Abstract
The ribonucleoprotein telomerase synthesizes telomeric DNA by copying an intrinsic RNA template. In most cancer cells, telomerase is highly activated. Here we report a telomerase-based antitumor strategy: expression of mutant-template telomerase RNAs in human cancer cells. We expressed mutant-template human telomerase RNAs in prostate (LNCaP) and breast (MCF-7) cancer cell lines. Even a low threshold level of expression of telomerase RNA gene constructs containing various mutant templates, but not the control wild-type template, decreased cellular viability and increased apoptosis. This occurred despite the retention of normal levels of the endogenous wild-type telomerase RNA and endogenous wild-type telomerase activity and unaltered stable telomere lengths. In vivo tumor xenografts of a breast cancer cell line expressing a mutant-template telomerase RNA also had decreased growth rates. Therefore, mutant-template telomerase RNAs exert a strongly dominant-negative effect on cell proliferation and tumor growth. These results support the potential use of mutant-template telomerase RNA expression as an antineoplastic strategy.
Telomeres are specialized functional complexes at the ends of chromosomes, consisting of the terminal stretch of chromosomal DNA and associated proteins. The telomeric DNA in most eukaryotes consists of tandem repeats of a simple sequence unit, which is TTAGGG in humans and other vertebrates (reviewed in ref. 1). DNA-sequence-specific binding and other proteins associate with telomeric DNA repeats to create a complex that “caps” the telomere, preserving its physical integrity so the cell can continue to divide. Recent evidence suggests that the telomeric DNA–protein complexes are dynamic and normally can switch stochastically between a nonfunctional uncapped state and a functional capped state (reviewed in ref. 2). Unless recapped in a timely fashion, an uncapped telomere can suffer degradation, inappropriate recombination, and end-to-end fusions manifested as anaphase bridges (2–4). Uncapped telomeres appear to act similarly to DNA damage in eliciting cell cycle exit arrest or apoptosis (5, 6). In yeast, only one chromosome break is sufficient to elicit cell cycle arrest in cells with intact checkpoint controls (7), and loss of a single telomere can cause a RAD9-dependent temporary cell cycle arrest (5). Similarly, it has been suggested that the uncapping of even one telomere may be sufficient to cause cell cycle arrest in human cells (8).
The ribonucleoprotein enzyme telomerase helps maintain capped telomeres, adding tandem telomeric DNA repeats to chromosome ends by copying a short template sequence within its RNA moiety TER (9). Such elongation of the telomeric DNA compensates for the inability of the DNA replication machinery to completely copy the tips of the chromosomal DNA. The higher-order complex nucleated on the telomeric DNA by sequence-specific binding proteins is also critical for maintaining functional capped telomeres. Mutating the telomeric DNA or its proteins can disrupt this complex, thereby uncapping the telomere. In a ciliate and two budding yeasts, such disruption has been achieved by mutating the sequences of only the terminal few telomeric repeats, generating “toxic” telomeres (i.e., telomeres containing deleterious terminal telomeric DNA sequences) (3, 9–13). These mutant telomeric DNA repeats were created by mutating the template sequence in TER. In these systems, rapid telomere fusions, massive failure of chromosome segregation, and decreased cellular proliferation resulted. For some template mutants in yeasts, the effects correlated with loss of binding affinity to the yeast telomeric DNA-sequence-specific protein Rap1p (12). That these effects were caused by disruption of the telomeric DNA–protein complex is supported by other experiments involving mutant telomere-binding proteins. For instance, mutating the DNA-binding domain of the yeast protein Cdc13p, which binds the terminal telomeric single-stranded DNA overhang, led to telomeric DNA degradation and cell cycle arrest (14). In human cells, overexpression of a truncated form of TRF2 [a human telomeric DNA-binding protein postulated to bind telomeric end regions (15)] caused telomeric fusions (4) and ATM- and p53-dependent apoptosis (6).
Telomerase is inactive in many adult human cell types but is highly activated in most human cancers. Telomerase activation promotes proliferation of cultured precancerous human cells (refs. 16–19, and reviewed in ref. 2). Experimental activation of telomerase (by ectopic overexpression of the core subunit hTERT), coupled with the expression of multiple oncogenes, induced malignant transformation of primary human diploid cells (20, 21). Thus, telomerase provides a potential antineoplastic therapeutic target, and its inhibition slows tumor cell proliferation in culture and in mouse xenograft models (22–25). However, in all these previous reports of perturbations of human telomerase, inhibition of cell proliferation required essentially complete loss of telomerase activity, via either an inhibitor or expression of a great excess of a mutated telomerase component(s) to swamp out the endogenous wild-type (WT) telomerase (24, 25).
Previous work on cancer cells has involved inhibiting telomerase activity, and thus shortening telomeres, as a means to uncap telomeres (22–25). Here we report a way of uncapping telomeres in human cancer cells without significant telomere shortening or loss of telomerase activity. We demonstrate that expressing various mutant-template human telomerase RNAs (MT-hTers) decreases cancer cell proliferation both in vitro and in vivo. We report that expression of even very low levels of MT-hTers, in either human breast or prostate cancer cells, decreases cellular viability and significantly increases apoptosis rates. Tumors in mice xenografted with human breast cancer cells expressing an MT-hTer were smaller than those generated from cells expressing the control WT template gene. Most importantly, a low threshold level of mutant-template telomerase expression was sufficient to cause these effects on cell and tumor growth. Thus it was unnecessary to block or overwhelm the expression of the endogenous telomerase RNA. MT-hTers therefore display true dominance over the WT telomerase RNA. To account for these findings, we propose that uncapping of as few as one telomere by the action of these mutant-template telomerases elicits cell cycle arrest and apoptosis in human cancer cells.
Methods
Plasmid Construction, Cell Lines, and Nucleic Acid Analyses.
hTER was PCR cloned from human genomic DNA, sequenced, and subcloned. MT-hTer constructs were generated from the hTER subclone by site-directed mutagenesis, and hTERT was cloned into pBabe-puro, as described in Supplemental Methods, which are published on the PNAS web site, www.pnas.org. Stable MCF-7 Tet-off and doxycycline-inducible LNCaP lines expressing WT-hTER or MT-hTer and stable LNCaP cell clones expressing hTERT in a pBabe-puro vector were obtained as described in Supplemental Methods. Analysis of hTER/Ter expression, telomere lengths, and telomerase activity are described in Supplemental Methods.
Cell Proliferation, Apoptosis, and Cell Cycle Assays.
3H-thymidine incorporation and colony-forming ability assays were performed as described in Supplemental Methods. Apoptosis was assessed by flow cytometric analysis of DNA after RNase A and propidium iodide treatment. To analyze the fraction of MCF-7 cells in G1/G0, S phase, and G2, cells were pulse labeled with BrdUrd for 4 hr, trypsinized, fixed, and analyzed by flow cytometry. Tumor growth of MCF-7 cells xenografted into nu/nu mice was measured twice weekly for 38 days and samples examined histologically, as described in Supplemental Methods.
Results
Expression of MT-hTer Constructs Occurs at Low Levels.
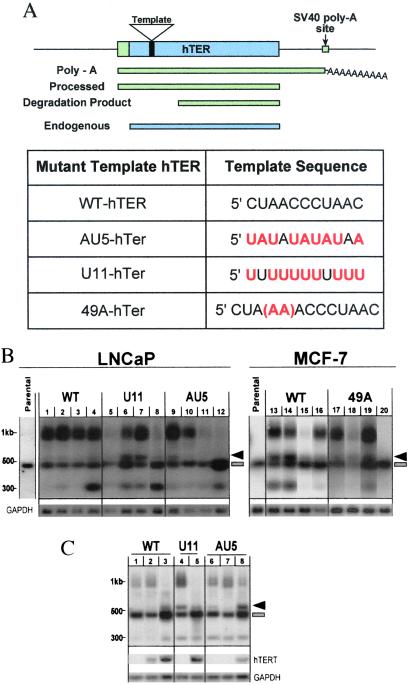
The choice of MT-hTers tested was based on corresponding mutations in the Tetrahymena thermophila telomerase RNA (3, 26) and/or on the predicted loss of DNA sequence-specific binding by the human telomere binding proteins TRF1 and TRF2 (27, 28). The templating domain of the human telomerase RNA (hTER, also known as hTR) is an 11-nt telomeric sequence in the 451-nt RNA (Fig. 1A). Here we report in full the results obtained with three of these MT-hTers: AU5-hTer, U11-hTer, and 49A-hTer (Fig. 1A). As will be described below, similar results were also obtained with four other mutant template hTers. The MT-hTers were expressed under the control of either a tetracycline-inducible system (AU5 and U11) or a tetracycline-repressible system (49A). The AU5-hTer and U11-hTer mutants were transfected into LNCaP, a prostate cancer cell line, and the 49A-hTer mutant into MCF-7, a breast cancer cell line. Both tumor cell lines are telomerase-positive. The WT-hTER gene was also transfected in parallel in both cell types. Four WT, seven U11, and five AU5 clonal LNCaP lines, and seven WT and twelve 49A clonal MCF-7 lines were analyzed in depth, and their proliferative and other properties were compared with the respective parental cell lines.
Figure 1.
Expression of mutant-template telomerase RNA. (A) The 11-nt template region is located near the 5′ end of the human telomerase RNA. Ectopic expression of the hTER construct leads to the production of three detectable species (green bars), in addition to the endogenous hTER (blue bar). The template sequences of WT, AU5, U11, and 49A telomerase RNAs are indicated below. (B) Northern blotting analysis of various WT-hTER- and MT-hTer-expressing clonal lines. The 451-nt endogenous hTER is present in every cell line examined (blue bar). Parental lines express only this endogenous species. Arrow, the mature functional MT- hTer or WT-hTER transcript. Broad band at ≈1 kb, polyadenylated species transcribed from the introduced hTER/hTer construct. Lower band at ≈300 nucleotides, degradation product from the introduced hTER/hTer construct. LNCaP clonal lines: lanes 1–4, WT-hTER expressing lines R10, R11, R12, and R19; lanes 5–8, U11-hTer expressing lines 11.1, 11.3, 11.6, and 11.19; lanes 9–12, AU5-hTer expressing lines 5.4, 5.5, 5.14, and 5.15. MCF-7 clonal lines: lanes 13–16, WT-hTER expressing lines K2, K3, P1, and A4; lanes 17–20, 49A-hTer expressing lines D2, G4, K6, and P1. (C) TERT is limiting for telomerase RNA levels. Northern blotting analysis of LNCaP clonal lines R10 (WT, lanes 1–3), 11.6 (U11, lanes 4–5), and 5.4 (AU5, lanes 6–8) after transfection with hTERT. Lane 1: clonal line R10-c (vector control); lane 2: clonal line R10–6 (low level of hTERT mRNA expression); lane 3: clonal line R10–3 (high level of hTERT mRNA expression); lane 4: clonal line 11.6–2 (hTERT expression not detected); lane 5: clonal line 11.6–1 (high level of hTERT mRNA); lane 6: clonal line 5.4-c (vector control); lane 7: 5.4–19 (hTERT expression not detected); lane 8: clonal line 5.4–5 (high level of hTERT mRNA).
We analyzed the levels and molecular forms of the transcripts of the introduced MT-hTer and WT-hTER genes, and of the endogenous WT-hTER gene (endo-hTER), by using Northern blotting analysis. The mature telomerase RNA species from the introduced gene construct (Fig. 1B, RNA species marked with arrow) was the expected ≈80 bases longer than the endogenous hTER because of the difference in the transcription initiation site of the minimal cytomegalovirus promoter and the endogenous hTER promoter (see Fig. 1A). The 3′ terminus of the mature WT-hTER or MT-hTer species was processed at the normal position (29) [the 5′ terminus is not processed (30)]. Notably, all stably selected cell lines induced to express the AU5-hTer or 49A-hTer gene had lower levels of mature MT-hTer RNA (Fig. 1B, arrow) than endogenous hTER (Fig. 1B, bar), with the MT-hTer level typically being several-fold lower. This was also the case for the 49A-hTer RNA levels in cells recultured after growth as tumors in mouse xenograft experiments (data not shown). The endogenous hTER levels remained relatively constant in nearly all cell lines (Fig. 1B and data not shown).
As reported previously (30), expression of the introduced telomerase RNA gene constructs produced two other RNA species besides the mature processed telomerase RNA (Fig. 1B). An ≈1-kb polyadenylated species detected by Northern blotting resulted from use of the simian virus 40 polyA signal in the expression construct (Fig. 1A). The polyadenylated status of this species was verified by Northern blot analysis of a polyA-specific RNA preparation and by reverse transcription–PCR (RT-PCR) analysis with oligo dT and hTER-specific primers (data not shown). An ≈300-nt degradation fragment (which, like the polyA species, was detected only when the introduced hTER/hTer construct was expressed) is a 3′ fragment lacking the templating sequence (ref. 30 and data not shown). The 1-kb species and ≈300-bp degradation product did not adversely affect cell proliferation despite accumulating to high levels in some lines, because proliferation rates of high expressor WT-hTER controls were indistinguishable from that of the parental line. Only the mature processed telomerase RNA appears to associate with active telomerase, as the polyA+ and degradation products do not coimmunoprecipitate with the telomerase RNP, which contains only the mature processed telomerase RNA (31). In addition, we stably transfected the hTERT gene (on a retroviral construct) into several WT-hTER- and MT-hTer-expressing LNCaP lines. In different clonal lines stably expressing various steady-state levels of hTERT mRNA, the higher the level of hTERT mRNA, the lower the ratio of polyA+ to mature processed telomerase RNA (Fig. 1C).
Decreased Cell Proliferation and Viability of Cells Expressing Low Levels of Mutant Telomerase RNAs.
Expression of each of the seven MT-hTer genes tested caused decreased cell proliferation rates, usually preventing these cell populations from reaching confluence as compared with the parental lines or the control WT-hTER-expression lines despite continuous passaging for 6 months. Monitoring population growth rates (recording the dilution factor used at each passage) during prolonged continuous serial passaging showed that over 150–200 days, the average inferred population doubling times of the LNCaP clonal lines were 3.16 +/− 0.10 days for the WT clonal lines, 4.20 +/− 0.43 days for the U11-hTer lines, and 5.75 +/− 1.05 days for the AU5-hTer lines (see Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org). However, cells are in logarithmic growth phase only at optimal densities and, whereas the WT cell populations reached confluence at each passage, the AU5 and U11 cell cultures only rarely reached confluence before passaging. Therefore these values most likely underestimate both the true population doubling rates and the difference between WT and the MT-hTer lines. Over a 60-day period of continuous passaging, the inferred average doubling times for MCF-7 cells were 4.20 +/− 1.19 days for 49A-hTer expressing cells and 1.82 +/− 0.24 days for the control WT-hTER expressing cells. Notably, even after 200 days of culturing, no fast-growing subpopulations that evaded the effects of the MT-hTer expression developed in either LNCaP or MCF-7 cells. Similar results were also obtained in MCF-7 cells expressing four additional different MT-hTers (51G, 53G, 50G, and 53A; see supplemental data for their template sequences), whose expression levels were also low (data not shown). In summary, nearly all WT-hTER clonal lines maintained rapid cell proliferation on long-term culturing and reached confluence at each passage. In contrast, expressing the mutant telomerase RNAs caused significant proliferative defects.
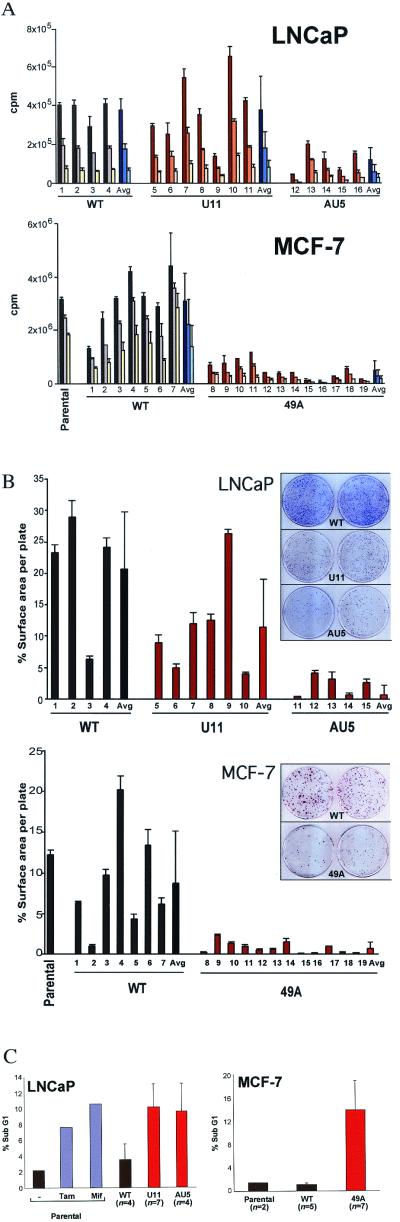
Relative 3H-thymidine incorporation rates (DNA synthesis) were measured to quantify the cellular proliferation of the MT-hTer clonal lines. Cells from each line were seeded at three different cell concentrations and pulsed for 18 h with 3H-thymidine. On average, the AU5 clonal lines incorporated 3-fold less and the 49A clonal lines 8-fold less 3H-thymidine than their respective WT-hTER controls or the parental cell line (Fig. 2A). Although the differences between 3H-thymidine incorporation rates of the U11 and WT clonal lines were not statistically significant (Fig. 2A), the U11 lines did show decreased long-term proliferation rates and increased apoptosis (see Fig. 5).
Figure 2.
MT-hTer expression decreases cell proliferation and viability. (A) 3H-thymidine incorporation. LNCaP clonal lines: 2.5 × 105, 1.25 × 105, and 6.25 × 104 cells were plated in triplicates and pulsed with 3H-thymidine. Columns 1–4, WT-hTER expressing lines R10, R11, R12, and R19; columns 5–11, U11-hTer expressing lines 11.1, 11.3, 11.6, 11.10, 11.12, 11.14, and 11.19; columns 12–16, AU5-hTer expressing lines 5.4, 5.5, 5.10, 5.14, and 5.15. Long-term proliferation of LNCaP lines is shown in Fig. 5. MCF-7 clonal lines: 3.0 × 105, 1.5 × 105, and 7.5 × 104 cells were plated in triplicate. Columns 1–7, WT-hTER expressing lines F4, A4, P1, K3, K2, K1, and H6; columns 8–19, 49A-hTer expressing lines C1, P1, M2, O1, K6, K5, K4, J2, G4, G3, D2, and D1. The averages (avg) for each group of clonal lines are also shown. (B) Colony-forming ability assays for LNCaP and MCF-7 clonal lines. For individual lines, the percent of the plate surface area covered by the cells is plotted. Representative plates are shown. LNCaP clonal lines: columns 1–4, WT-hTER expressing lines R10, R11, R12, and R19; columns 5–10, U11-hTer expressing lines 11.1, 11.3, 11.6, 11.10, 11.14, and 11.19; columns 11–15, AU5-hTer expressing lines 5.4, 5.5, 5.10, 5.14, and 5.15. MCF-7 clonal lines: columns 1–7, WT-hTER expressing lines F4, A4, P1, K3, K2, K1, and H6; columns 8–19, 49A-hTer expressing lines C1, P1, M2, O1, K6, K5, K4, J2, G4, G3, D2, and D1. The averages (avg) for each group of clonal lines are also shown. (C) Increased apoptosis caused by MT-hTer expression. Fraction of cells with subG1 DNA levels (determined by flow cytometry; the flow cytometry histograms are shown in Fig. 6). (Left) LNCaP: parental (−), no treatment; tam, tamoxifen treated; mif, mifepristone treated. The average percent subG1 of WT, U11, or AU5-hTer expressing LNCaP clonal lines are also shown (n = number of clonal lines). (Right) Flow cytometry analysis of parental, WT, or 49A-hTer expressing MCF-7 clonal lines.
These findings were corroborated by colony plating assays. The AU5 and 49A cells produced fewer and, on average, smaller colonies than the WT-hTER controls or parental lines (Fig. 2B). On the basis of the measurements of percent of each plate surface covered by colonies (reflecting both colony number and sizes), the average colony-forming efficiencies of AU5 and 49A lines were, respectively, 30- and 12-fold lower than those of their control WT lines (Fig. 2B). The difference between the U11 and WT lines was not statistically significant. Microscopic examination of the 49A colonies, but not the WT colonies, showed frequent gaps between the cells in a colony (data not shown), consistent with cell death.
Cell Cycle Effects and Apoptosis of Cells Expressing Low Levels of Mutant Telomerase RNAs.
Flow cytometry showed that the fraction of cells with sub-G1 levels of DNA, indicative of apoptosis, was significantly increased in AU5 and U11 compared with the WT-hTER or parental control LNCaP lines [Fig. 2C and Fig. 6 (which is published as supplemental data on the PNAS web site)]. The sub-G1 peak levels observed were similar to those of LNCaP cell cultures treated with tamoxifen or mifepristone, chemotherapeutic agents known to induce apoptosis in LNCaP cells (32). Similarly, apoptosis was significantly increased in the 49A clonal lines compared with WT-hTER and parental MCF-7 controls (Fig. 2C). In addition, cell cycle analysis by using BrdUrd incorporation showed that the fraction of 49A cells in G1 (or G0) was significantly increased, and the fraction in S-phase was correspondingly decreased, compared with the WT control lines (Fig. 7, which is published as supplemental data on the PNAS web site). Similar cell cycle effects and increased apoptosis were also seen with multiple independent clonal MCF-7 lines expressing either of two other MT-hTer genes (53G and 51G; data not shown). We conclude that both apoptosis and G1 (or G0) cell cycle arrest contribute to the decreased cell proliferation caused by MT-hTer expression.
Tumor Growth of Breast Cancer Cells Expressing Low Levels of Mutant-Template Telomerase RNA.
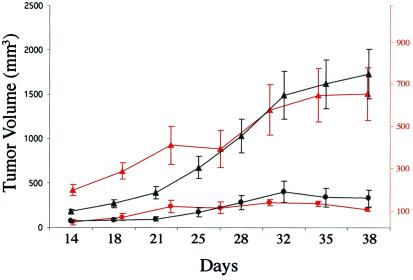
Stable transfectant MCF-7 breast cancer cell lines expressing either the 49A-MT-hTer (clonal line C1) or the control WT-hTER gene (clonal line K3) were xenografted into nude mice and assessed for tumor growth over 38 days. In two independent experiments, the average growth rate of the 49A-hTer tumors was 7-fold less than that of the WT-hTER control tumors (Fig. 3). Some regression in size was observed at late time points for the 49A-hTer, but not the WT-hTER, tumors. By immunohistological analysis, in the 49A compared with the WT-hTER tumors, the mitotic index was decreased (average mitotic index 6.3 versus 13.7 per 1,000 cells), and the apoptotic index increased (average apoptotic index 49.0 versus 14.4 per 1,000 cells). In summary, MT-hTer expression increased apoptosis and decreased tumor growth rates in this in vivo system, as well as in the cells cultured in vitro.
Figure 3.
49A-hTer expressing cells form smaller tumors in mice with increased apoptosis. Nude mice were injected with MCF-7 cells expressing either WT-hTER (clonal line K3; triangles) or 49A-hTer (clonal line C1; circles), and tumor volume was measured twice weekly for 38 days. Results from two independent experiments are shown (first, black; second, red).
Bulk Telomere Length and Telomerase Activity Are Maintained Despite Loss of Cell Viability.
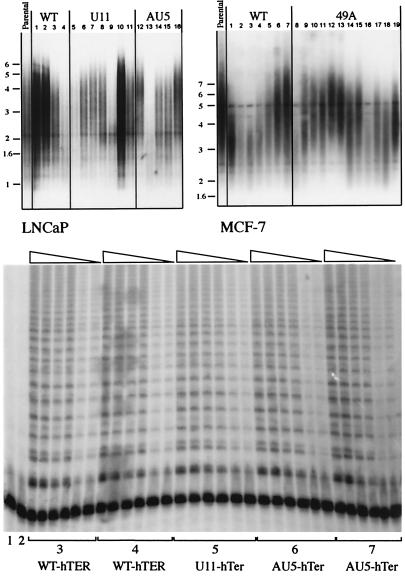
Southern blotting analysis revealed no significant difference in average telomere lengths between the MT-hTER- versus the WT-hTER-expressing LNCaP and MCF-7 cells. Although telomere lengths and MT-hTer levels varied among clonal lines within each group (Fig. 4A), for the individual clonal lines we found no correlation between mean telomere length, 3H-thymidine incorporation rate, or expression level of the introduced MT-hTer or WT-hTER. WT telomerase activity, measured in standard telomerase activity assays (TRAP assays), was retained in all cell lines regardless of whether WT-hTER or MT-hTer was introduced (Fig. 4B and data not shown). This was consistent with the retention of normal levels of the endogenous WT-hTER (Fig. 1B).
Figure 4.
MT-hTer expression does not change bulk telomere lengths or cause loss of telomerase activity. (A) Southern blotting analysis of TRF lengths. Genomic DNA was isolated 4 months after induction of expression of WT- hTER or MT-hTer expression. (Left) LNCaP clonal lines: lanes 1–4, WT-hTER expressing clonal lines R10, R11, R12, and R19; lanes 5–11, U11-hTer expressing lines 11.1, 11.3, 11.6, 11.10, 11.12, 11.14, and 11.19; lanes 12–16, AU5-hTer expressing lines 5.4, 5.5, 5.10, 5.14, and 5.15. Right: MCF-7 clonal lines: lanes 1–7, WT-hTER expressing lines F4, A4, P1, K3, K2, K1, and H6; lanes 8–19, 49A-hTer expressing lines C1, P1, M2, O1, K6, K5, K4, J2, G4, G3, D2, and D1. There was no correlation between TRF lengths and proliferation rates. (B) Telomerase activity assays (TRAP) were carried out on representative LNCaP clonal cell lines. Lane 1: lysis buffer only; lane 2: heat-inactivated WT sample; lanes 3: R11; lanes 4: R19; 5: 11.6; 6: 5.14; 7: 5.15. Each set of lanes shows 10-fold dilutions (1, 1:10 and 1:100) of each cell extract assayed; each sample was loaded in duplicate in two adjacent lanes.
Discussion
Here we report the surprising finding that expressing even low levels of mutant-template telomerase RNA decreases cancer cell viability and breast tumor growth in a model xenograft animal system. Apoptosis rates and cell cycle arrest were both increased. Most notably, the mutant-template telomerase RNA showed strong dominance over WT with respect to cellular phenotype, despite much higher levels of the WT RNA and no abrogation of WT telomerase activity. These dominant–negative effects were exerted without apparently changing bulk telomere length.
These findings contrast with previous reports of inhibiting cancer cell proliferation by inhibiting telomerase, which was associated with net telomere shortening (22, 24, 25). Overexpressing catalytically inactive mutant hTERT caused telomere shortening and senescence or apoptosis (24, 25), but the inactive hTERT protein was always present in large excess over the endogenously expressed WT-hTERT, which was therefore likely outcompeted for limiting components of the telomerase complex. Similarly, in previous experiments by using mutant-template telomerase RNAs in the ciliate T. thermophila and in budding yeasts, deleterious effects on cell proliferation also required complete or near-complete replacement of the WT RNA by mutant-template RNA (3, 9–13, 33). We propose that these previously described “dominant–negative” phenotypic effects, which to date have involved experimentally “swamping out” the WT components in the telomerase RNP macromolecular complex by mutant components, be termed “overexpression negative” effects. In contrast, except for the U11 lines, all of the independently transfected clonal lines analyzed (over 60 lines) had steady-state levels of processed MT-hTer RNA (and often the polyA species as well; see Fig. 1B, lanes 5, 11, 12, and 20), which were consistently low and never exceeded that of WT endogenous telomerase RNA, even after induction of gene expression. Hence the MT-hTer RNA was unlikely to have outcompeted the endogenous WT telomerase RNA. Thus, the human cancer cells tested here appear to be more sensitive to MT-telomerase RNAs than ciliate or yeast cells. Although different mutant hTer sequences were tested in the two cancer cell lines used here, the results were very similar between the two lines, suggesting a common mechanistic basis for the observed effects. Previously, Marusic et al. (34) studied a single-base substitution template mutant in HT-1080 cells. Although it was expressed at similar levels to the endogenous RNA, and mutant repeats were detectably incorporated into telomeres, cell viability was only slightly decreased. The contrastingly potent and similar effects caused by low expression of all seven template mutants tested in our work may be attributable to our use of different template mutations, cell types, and/or expression vector.
A striking and consistent finding with the large number of clonal MT-hTer-expressing lines analyzed (total >60) was that a positive RT-PCR scoring for MT-hTer expression always correlated with cell proliferation inhibition. Conversely, MT-hTer lines that scored negative by RT-PCR grew comparably to parental and WT-hTER lines (data not shown). Hence, only a low threshold level of MT-hTer expression was necessary for the strong cell proliferation and apoptosis effects in the two epithelial cancer cell lines investigated. Interestingly, in the WT and U11 lines, which had, respectively, no and relatively little change in cell proliferation rates, average steady-state RNA levels were higher than in the other six MT-hTer lines (Fig. 1B and data not shown). Although the initial stable selection of the clonal lines with geneticin was performed in a supposedly uninduced state, all of the RT-PCR positive lines showed “leaky” expression even without induction (data not shown), like that reported previously for other genes with these Tet-Off and Tet-On systems (35). Therefore, we suggest that the negative effects of MT-hTers (excepting U11) combined with the low leakage level of these systems led to the preferential selection of low expressors of these MT-hTers.
The negative effects on cell proliferation caused by MT-hTer expression are unlikely to be caused by nonspecific toxic effects related to the construct or to any effects of MT-hTers on processes other than telomerase action. We found no significant differences in the rates of proliferation, 3H-thymidine incorporation, or apoptosis between cells expressing the WT-hTER gene construct and the parental cell line controls, ruling out the possibility of nonspecific toxic effects of the WT-hTER construct. hTER is not known to have any function besides that in telomerase. The only difference between the WT-hTER and MT-hTer constructs was the mutated template sequence: 5′ CUAACCCUAAC 3′ for the WT template, and 5′ UUUUUUUUUUU 3′, 5′ UAUAUAUAUAA 3′, 5′ CUAAAACCCUAAC 3′, 5′ GGAAGGCUAAC 3′, 5′ GGAAGGGGAAC 3′, 5′ CAAAGCCUAAC 3′ and 5′ CAAAGCCAAAC 3′ for the seven different MT-hTers. Because each is very different from the others, it is unlikely that chance similarity to a cellular RNA species caused the observed apoptotic and proliferative effects of all seven MT-hTers.
We propose that uncapping of only one or a few telomeres per cell by the MT-hTer-containing telomerase can trigger a DNA damage response. This model can account for the low threshold of MT-hTer expression required to cause cellular effects in human cells and is consistent with findings in yeast showing that only one double-stranded DNA break, or loss of only one functional telomere per cell, elicits cell cycle arrest (5, 7, 8, 36). High sensitivity to MT-hTers, which was observed despite the relaxed checkpoint pathways and resistance to apoptosis likely to exist in these cancer cells, can account for the strong dominance and low expression threshold needed to cause cell cycle arrest and apoptosis. In T. thermophila telomerase, mutations equivalent to AU5 and U11 caused the expected sequences to be synthesized in vitro (26), and the equivalent of 49A caused the corresponding repeats to be added in vivo (3). In two budding yeasts and T. thermophila, addition of only a few mutant repeats by a mutant-template TER telomerase is sufficient to interfere with cell proliferation (3, 11–13, 33, 37). In the human cancer cells analyzed here, such mutant repeats added to a telomere are predicted to perturb binding of the DNA sequence-specific proteins TRF1 and/or the capping protein TRF2 and hence to cause uncapping. As the cells containing such telomeres ceased to proliferate or underwent apoptosis, mutant telomeres would remain greatly underrepresented, accounting for the unchanged bulk telomere length.
The inhibition of proliferation by the telomerase template mutants reported here exploits the activation of telomerase that characterizes most human tumor cells. It converts the active telomerase pathway, normally advantageous for tumor cell proliferation, into a process detrimental to cancer cells. Tumors expressing an MT-hTer grew more slowly, with higher apoptotic rates, than controls. Therefore, use of MT-hTer genes or agents that mimic their effects may be useful in antitumor therapy.
Supplementary Material
Acknowledgments
We thank Thea Tlsty for valuable discussions and comments on the manuscript and Shivani Nautiyal for critical reading of the manuscript. This work was supported by grants from the Department of Defense, University of California, San Francisco (UCSF), Cancer Center Breast Oncology Program, Breast Cancer Research Program (California), the Steven and Michelle Kirsch Foundation, a UCSF Prostate Cancer Developmental Grant, a Special Program of Research Excellence grant from the National Institutes of Health, and a National Science Foundation Predoctoral Fellowship to M.A.R. M.M.K. is supported by a Medical Scientist Training Program grant from the National Institutes of Health.
Abbreviations
- MT-hTer
mutant-template human telomerase RNA
- WT
wild type
- RT-PCR
reverse-transcription–PCR
Footnotes
See commentary on page 7649.
References
- 1.Zakian V A. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E H. Nature (London) 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 3.Kirk K E, Harmon B P, Reichardt I K, Sedat J W, Blackburn E H. Science. 1997;275:1478–1481. doi: 10.1126/science.275.5305.1478. [DOI] [PubMed] [Google Scholar]
- 4.van Steensel B, Smogorzewska A, de Lange T. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 5.Sandell L L, Zakian V A. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 6.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 7.Bennett C B, Lewis A L, Baldwin K K, Resnick M A. Proc Natl Acad Sci USA. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy M Z, Allsopp R C, Futcher A B, Greider C W, Harley C B. J Mol Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 9.Yu G L, Bradley J D, Attardi L D, Blackburn E H. Nature (London) 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 10.Gilley D, Lee M S, Blackburn E H. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 11.McEachern M J, Blackburn E H. Nature (London) 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 12.Krauskopf A, Blackburn E H. Nature (London) 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 13.Smith C D, Blackburn E H. J Cell Biol. 1999;145:203–214. doi: 10.1083/jcb.145.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes T R, Weilbaecher R G, Walterscheid M, Lundblad V. Proc Natl Acad Sci USA. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 16.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 17.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature (London) 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 18.Zhu J, Wang H, Bishop J M, Blackburn E H. Proc Natl Acad Sci USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Chang E, Cherry A M, Bangs C D, Oei Y, Bodnar A, Bronstein A, Chiu C P, Herron G S. J Biol Chem. 1999;274:26141–26148. doi: 10.1074/jbc.274.37.26141. [DOI] [PubMed] [Google Scholar]
- 20.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Nature (London) 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 21.Elenbaas B, Spirio L, Koerner F, Fleming M D, Zimonjic D B, Donaher J L, Popescu N C, Hahn W C, Weinberg R A. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo S, Kondo Y, Li G, Silverman R H, Cowell J K. Oncogene. 1998;16:3323–3330. doi: 10.1038/sj.onc.1201885. [DOI] [PubMed] [Google Scholar]
- 23.Naasani I, Seimiya H, Yamori T, Tsuruo T. Cancer Res. 1999;59:4004–4011. [PubMed] [Google Scholar]
- 24.Hahn W C, Stewart S A, Brooks M W, York S G, Eaton E, Kurachi A, Beijersbergen R L, Knoll J H, Meyerson M, Weinberg R A. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Mar V, Zhou W, Harrington L, Robinson M O. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ware T L, Wang H, Blackburn E H. EMBO J. 2000;19:3119–3131. doi: 10.1093/emboj/19.12.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 28.Broccoli D, Smogorzewska A, Chong L, de Lange T. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 29.Zaug A J, Linger J, Cech T R. Nucleic Acids Res. 1996;24:532–533. doi: 10.1093/nar/24.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell J R, Cheng J, Collins K. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell J R, Wood E, Collins K. Nature (London) 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 32.Fathy El Etreby M, Liang Y, Lewis R W. Prostate. 2000;43:31–42. doi: 10.1002/(sici)1097-0045(20000401)43:1<31::aid-pros5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 33.McEachern M J, Iyer S, Fulton T B, Blackburn E H. Proc Natl Acad Sci USA. 2000;97:11409–11414. doi: 10.1073/pnas.210388397. . (First Published October 3, 2000; 10.1073/pnas.210388397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marusíc L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossi F M, Guicherit O M, Spicher A, Kringstein A M, Fatyol K, Blakely B T, Blau H M. Nat Genet. 1998;20:389–393. doi: 10.1038/3871. [DOI] [PubMed] [Google Scholar]
- 36.McEachern M J, Blackburn E H. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 37.Krauskopf A, Blackburn E H. Proc Natl Acad Sci USA. 1998;95:12486–12491. doi: 10.1073/pnas.95.21.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.