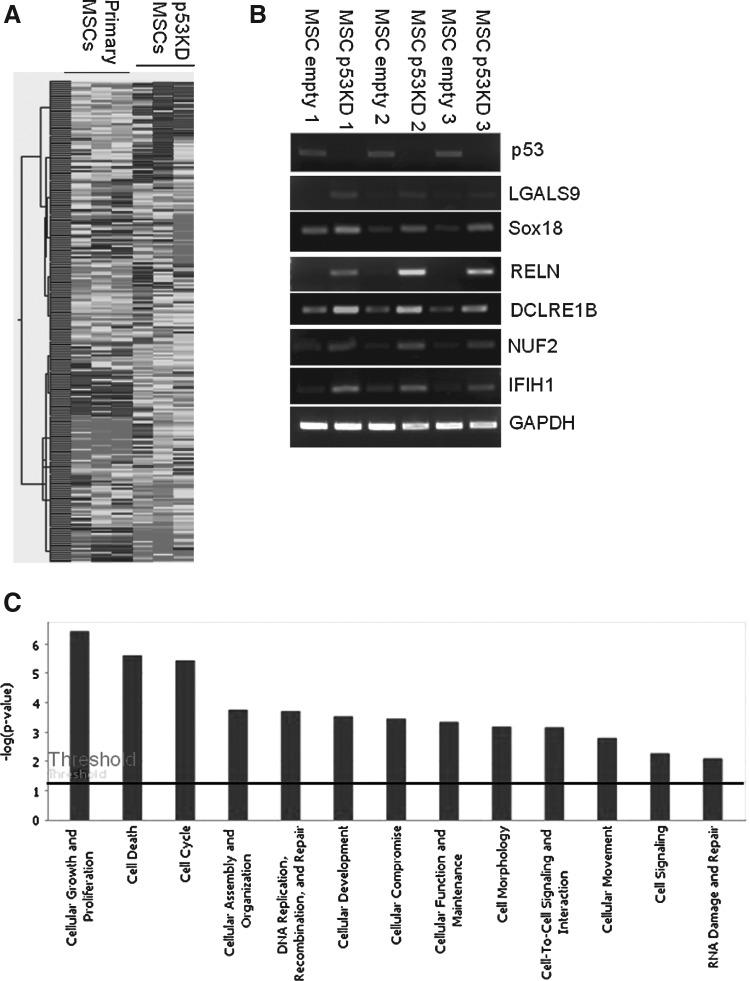
Comparison of gene expression profile between primary and p53 knockdown MSCs. (A) Comparison of gene expression profile between primary MSCs at passage 5 and p53 knockdown MSCs from 3 patients by microarray. (B) Verification of microarray data by reverse transcription (RT)-PCR. (C) Gene Ontology Classification of upregulated genes in p53 knockdown MSCs compared with control MSCs by Ingenuity Pathways Analysis. High-expression gene categories in p53 knockdown MSCs include cellular growth and proliferation, cell cycle and DNA replication, recombination, and repair.
Identification of immortalization-related genes by microarray analysis
To understand the molecular basis underlying immortalization of MSCs, we compared gene expression profile between primary and immortalized MSCs by microarray studies. The correlation coefficient between primary and immortalized MSCs was 0.927, showing their gene expression profile is highly similar.
However, some differences were also observed: compared with primary MSCs, 746 genes changed by more than 2-fold in immortalized MSCs including 413 upregulated genes and 333 downregulated genes (Fig. 5A and Supplementary Tables S1 and S2). RT-PCR assays of selected genes were consistent with microarray data (Fig. 5B). Among the upregulated genes in immortalized MSCs, the principal categories were cell cycle, and DNA replication, recombination, and repair genes (Fig. 5C and Supplementary Fig. S2). Compared with p53 knockdown MSCs, more genes related to cellular growth and proliferation, and cell cycle were upregulated in immortalized MSCs (Table 1 and Supplementary Tables S3 and S4), which provides a molecular explanation for why p53 knockdown alone is not sufficient but combination of p53 knockdown and TERT overexpression is sufficient to immortalize MSCs.
FIG. 5.
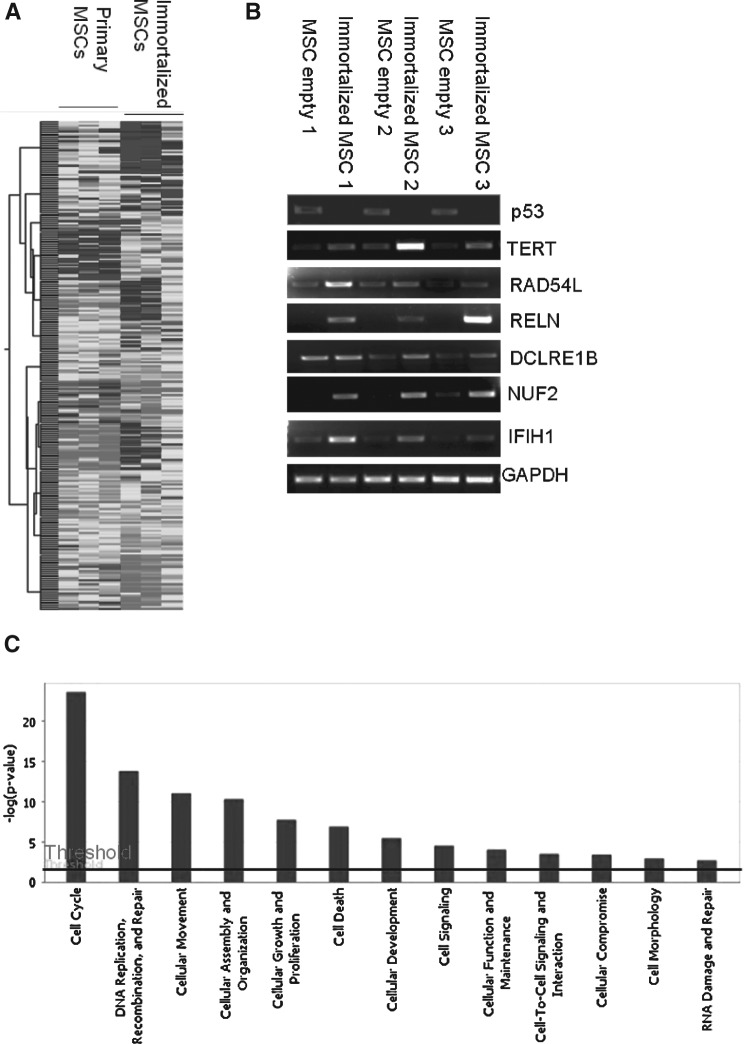
Comparison of gene expression profile between primary and immortalized MSCs. (A) Comparison of gene expression profile between primary MSCs at passage 5 and immortalized MSCs from 3 patients by microarray. (B) Verification of microarray data by RT-PCR. (C) Gene Ontology Classification of upregulated genes in immortalized MSCs compared with control MSCs by Ingenuity Pathways Analysis. More genes related to cell cycle, DNA replication, recombination, and repair were upregulated in MSCs immortalized by combination of p53 knockdown and hTERT overexpression than those with p53 knockdown alone.
Table 1.
Top 20 Upregulated Genes in Immortalized Mesenchymal Stem Cells, But Not in p53 Knockdown-Mesenchymal Stem Cells, Compared with Primary Mesenchymal Stem Cells
| Accession | Symbol | Fold | Definition |
|---|---|---|---|
| NM_014622.4 | LOH11CR2A | 18.01 | Homo sapiens loss of heterozygosity, 11, chromosomal region 2, gene A (LOH11CR2A), transcript variant 1, mRNA. |
| NM_152468.3 | TMC8 | 16.66 | Homo sapiens transmembrane channel-like 8 (TMC8), mRNA. |
| NM_014438.3 | IL1F8 | 16.35 | Homo sapiens interleukin 1 family, member 8 (eta) (IL1F8), transcript variant 1, mRNA. |
| NM_004662.1 | DNAH9 | 15.93 | Homo sapiens dynein, axonemal, heavy polypeptide 9 (DNAH9), transcript variant 1, mRNA. |
| NM_001012505.1 | FOXP1 | 14.5 | Homo sapiens forkhead box P1 (FOXP1), transcript variant 2, mRNA. |
| XM_936944.1 | BIRC1 | 13.88 | PREDICTED: Homo sapiens baculoviral IAP repeat-containing 1 (BIRC1), mRNA. |
| NM_018369.1 | DEPDC1B | 12.26 | Homo sapiens DEP domain containing 1B (DEPDC1B), mRNA. |
| NM_001080848.1 | CSAG2 | 10.9 | Homo sapiens CSAG family, member 3B (CSAG3B), mRNA. |
| NM_000394.2 | CRYAA | 10.63 | Homo sapiens crystallin, alpha A (CRYAA), mRNA. |
| NM_012190.2 | ALDH1L1 | 10.37 | Homo sapiens aldehyde dehydrogenase 1 family, member L1 (ALDH1L1), mRNA. |
| NM_198537.1 | FLJ44968 | 10.35 | Homo sapiens FLJ44968 protein (FLJ44968), mRNA. |
| NM_005682.4 | GPR56 | 10.34 | Homo sapiens G protein-coupled receptor 56 (GPR56), transcript variant 1, mRNA. |
| NM_014985.2 | CEP152 | 9.373 | Homo sapiens centrosomal protein 152 kDa (CEP152), mRNA. |
| NM_001006641.1 | SLC25A25 | 9.108 | Homo sapiens solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 (SLC25A25), nuclear gene encoding mitochondrial protein, transcript variant 2, mRNA. |
| NM_001986.1 | ETV4 | 8.661 | Homo sapiens ets variant gene 4 (E1A enhancer binding protein, E1AF) (ETV4), mRNA. |
| NM_000698.2 | ALOX5 | 8.651 | Homo sapiens arachidonate 5-lipoxygenase (ALOX5), mRNA. |
| NM_003579.2 | RAD54L | 8.471 | Homo sapiens RAD54-like (S. cerevisiae) (RAD54L), mRNA. |
| NM_002343.2 | LTF | 8.053 | Homo sapiens lactotransferrin (LTF), mRNA. |
| NM_014191.1 | SCN8A | 7.918 | Homo sapiens sodium channel, voltage gated, type VIII, alpha (SCN8A), mRNA. |
| NM_130855.2 | PTPRS | 7.341 | Homo sapiens protein tyrosine phosphatase, receptor type, S (PTPRS), transcript variant 4, mRNA. |
It was in line with upregulation of FOXM1 by p53 knockdown by 5.47-fold [21], suggesting that p53 negatively regulated oncogenic transcription factor FOXM1. RAD54L involved in the homologous recombination and repair of DNA [22] was upregulated by 8.5-fold. The cell-cycle-related gene PML [23] was upregulated by 6-fold. CDC20 as a regulatory protein in the cell cycle [24] was upregulated by 4.7-fold. In cell cycle checkpoint genes, commonly upregulated genes in p53 knockdown and immortalized MSCs included BRCA1 and CENPF, specifically upregulated genes in immortalized MSCs included PML, CDC45L, GTSE1, CCNA2, KNTC1, FANCG, and CHEK1. In 65 upregulated cell cycle genes in immortalized MSCs, 21 genes were also commonly upregulated in p53 knockdown MSCs and 44 genes were specific for immortalized MSCs, including LOH11CR2A, RAD54L, CDC25C, CDC20, CEP55, and CDC45L (Supplementary Table S3). These data suggest that immortalization of MSCs is associated with upregulation of cell cycle regulation and DNA replication genes.
Discussion
MSCs are one of the most promising resources for cell and gene therapy because of versatile plasticity in vitro and in vivo [25]. However, limited life span with primary MSCs greatly hampers their use in clinical application and basic research. To overcome this problem, one of the approaches is to immortalize MSCs.
Although ectopic expression of hTERT extended life span in a number of cell types by prolonging the telomere [4–6], it is controversial that overexpression of hTERT alone is sufficient to immortalize MSCs. Some showed that TERT alone is not sufficient to immortalize MSCs, additional viral genes are required [8,9,26], while others demonstrated opposite results [10,11]. Our data were consistent with the former. Although combination of TERT overexpression and viral oncogenes [8,9,26] or E7/E7 only [27] has been used to achieve immortalization of human MSCs, the use of viral genes definitely poses more safety issues. To avoid the use of viral genes to immortalize MSCs, the novel strategy of p53 knockdown and TERT overexpression only by modulating human genes was developed in this study.
It was reported that p53 knockdown extended life span but was not sufficient to immortalize human ovarian surface epithelial cells, suggesting that blocking p53 is able to delay the senescence. Combination of p53 knockdown and hTERT overexpression was sufficient to immortalize primary human ovarian surface epithelial cells [13]. The tumor suppressor p53 is an important regulator including cell cycle and differentiation. Our data showed that p53 knockdown promoted proliferation and enhanced colony formation of hMSCs. Interestingly, our data showed that p53 knockdown improved osteogenesis and adipogenesis of human MSCs, which was consistent with previous reports in mouse differentiation model [19,28], suggesting that p53 is involved in differentiation of MSCs as a negative regulator. Compared with control MSCs, p53 knockdown enhanced the expression of PPARγ and CEBPα, 2 master regulators of adiopogenesis, which regulate their downstream molecules during adipocyte differentiation. This might represent a mechanistic basis of p53 knockdown on adipogenesis of MSCs.
In MSCs, we first showed that p53 knockdown and TERT overexpression maintained MSC morphology for more than 13 months of continuous culture. The immortalized cells displayed similar surface antigen profile to parental primary MSCs and had MSC differentiation potential, showing that combination of p53 knockdown and hTERT overexpression is sufficient to immortalize MSCs.
Cellular senescence is a complex process that so far remains poorly understood. Such senescence appears to involve increased levels of cyclin-dependent kinase inhibitor, oxidative stress, and DNA damage [29–31]. Ontogenetically, cell cycle genes, replication of DNA, and mitosis were less apparent in cells undergoing senescence [1]. To understand the little known molecular basis underlying immortalization of MSCs so far, gene expression profiles between primary MSCs and p53 knockdown or immortalized MSCs were compared to identify immortalization-related genes. Our data showed that p53 knockdown alone upregulated 21 cell cycle genes that are not sufficient to immortalize MSCs, and immortalization of MSCs needs additional 44 upregulated cell cycle genes by combination of p53 knockdown and TERT overexpression, including RAD54L, CDC25C, CDC20, CEP55, and CDC45L. The upregulation of these genes makes MSCs bypass cell cycle crisis and maintain stemness of MSCs. However, the molecular signatures linked to the immortalization of MSCs remain to be determined.
In summary, all these lines of evidence strongly suggest that immortalization of MSCs is coordinated or associated with upregulation of cell cycle and DNA repair genes, which are required for cell crisis bypass. The immortalization of MSCs and molecular signature of immortalized MSCs will provide insights into cell-based assays of MSCs.
Supplementary Material
Acknowledgments
This work is supported by Biomedical Research Council Grant R-175-000-085-305, National Medical Research Council NMRC/1138/2007, and A*STAR. The funders have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Wagner W. Horn P. Castoldi M. Diehlmann A. Bork S. Saffrich R. Benes V. Blake J. Pfister S. Eckstein V. Ho AD. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter MA. Wynn RF. Jowitt SN. Wraith JE. Fairbairn LJ. Bellantuono I. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem cells. 2004;22:675–682. doi: 10.1634/stemcells.22-5-675. [DOI] [PubMed] [Google Scholar]
- 3.Rubio D. Garcia-Castro J. Martín MC. de la Fuente R. Cigudosa JC. Lloyd AC. Bernad A. Spontaneous human adult stem cell transformation. Cancer Res. 2005;65:3035–3039. doi: 10.1158/0008-5472.CAN-04-4194. [DOI] [PubMed] [Google Scholar]
- 4.Vaziri H. Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 5.Bodnar AG. Ouellette M. Frolkis M. Holt SE. Chiu CP. Morin GB. Harley CB. Shay JW. Lichtsteiner S. Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 6.Simonsen JL. Rosada C. Serakinci N. Justesen J. Stenderup K. Rattan SI. Jensen TG. Kassem M. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol. 2002;20:592–596. doi: 10.1038/nbt0602-592. [DOI] [PubMed] [Google Scholar]
- 7.Tsai CC. Chen CL. Liu HC. Lee YT. Wang HW. Hou LT. Hung SC. Overexpression of hTERT increases stem-like properties and decreases spontaneous differentiation in human mesenchymal stem cell lines. J Biomed Sci. 2010;17:64. doi: 10.1186/1423-0127-17-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto T. Aoyama T. Nakayama T. Nakamata T. Hosaka T. Nishijo K. Nakamura T. Kiyono T. Toguchida J. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem Biophys Res Commun. 2002;295:354–361. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- 9.Mori T. Kiyono T. Imabayashi H. Takeda Y. Tsuchiya K. Miyoshi S. Makino H. Matsumoto K. Saito H, et al. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol Cell Biol. 2005;25:5183–5195. doi: 10.1128/MCB.25.12.5183-5195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihara K. Imai C. Coustan-Smith E. Dome JS. Dominici M. Vanin E. Campana D. Development and functional characterization of human bone marrow mesenchymal cells immortalized by enforced expression of telomerase. Br J Haematol. 2003;120:846–849. doi: 10.1046/j.1365-2141.2003.04217.x. [DOI] [PubMed] [Google Scholar]
- 11.Bentzon JF. Stenderup K. Hansen FD. Schroder HD. Abdallah BM. Jensen TG. Kassem M. Tissue distribution and engraftment of human mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene. Biochem Biophys Res Commun. 2005;330:633–640. doi: 10.1016/j.bbrc.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 12.Rubio R. García-Castro J. Gutiérrez-Aranda I. Paramio J. Santos M. Catalina P. Leone PE. Menendez P. Rodríguez R. Deficiency in p53 but not retinoblastoma induces the transformation of mesenchymal stem cells in vitro and initiates leiomyosarcoma in vivo. Cancer Res. 2010;70:4185–4194. doi: 10.1158/0008-5472.CAN-09-4640. [DOI] [PubMed] [Google Scholar]
- 13.Yang G. Rosen DG. Mercado-Uribe I. Colacino JA. Mills GB. Bast RC., Jr Zhou C. Liu J. Knockdown of p53 combined with expression of the catalytic subunit of telomerase is sufficient to immortalize primary human ovarian surface epithelial cells. Carcinogenesis. 2007;28:174–182. doi: 10.1093/carcin/bgl115. [DOI] [PubMed] [Google Scholar]
- 14.Funes JM. Quintero M. Henderson S. Martinez D. Qureshi U. Westwood C. Clements MO. Bourboulia D. Pedley RB. Moncada S. Boshoff C. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc Natl Acad Sci USA. 2007;104:6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekiya I. Vuoristo JT. Larson BL. Prockop DJ. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci USA. 2002;99:4397–4402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu TM. Martina M. Hutmacher DW. Hui JH. Lee EH. Lim B. Identification of common pathways mediating differentiation of bone marrow and adipose tissues derived human mesenchymal stem cells (MSCs) into three mesenchymal lineages. Stem cells. 2007;25:250–260. doi: 10.1634/stemcells.2006-0394. [DOI] [PubMed] [Google Scholar]
- 17.Johnstone B. Hering TM. Caplan AI. Goldberg VM. Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 18.Vogelstein B. Lane D. Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 19.Molchadsky A. Shats I. Goldfinger N. Pevsner-Fischer M. Olson M. Rinon A. Tzahor E. Lozano G. Zipori D. Sarig R. Rotter V. p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One. 2008;3:e3707. doi: 10.1371/journal.pone.0003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi S. Gronthos S. Chen S. Reddi A. Counter CM. Robey PG. Wang CY. Bone formation by human postnatal bone marrow stromal stem cells is enhanced by telomerase expression. Nat Biotechnol. 2002;20:587–591. doi: 10.1038/nbt0602-587. [DOI] [PubMed] [Google Scholar]
- 21.Pandit B. Halasi M. Gartel AL. p53 negatively regulates expression of FoxM1. Cell Cycle. 2009;8:3425–3427. doi: 10.4161/cc.8.20.9628. [DOI] [PubMed] [Google Scholar]
- 22.Swagemakers SM. Essers J. de Wit J. Hoeijmakers JH. Kanaar R. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 23.Mu ZM. Le XF. Vallian S. Glassman AB. Chang KS. Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis. 1997;18:2063–2069. doi: 10.1093/carcin/18.11.2063. [DOI] [PubMed] [Google Scholar]
- 24.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Pittenger MF. Mackay AM. Beck SC. Jaiswal RK. Douglas R. Mosca JD. Moorman MA. Simonetti DW. Craig S. Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 26.Liu TM. Guo XM. Tan HS. Hui JH. Lim B. Lee EH. Zinc-finger protein 145, acting as an upstream regulator of SOX9, improves the differentiation potential of human mesenchymal stem cells for cartilage regeneration and repair. Arthritis Rheum. 2011;63:2711–2720. doi: 10.1002/art.30430. [DOI] [PubMed] [Google Scholar]
- 27.Osyczka AM. Nöth U. O'Connor J. Caterson EJ. Yoon K. Danielson KG. Tuan RS. Multilineage differentiation of adult human bone marrow progenitor cells transduced with human papilloma virus type 16 E6/E7 genes. Calcif Tissue Int. 2002;71:447–458. doi: 10.1007/s00223-001-1090-2. [DOI] [PubMed] [Google Scholar]
- 28.Tataria M. Quarto N. Longaker MT. Sylvester KG. Absence of the p53 tumor suppressor gene promotes osteogenesis in mesenchymal stem cells. J Pediatr Surg. 2006;41:624–632. doi: 10.1016/j.jpedsurg.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Ho AD. Wagner W. Mahlknecht U. Stem cells and ageing. The potential of stem cells to overcome age-related deteriorations of the body in regenerative medicine. EMBO Rep. 2005;6:S35–S38. doi: 10.1038/sj.embor.7400436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janzen V. Forkert R. Fleming HE. Saito Y. Waring MT. Dombkowski DM. Cheng T. DePinho RA. Sharpless NE. Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 31.Kiyono T. Foster SA. Koop JI. McDougall JK. Galloway DA. Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


