Abstract
This work demonstrates the use of a continuous online monitoring system for tracking systemic inflammation biomarkers during cardiopulmonary bypass (CPB) procedures. The ability to monitor inflammation biomarkers during CPB will allow surgical teams to actively treat inflammation and reduce harmful effects on postoperative morbidity and mortality, enabling improved patient outcomes. A microfluidic device has been designed which allows automation of the individual processing steps of a microbead immunoassay to allow continuous tracking of antigen concentrations. Preliminary experiments have demonstrated that the results produced by the micro-immunoassay are comparable to results produced from a standard ELISA (r=0.98). Additionally, integration of the assay with a simulated CPB circuit has been demonstrated with temporal tracking of C3a concentrations within blood continuously sampled from the circuit. The presented work describes the motivation, design challenges, and preliminary experimental results of this project.
Introduction
The purpose of this work is to develop an automated assay which can continuously monitor systemic inflammation during and following cardiac surgical procedures, especially those involving cardiopulmonary bypass (CPB). Numerous studies have investigated the complex systemic inflammatory responses and comorbidity resulting from cardiac surgery, particularly when CPB is used (1, 2). This “systemic inflammatory response syndrome” is characterized by complement, neutrophil, and platelet activation, and release of pro-inflammatory cytokines. These systemic inflammatory responses are attributed to numerous surgical and clinical factors, including: the exposure of blood to the nonphysiologic surfaces of the heart-lung circuit, ischemia/reperfusion injury of the involved tissues as blood flow is stopped (resulting in tissue ischemia) and restarted (resulting in reperfusion injury), surgical trauma and vacuum assisted venous drainage, and hypothermia (3). The systemic inflammatory syndrome is the primary cause of many postoperative complications resulting in vital organ dysfunction, multi-organ failure, and even death (1–3). Furthermore, the intensity of the inflammatory response is directly correlated with the severity of CPB-related morbidity (4). Advanced time-course inflammation studies using simulated CPB circuits have used sample periods of 15 minutes or greater over the course of 2 hours (5). Studies in cardiac patients undergoing CPB typically use longer sample periods of 1 hour and greater over a total monitoring period of 24–48 hours, including during both the cardiac surgical procedures and post operative monitoring (5). Two major bottlenecks to performing more detailed studies with shorter sample periods, for better temporal characterization of immune activation, are sample volume and manpower to run and analyze immunoassays. Since blood is typically collected using vacutainer tubes (3–5 ml/sample), conventional assays require several milliliters of blood per sample, and the samples are usually analyzed hours, days or even weeks post-surgery. Therefore, these types of studies are not capable of real-time monitoring of the inflammatory processes during the surgical procedures.
The microanalytical instrument currently being developed is designed to address these bottlenecks by requiring only a very small sample flow rate (µl/min sample usage), while permitting continuous measurements. It is believed that a detailed determination of a patient’s blood plasma cytokine and complement concentrations while undergoing CPB procedures will be one of the fundamental steps in addressing inflammation related morbidity by allowing clinicians to investigate how changes in surgical procedures, equipment, patient clinical management and intra-operative substitution of pharmacological agents affect CPB related immune activation and related morbidity
Immunoassays such as the enzyme linked immunosorbent assay (ELISA) and immunofluorocytometry are commonly used for biomarker quantification in both research and clinical settings. These assays can be used to directly measure the concentration of a specific protein in blood plasma, such as complements and cytokines, seen during the inflammation process. They offer numerous benefits including high sensitivity and selectivity, a wide concentration detection range, repeatability, and are generally available as off-the-shelf assay kits for scores of commonly investigated analytes. However, these assays are inherently discrete in terms of both their implementation as well as the data they produce. This limitation is inherent to these assays since they rely on a series of bench-top serial preparation steps for each sample analyzed. The sample rate of both ELISA and immunofluorocytometry is further limited when the volume of sample fluid available for testing is limited. These assays generally require at least 50–100 µl of fluid for each sample. For tracking of time-varying antigen concentrations, many samples are required, so the total sampled volume adds up very quickly and can result in excessive depletion of the sample fluid, especially in neonates or infants since the total blood volume of a 3 kg neonate is only ~240 ml.
Microfluidic devices have been developed which are analogous to traditional immunoassays, but offer benefits such as higher throughput and smaller required sample and reagent volumes due to microchip integration (6–8). Many of these devices use optical detection schemes based on fluorescent labels (8) or chemiluminescent reactions using horseradish peroxidase (9, 10). Other microfluidic biosensor designs have replaced these optical detection schemes with electrochemical detection schemes. Electrochemical detection techniques generally involve measurement of the alternating current impedance change (11), or DC current (12) proportional to the concentration of analyte adsorbed on a sensing element with specificity to that particular analyte. These microfluidic detection technologies have resulted in both higher throughput and significantly increased sensitivity immunoassays. For many applications, the data provided by these microfluidic assays are sufficient. This is especially true when the analyte concentration is not time-varying in the short term, such as in detection of disease (7) or environmental toxins (9). These assays use reduced sample volumes as compared to traditional methods, but they still function as discrete-sample assays. In applications where the analyte concentration is changing with time and must be analyzed repeatedly, these assays must be processed successively, which results in a sample rate much lower than would be desirable, and they therefore lack the ability to provide real-time data. Some designs have applied discrete sampling methods to continuous monitoring by adding automated controls to quickly repeat the measurement (13). As an alternative method, this manuscript details a continuous flow microfluidic assay which has the potential to provide very high sampling rates and requires no external controls.
Methods
Assay Principle
The assay is based on a sandwich immunoassay using antibody-coated microbeads which capture the antigen of interest when incubated within the sample fluid. By tightly controlling the incubation time, the amount of antigen captured by each microbead is directly correlated to the antigen concentration of the sample. Detection is enabled by incubating the microbeads with a fluorescently tagged secondary antibody. Thus, the fluorescence intensity of each microbead is directly correlated to the antigen sample concentration. Figure 1 shows a schematic of the microbead incubation scheme. The microbeads used in the assay are coated with a monoclonal antibody specific to the antigen of interest. During the first incubation stage, the antigen binds to these antibodies. In the second incubation, a fluorescently tagged monoclonal antibody, specific to a different epitope of the antigen, binds to the antigen on each bead. The fluorescence of each bead is measured using flow cytometry to determine the sample concentration. By averaging the fluorescence of multiple microbeads over short periods of time, the bead-to-bead error is reduced, allowing precise measurements to be made.
Figure 1.
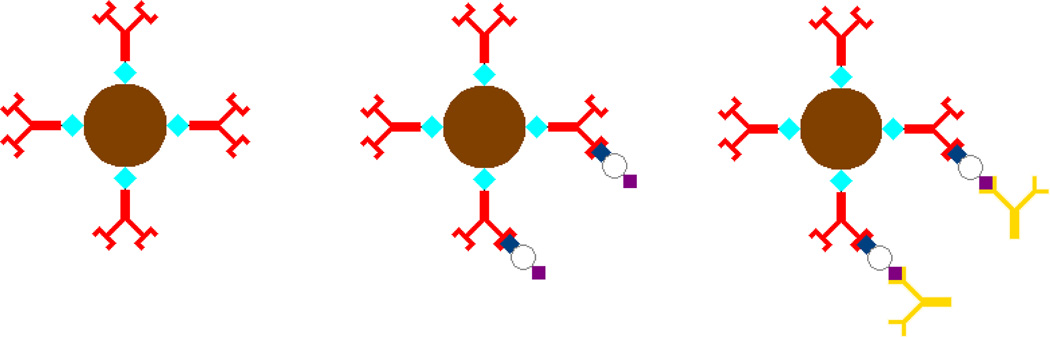
Schematic of microbead incubation. (1) Prior to assay, the magnetic microbeads are conjugated with an antigen-specific antibody. (2) After the first incubation stage, the antigen is bound to each bead in an amount proportional to the sample concentration. (3) In the second incubation stage, the microbeads are incubated with a fluorescently tagged secondary antibody to enable fluorescence detection of the antigen concentration.
While microbead assays can be performed using standard bench-top laboratory techniques, they also lend themselves to automation and miniaturization using microfluidics. By using microbeads with a magnetic core, magnets can be used to affect the trajectory of the microbeads within flow streams to direct the bead trajectory through the device. As shown in (Figure 2), the microdevice uses a special separation method where the magnetic force on the paramagnetic core bead pulls the microbeads from their carrier stream into an adjacent receiving sample stream, allowing serial incubation of the beads in different solutions. This magnetic separation scheme allows automated sequential incubation of microbeads in reagent fluids. In the first stage of the device, the beads are pulled into and incubated with the plasma sample. In a second, identical stage, they are incubated with the fluorescently tagged secondary antibody. A 60 cm long spiral shaped channel on each device layer allows to flow in the sample long enough to allow sufficient incubation time before the microbeads are transferred to their next stage (Figure 2). The incubation time used in this experiment was 2.5 minutes. After the two incubation stages, the microbeads are magnetically transferred into a wash buffer and are ready for fluorescence detection. This operating principle has been detailed in depth in previous publications (14, 15) demonstrating the ability to conduct autonomous immunoassays requiring no user input beyond sample perfusion through the devices. This work expands on these previous studies by detailing the automated and continuous flow nature of the microdevice which enables temporal concentration tracking of the analyte C3a from blood sampled directly from a simulated cardiopulmonary bypass circuit.
Figure 2.
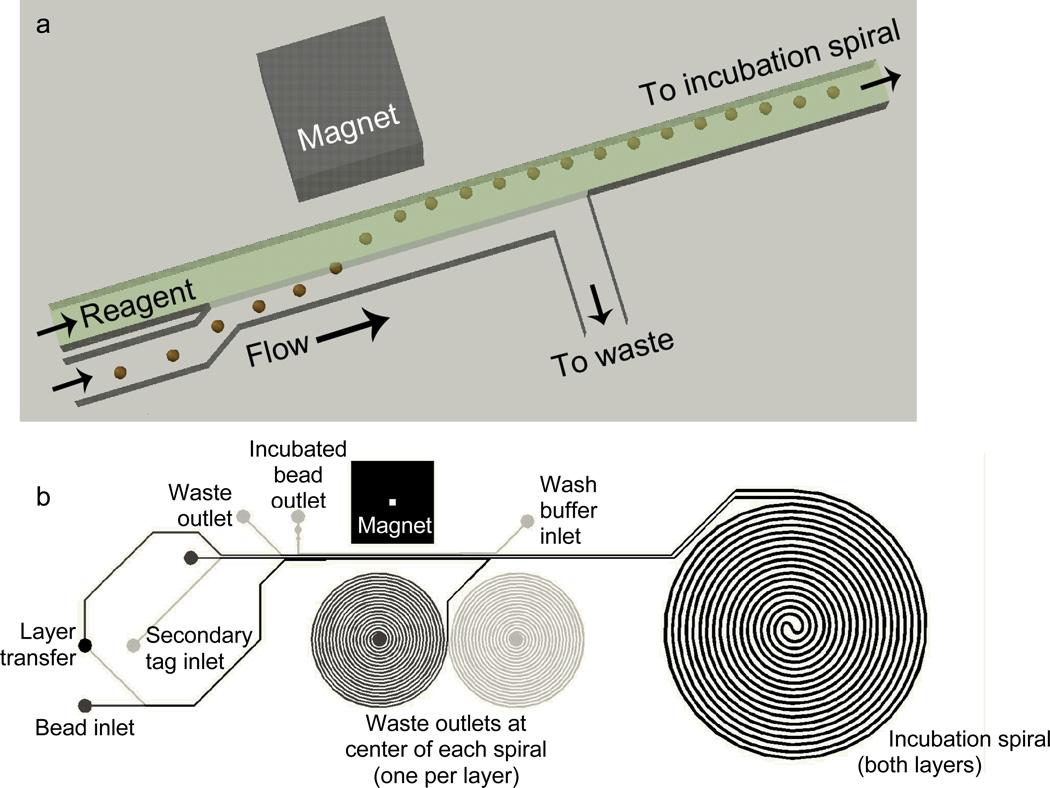
(a) Schematic of magnetically actuated bead transfer. The magnet pulls the beads from the carrier stream into the reagent stream, and the carrier stream is diverted to waste so that only the reagent stream with the microbeads continues to an incubation spiral. The device uses two of these separation regions on two aligned and bonded device layers, first to transfer the microbeads into the plasma sample, and second to transfer them into the fluorescently labeled secondary antibody. (b) CAD drawing showing the entire microfluidic channel layout. Black lines are on the upper layer and gray lines are on the lower layer. The large spiral (incubation spiral) is identical on both layers. The smaller spirals are incorporated only to balance hydrodynamic resistance for flow control.
Materials and Reagent Preparation
The microimmunoassay uses 8 µm, paramagnetic streptavidin-coated beads (Bang’s Laboratories, Inc., Fisher, IN). Antibody-coated beads were prepared by washing the beads twice in Phosphate Buffered Saline (PBS), and then incubating the beads with a biotinylated primary monoclonal anti-C3a antibody (Assay Designs Ann Arbor, MI, catalog number GAU017-01B) at room temperature for 1 hour in a microcentrifuge tube. An excess of antibody was used to ensure complete conjugation of the antibody onto the surface of the beads. In this case 25 µl of bead solution was incubated with 5 µl of antibody solution. The beads were then wash twice and resuspended in 500 ml of Ficoll-Paque PLUS (GE Healthcare), and stored at 4°C. The fluorescently labeled secondary antibody (Assay Designs Ann Arbor, MI, catalog number GAU013-16) was prepared using a phycoerythrin (PE) conjugation kit (Prozyme, Inc., San Leandro, CA) following the manufacturer’s instruction. After resuspending 50 µg of the antibody in 1 ml of PBS, a 3 to 1 dilution with PBS was used as the final labeling concentration used within the microdevice. PBS was used for the wash inlet.
Microdevice Fabrication
The microdevice was fabricated by the standard soft lithography process (16, 17) with an SU-8 master mold with a feature depth of 20 µm. Briefly, in this process a silicone based elastomer (PDMS) is cast over a lithographically defined mold with the desired microfluidic features shown in Figure 2(b). A cross-linking agent solidifies the PDMS, and the resulting channels are sealed by bonding the microfluidic surface to a flat glass or PDMS surface. For this device, the two layers were cast separately. The device inlet and outlet ports were punched with a 19 gauge needle. The top layer was then bonded to the top surface of the bottom layer, aligning the transfer hole which carries fluid between the layers. The lower layer was then bonded to a 75 mm×25 mm glass microscope slide. Bonding was accomplished by treating the PMDS and glass with corona discharge for approximately 10 seconds, pressing the PDMS to the glass, and placing the device in a 125°C oven for 1 hour. A slot for the actuation magnet was cut by hand with a razor blade. The magnet (K&J Magnetics, Jamison, PA, number B444) was pressed into the slots such that the field was aligned with the bead trajectory.
Benchmarking Assay
The microdevice assay was also benchmarked against a standard commercial ELISA assay for C3a at a 1:1000 sample dilution (Quidel, San Diego, CA). Blood plasma samples collected from the CPB circuit and known calibration standards were assayed using both a standard ELISA and the microdevice immunofluorocytometry method and the results were compared. The microdevice assay was performed by infusing all reagents and the sample through the microdevice using syringe pumps set to 1 µl/min. Each standard and sample were infused for 10 minutes to flush the device, and then fluorescence detection was conducted for 5 minutes. The detection system was based on an epifluorescent microscopy platform (Nikon TE2000U) coupled to an argon ion laser (Modu-Laser, Stellar-Pro-CE) and a photomultiplier tube (PMT) (C&L Instruments, Hershey, PA Model DPC-BA). The output of the PMT was recorded and a peak finding algorithm was used to measure the height of peaks caused by passing incubated beads in the outlet channel of the device as previously described (14). A filter block with a 480 nm bandpass excitation filter and a 488 nm dichroic mirror was used for excitation, along with a 580 nm bandpass emission filter.
Simulated CPB
In order to recreate the conditions of CPB perfusion during cardiac surgery as closely as possible, a replica CPB system is used with donor blood (18). The simulated extracorporeal circuit consisted of an HL-20 heart-lung machine with a multiflow HL-20 Roller blood pump (Jostra HL-20, Jostra, Austin, TX), a hollow fiber pediatric oxygenator with an integrated heat exchanger module (Capiox RX 05RW Terumo Corporation, Tokyo, Japan), a 32 mm Capiox pediatric arterial filter (CX*AF02; Terumo Corporation, Tokyo, Japan), a pediatric venous reservoir with an integrated cardiotomy filter (Capiox cardiotomy reservoir CX*CR10NX), and a MAQUET Heater-Cooler (Jostra Heater-Cooler Unit HCU 30 Houston, TX). In order to simulate the systemic vasculature resistance from a patient, a Hoffman clamp was tightened distal to a second reservoir which simulated our ‘pseudo patient’ at clinically relevant circuit pressure and pump flow rate. Pulmonary and aortic resistances were controlled via a series of pinch clamps. A schematic of the extracorporeal circuit is shown in Figure 3. The CPB circulation loop was primed with 500 ml of heparinized fresh human blood (19), drawn from healthy adult volunteers and brought into the lab within 30 min of collection due to previous studies which showed that stored donor blood did not replicate the inflammatory conditions of patient blood during surgery (20). This project was approved by the Penn State Hershey College of Medicine’s institutional review board. The blood was hemodiluted to 27%–30% hematocrit in Lactated Ringer’s solution and heparinized up to 5000 IU/blood bag (each blood bag contains between 300–500 ml of blood) consistent with surgical anticoagulation protocols. The blood was then circulated at a rate of 500 ml/min at an arterial circuit pressure of 100 mmHg. Nonpulsatile perfusion was performed and the temperature was set at normothermia (35°C) for the duration of the experiments. After complete mixing of the blood and prime fluid was accomplished, the arterial port of the membrane oxygenator was used as a source of sample blood (Figure 3). This location provides blood at a positive pressure and is continually replenished from the (simulated) patient blood supply.
Figure 3.
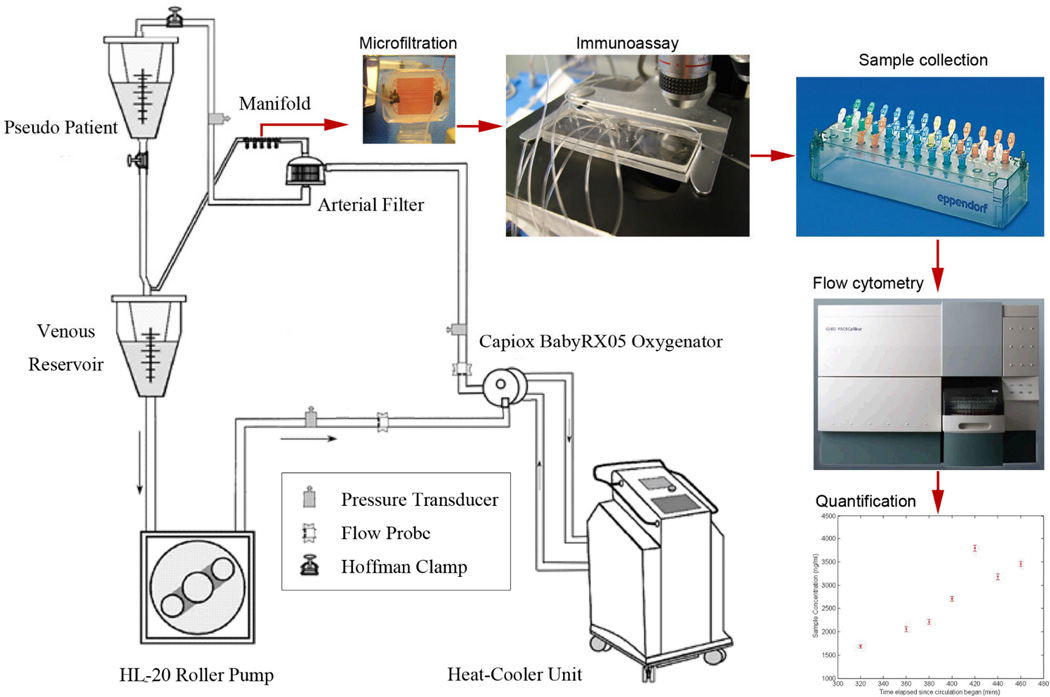
Diagram of the experimental procedure. Blood sampled from the simulated CPB circuit is diverted into the microfiltration device. The filtered plasma is pumped into the immunoassay device along with the microbeads and other reagents. The incubated microbeads are collected in batches. Each microbead sample is fluorescently labeled and quantified using a flow cytometer. Concentration quantification is based on comparison with a calibration curve.
Device Infusion with simulated CPB sample stream
All of the assay reagents are infused using high precision syringe pumps (PicoPlus 22, Harvard Apparatus). The blood plasma sample was supplied from the CPB circuit using a previously described inline microfiltration device (18). This device uses tangential-flow filtration in a microfluidic format to continuously separate cells from blood plasma. Since the sample is sourced from an external system, a syringe pump cannot be used to infuse it into the microdevice. The sample infusion system instead used a small peristaltic pump (Instech Laboratories model P720, Plymouth Meeting, PA). Unlike a syringe pump, the peristaltic pump has imprecise flow control, and its flow rate varies with the flow resistance into which it is pumping. Furthermore, the output pressure is not continuous but is instead periodic. Since the operation of the microfluidic device relies on a precise and steady sample flow, the pump must be complemented with a flow control and modulation system. This system is composed of a solenoid-actuated valve and a pressure sensor. Since the other inlet streams are infused at highly controlled flow rates by syringe pumps, the sample flow rate can be maintained by controlling the inlet pressure to a predetermined setpoint. A microcontroller circuit was used to monitor the input pressure and actuate the valve as necessary to bleed off excess pressure in order to hold the device inlet pressure constant.
When sampling plasma from the CPB circuit, only the primary antigen capture (1st incubation stage) was completed within the microdevice. Following incubation, the beads were collected from the microdevice in 20 minute batches. Fluorescence labeling of the beads and flow cytometry fluorescence detection were conducted separately. This choice was made because the fluorescence quantification could not be conducted right at the CPB circuit location and there were concerns the bead fluorescence intensity may diminish prior to flow cytometry. However, in order to maintain proper bead trafficking through the device and mimic full device operation,.the second device layer was infused with PBS. The syringes were loaded with reagents and placed into syringe pumps set to a flow rate of 1 µl/min, giving an incubation time of 2.5 minutes in each layer. The microfiltration device was attached to the simulated CPB circuit as described above, and its outlet was attached to the peristaltic pump. The pump was used to draw blood plasma from the microfiltration device and infuse it into the immunoassay device. The overall equipment arrangement and experimental process is detailed in Figure 3.
The incubated beads were collected in batches from the incubated bead outlet port of the microdevice every 20 minutes. This was done using a short piece of tubing running from the outlet port into a microcentrifuge tube. Each sample of incubated beads was labeled and refrigerated at 4°C. Measurements began after 300 minutes of CPB circulation time, and samples were collected every 20 minutes for the next 160 minutes. A calibration curve was constructed off-site by perfusing the same microfluidic device which was used with the CPB circuit with calibration standards using an identical infusion setup. The incubated beads from the collected CPB samples and the calibration standards were fluorescently labeled at the same time. The beads were incubated in microcentrifuge tubes with the prepared fluorescently tagged secondary antibody for 2.5 minutes before washing 2 times with PBS. The calibration standards and samples were then interrogated using a BD FACSCalibur flow cytometer.
Results and Discussion
The benchmarking assay was performed to show that measurements of blood plasma using the microdevice were accurate when compared against a calibration curve produced with doped standards. Following these analyses, a pairwise scatterplot was constructed as shown in Figure 4, with the standard ELISA concentration on the x-axis and the novel measurement on the y-axis. A linear fit of the correlation line shows a slope of 1.02 and a correlation coefficient between the two measurement methods of r=0.98. It is therefore expected that continuous monitoring data taken with the same microdevice design should be accurate when calibrated against a standard curve.
Figure 4.
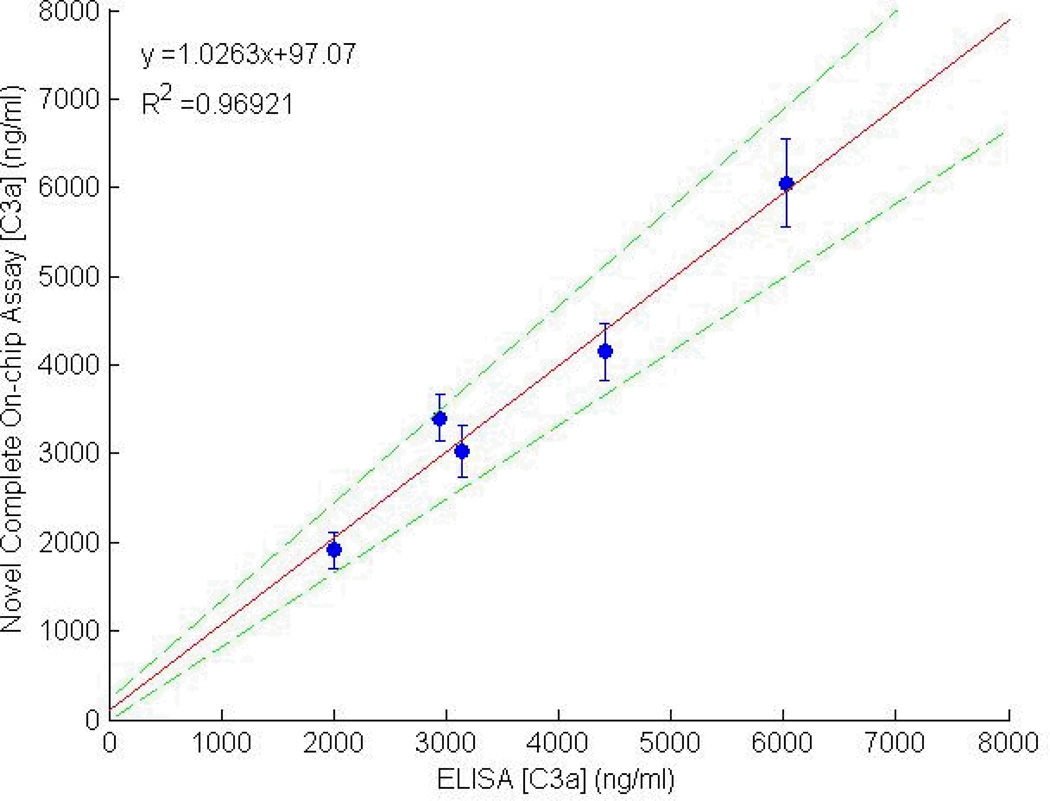
Pair-wise scatterplot comparing C3a measurements made with the microfluidic assay with those made using a standard ELISA plate. The error bars represent the fluorescence variation within the bead population (n=1000) analyzed in the microdevice. The dashed lines represent the 95% confidence interval of the data set.
The continuous monitoring experiment demonstrates proof of concept for the major technical challenges of an integrated sampling and analysis system. First, it coupling of the immunoassay device with a continuous plasma filtration device was demonstrated. The immunoassay device is unable to process whole blood directly from the circulation circuit because whole blood has the potential to disrupt the bead movement and antigen capture and/or clot or block small channels. Additionally, it is important to control and stabilize the flow into the immunoassay device for a period of time sufficient for CPB monitoring. During the simulated CPB circulation, the device operated for just under 3 hours. However, in previous laboratory testing the flow controller has been able to provide a continuous and steady flow rate for over 4 hours (data not shown). Also important is the ability of the microbeads to stay suspended for the duration of the experiment. If the microbeads settle to the bottom of the infusion syringe they will not be available for the assay. By using Ficoll-Paque Plus (GE Healthcare, Wakesha, WI) as a buffer, the density of the microbeads and carrier solution are nearly matched, so that the microbeads remain suspended for many hours. Finally, this experiment confirms the ability of the device to track time varying sample concentrations of an important inflammatory biomarker, complement C3a (Figure 5). While this data is taken in discrete batches, the microdevice can be integrated with a fluorescence detector to continuously measure the sample concentration. The integrated system will allow fully automated monitoring of inflammatory biomarkers. This microdevice assay should enable measurements with propagation delays of less than 20 minutes.
Figure 5.
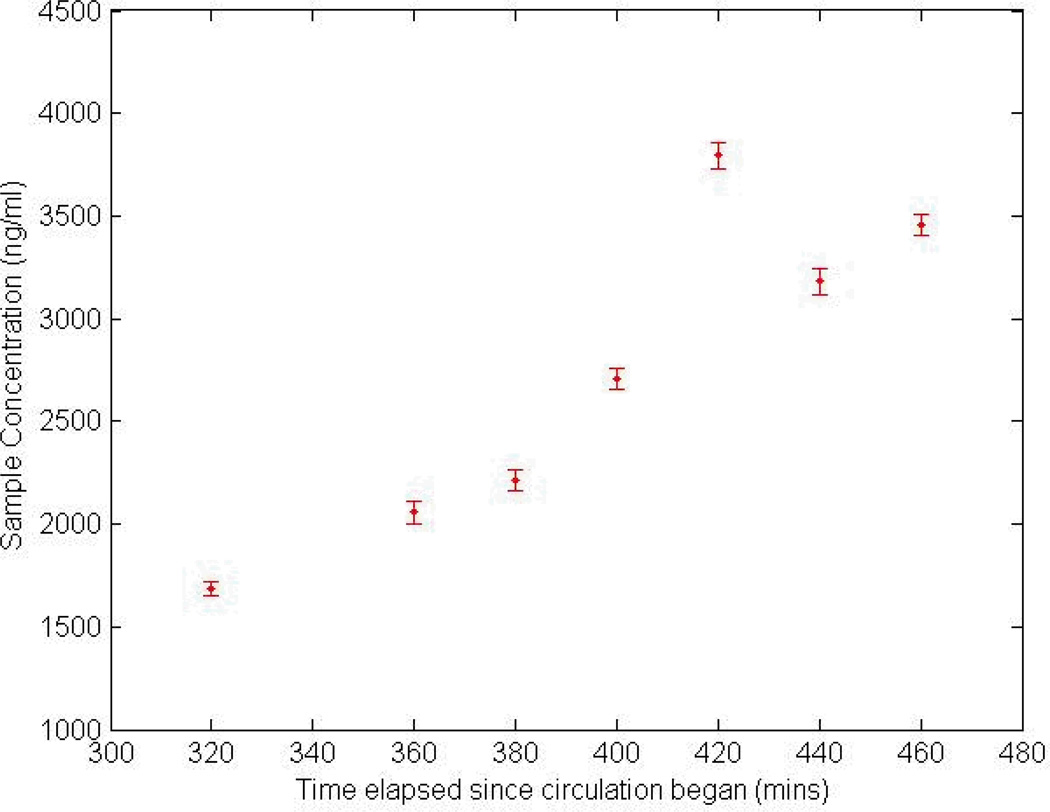
Temporal tracking of C3a concentration in an integrated plasma filtration and bead incubation device. Fluorescence labeling and flow cytometry of beads were conducted subsequent to antigen labeling of the beads in the microdevice. The 2nd time point was corrupted due to a problem with the flow cytometer and is excluded from the plot.
There are several significant advantages of continuous measurements of systemic inflammation during CPB as compared to conventional techniques which will be useful to the physicians. First, pharmacological agent selection including anesthetic drugs, in conjunction with different modes of perfusion has significant effects on systemic inflammation (21–28), and continuous monitoring of inflammatory biomarkers will benefit several ongoing surgical studies of the immune response to the use of certain anesthetic agents and anti-inflammatory steroids. Online measurements would permit a study of the effects of different pharmacological agents and provide indications for intraoperative substitution of agents. Second, most children’s hospitals in the US use blood ultrafilitration (UF) or modified UF to hemoconcentrate the circulating blood at the end of surgery which also removes pro-inflammatory cytokines (29–34). During UF, cytokine concentrations are not measured and UF is ended once the blood hematocrit level reaches a certain value. One study which would be enabled by the development of a continuous inflammatory protein monitor is examining how ending UF based on cytokine concentrations instead of just by blood hematocrit level, as is conventionally done, affects postoperative morbidity. Third, continuous monitoring of systemic inflammation will also allow the surgical team to determine any significant changes in inflammation levels during surgery to anticipate surgical complications in patients and give early indications of patients who may need more aggressive post-surgical support and recovery. The cause of inflammation can be more precisely defined, and intraoperative adjustments (enabled by novel clinical studies) such as anesthetic substitution, steroid administration or more aggressive plasma UF will be possible (35, 36).
Conclusion
This work demonstrates the first steps towards producing a continuous biochemical monitoring system for tracking CPB related inflammation during cardiac surgery. While the data from this continuous monitoring experiment show the expected increase in complement C3a concentration as result of mechanical perfusion, a comparative study monitoring the simulated CPB setup with the microfluidic assay and ELISA will be an important step in validating the micro-immunoassay. Future work will also include monolithic integration of the microfiltration and microimmunoassay modules to allow in-situ blood filtration and biomarker concentration assessments. Additionally, a multiplexing system will be incorporated so that multiple biomarkers can be measured simultaneously using the same blood sample stream. A validated assay capable of measuring multiple significant inflammatory biomarkers will provide critical information to surgical teams to reduce inflammation during CPB procedures and mitigate the harmful effects of inflammation.
Acknowledgments
This work was funded by grants from the NIH Heart Lung Blood Institute grant No. 1R21HL084367-01A1 and the Wallace H. Coulter Foundation Early Career Translational Research Awards in Biomedical Engineering. LAS received support from an NIH Biotechnology Fellowship Grant No. T32 GM008339-19.
Footnotes
Presented in part at the 8th International Conference on Pediatric Mechanical Circulatory Support Systems and Pediatric Cardiopulmonary Perfusion held on June 13 – 16, 2012, in Istanbul, Turkey
References
- 1.Kim T, Arnaoutakis GJ, Bihorac A, et al. Early Blood Biomarkers Predict Organ Injury and Resource Utilization Following Complex Cardiac Surgery. Journal of Surgical Research. 2009;168(2):168–172. doi: 10.1016/j.jss.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan CK, Newburger JW, McGrath E, et al. The Relationship Between Inflammatory Activation and Clinical Outcome After Infant Cardiopulmonary Bypass. Anesthesia and Analgesia. 2010;111(5):1244–1251. doi: 10.1213/ANE.0b013e3181f333aa. [DOI] [PubMed] [Google Scholar]
- 3.Westaby S, et al. Organ Dysfunction after cardiopulmonary bypass: a systemic inflammatory reaction initiated by the extracorporeal circuit. Intensive Care Med. 1987;13:89–95. doi: 10.1007/BF00254791. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg JB, Kapelanski DP, Olson JD, et al. CYTOKINE AND COMPLEMENT LEVELS IN PATIENTS UNDERGOING CARDIOPULMONARY BYPASS. Journal of Thoracic and Cardiovascular Surgery. 1993;106(6):1008–1016. [PubMed] [Google Scholar]
- 5.Fung M, Loubser PG, Undar A, et al. Inhibition of complement neutrophil, and platelet activation by an anti-factor D monoclonal antibody in simulated cardiopulmonary bypass circuits. Journal of Thoracic and Cardiovascular Surgery. 2001;122(1):113–122. doi: 10.1067/mtc.2001.114777. [DOI] [PubMed] [Google Scholar]
- 6.Sato K, Yamanaka M, Hagino T, et al. Microchip-based enzyme-linked immunosorbent assay (microELISA) system with thermal lens detection. Lab on a Chip. 2004;4(6):570–575. doi: 10.1039/b411356j. [DOI] [PubMed] [Google Scholar]
- 7.Cheng XH, Irimia D, Dixon M, et al. A microfluidic device for practical label-free CD4+T cell counting of HIV-infected subjects. Lab on a Chip. 2007;7(2):170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kartalov EP, Zhong JF, Scherer A, et al. High-throughput multi-antigen microfluidic fluorescence immunoassays. Biotechniques. 2006;40(1):85–90. doi: 10.2144/000112071. [DOI] [PubMed] [Google Scholar]
- 9.Ahn KC, Lohstroh P, Gee SJ, et al. High-throughput automated luminescent magnetic particle-based immunoassay to monitor human exposure to pyrethroid insecticides. Analytical Chemistry. 2007;79(23):8883–8890. doi: 10.1021/ac070675l. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharyya A, Klapperich CM. Design and testing of a disposable microfluidic chemiluminescent immunoassay for disease biomarkers in human serum samples. Biomedical Microdevices. 2007;9(2):245–251. doi: 10.1007/s10544-006-9026-2. [DOI] [PubMed] [Google Scholar]
- 11.Barton AC, Davis F, Higson SPJ. Labeless immunosensor assay for prostate specific antigen with picogram per milliliter limits of detection based upon an ac impedance protocol. Analytical Chemistry. 2008;80(16):6198–6205. doi: 10.1021/ac800491m. [DOI] [PubMed] [Google Scholar]
- 12.Wang HX, Meng S, Guo K, et al. Microfluidic immunosensor based on stable antibody-patterned surface in PMMA microchip. Electrochemistry Communications. 2008;10(3):447–450. [Google Scholar]
- 13.Yoo SK, Kim YM, Yoon SY, et al. Bead Packing and Release Using Flexible Polydimethylsiloxane Membrane for Semi-Continuous Biosensing. Artificial Organs. 2011;35(7):E136–E144. doi: 10.1111/j.1525-1594.2011.01240.x. [DOI] [PubMed] [Google Scholar]
- 14.Sasso LA, Undar A, Zahn JD. Autonomous magnetically actuated continuous flow microimmunofluorocytometry assay. Microfluidics and Nanofluidics. 2010;9(2):253–265. doi: 10.1007/s10404-009-0543-1. PMCID: PMC2916684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasso LA, Johnston IH, Zheng M, et al. Automated microfluidic processing platform for multiplexed magnetic bead immunoassays. Microfluidics and Nanofluidics. 2012 doi: 10.1007/s10404-012-0980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffy DC, McDonald JC, Schueller OJA, et al. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Analytical Chemistry. 1998;70(23):4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 17.Xia YN, Whitesides GM. Soft lithography. Annual Review of Materials Science. 1998;28:153–184. [Google Scholar]
- 18.Aran K, Fok A, Sasso LA, et al. Microfiltration platform for continuous blood plasma protein extraction from whole blood during cardiac surgery. Lab on a Chip. 2011;11(17):2858–2868. doi: 10.1039/c1lc20080a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Palanzo D, Undar A. Current ultrafiltration techniques before, during and after pediatric cardiopulmonary bypass procedures. Perfusion. 2012 doi: 10.1177/0267659112450061. [DOI] [PubMed] [Google Scholar]
- 20.Aran K, Fok A, Guan YL, et al. Differential Immune Activation During Simulated Cardiopulmonary Bypass Procedure Using Freshly Drawn and Week-Old Blood-A Pilot Study. Artificial Organs. 2010;34(11):1048–1053. doi: 10.1111/j.1525-1594.2010.01122.x. [DOI] [PubMed] [Google Scholar]
- 21.Bach F, Grundmann U, Bauer M, et al. Modulation of the inflammatory response to cardiopulmonary bypass by dopexamine and epidural anesthesia. Acta Anaesthesiologica Scandinavica. 2002;46(10):1227–1235. doi: 10.1034/j.1399-6576.2002.461010.x. [DOI] [PubMed] [Google Scholar]
- 22.DQ C, YP C, DQ Z. Effects of ketamine on cardiopulmonary bypass-induced interlekin-6 and interleukin-8 response and its significance. Hunan Yi Ke Da Xue Xue Bao. 2001;26:350–352. [PubMed] [Google Scholar]
- 23.El Azab SR, Rosseel PMJ, De Lange JJ, et al. Effect of sevoflurane on the ex vivo secretion of TNF-alpha during and after coronary artery bypass surgery. European Journal of Anaesthesiology. 2003;20(5):380–384. doi: 10.1017/s0265021503000577. [DOI] [PubMed] [Google Scholar]
- 24.Giraud O, Molliex S, Rolland C, et al. Halogenated Anesthetics reduce interleukin-1 beta-induced cytokine secretion by rat alveolar type II cells in primary culture. Anesthesiology. 2003;98(1):74–81. doi: 10.1097/00000542-200301000-00015. [DOI] [PubMed] [Google Scholar]
- 25.Sun J, Wang XD, Liu H, et al. Ketamine suppresses endotoxin-induced NF-kappa B activation and cytokines production in the intestine. Acta Anaesthesiologica Scandinavica. 2004;48(3):317–321. doi: 10.1111/j.0001-5172.2004.0312.x. [DOI] [PubMed] [Google Scholar]
- 26.Takala RSK, Soukka HR, Salo MS, et al. Pulmonary inflammatory mediators after sevoflurane and thiopentone anaesthesia in pigs. Acta Anaesthesiologica Scandinavica. 2004;48(1):40–45. doi: 10.1111/j.1399-6576.2004.00266.x. [DOI] [PubMed] [Google Scholar]
- 27.Tomasdottir H, Hjartarson H, Ricksten A, et al. Tumor necrosis factor gene polymorphism is associated with enhanced systemic inflammatory response and increased cardiopulmonary morbidity after cardiac surgery. Anesthesia and Analgesia. 2003;97(4):944–949. doi: 10.1213/01.ANE.0000078574.76915.11. [DOI] [PubMed] [Google Scholar]
- 28.Undar A, Eichstaedt HC, Clubb FJ, et al. Anesthetic induction with ketamine inhibits platelet activation before, during, and after cardiopulmonary bypass in baboons. Artificial Organs. 2004;28(10):959–962. doi: 10.1111/j.1525-1594.2004.07377.x. [DOI] [PubMed] [Google Scholar]
- 29.Berdat PA, Eichenberger E, Ebell J, et al. Elimination of proinflammatory cytokines in pediatric cardiac surgery: Analysis of ultrafiltration method and filter type. Journal of Thoracic and Cardiovascular Surgery. 2004;127(6):1688–1696. doi: 10.1016/j.jtcvs.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 30.Chew MS, Brandslund I, Brix-Christensen V, et al. Tissue injury and the inflammatory response to pediatric cardiac surgery with cardiopulmonary bypass - A descriptive study. Anesthesiology. 2001;94(5):745–753. doi: 10.1097/00000542-200105000-00010. [DOI] [PubMed] [Google Scholar]
- 31.Hickey E, Karamlou T, You J, et al. Effects of circuit miniaturization in reducing inflammatory response to infant cardiopulmonary bypass by elimination of allogeneic blood products. Annals of Thoracic Surgery. 2006;81(6):S2367–S2372. doi: 10.1016/j.athoracsur.2006.02.071. [DOI] [PubMed] [Google Scholar]
- 32.Liu JP, Ji BY, Long C, et al. Comparative effectiveness of methylprednisolone and zero-balance ultrafiltration on inflammatory response after pediatric cardiopulmonary bypass. Artificial Organs. 2007;31(7):571–575. doi: 10.1111/j.1525-1594.2007.00423.x. [DOI] [PubMed] [Google Scholar]
- 33.Sever K, Tansel T, Basaran M, et al. The benefits of continuous ultrafiltration in pediatric cardiac surgery. Scandinavian Cardiovascular Journal. 2004;38(5):307–311. doi: 10.1080/14017430410021480. [DOI] [PubMed] [Google Scholar]
- 34.Williams GD, Ramamoorthy C, Chu L, et al. Modified and conventional ultrafiltration during pediatric cardiac surgery: Clinical outcomes compared. J Thorac Cardiovasc Surg. 2006;132(6):1291–1298. doi: 10.1016/j.jtcvs.2006.05.059. [DOI] [PubMed] [Google Scholar]
- 35.Fujita F, Ishihara M, Kusama Y, et al. Effect of modified ultrafiltration on inflammatory mediators, coagulation factors, and other proteins in blood after an extracorporeal circuit. Artificial Organs. 2004;28(3):310–313. doi: 10.1111/j.1525-1594.2004.47230.x. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Hoschtitzky A, Allen ML, et al. An analysis of oxygen consumption and oxygen delivery in euthermic infants after cardiopulmonary bypass with modified ultrafiltration. Annals of Thoracic Surgery. 2004;78(4):1389–1396. doi: 10.1016/j.athoracsur.2004.02.032. [DOI] [PubMed] [Google Scholar]


