Summary
Adenocarcinoma is the most common type of lung cancer, the leading cause of cancer deaths in the world. Early detection is the key to improve the survival of lung adenocarcinoma patients. We have previously shown that microRNAs were stably present in sputum and could be applied to diagnosis of lung cancer. The aim of this study was to develop a panel of microRNAs that can be used as highly sensitive and specific sputum markers for early detection of lung adenocarcinoma. This study contained three phases: (1) marker discovery using microRNA profiling on paired normal and tumor lung tissues from 20 patients with lung adenocarcinoma; (2) marker optimization by real-time RT-qPCR on sputum of a case-control cohort consisting of 36 cancer patients and 36 health individuals; and (3) validation on an independent set of 64 lung cancer patients and 58 cancer-free subjects. From the surgical tissues, seven microRNAs with significantly altered expression were identified, of which “four” were overexpressed and “three” were underexpressed in all 20 tumors. On the sputum samples of the case-control cohort, four (miR-21, miR-486, miR-375, and miR-200b) of the seven microRNAs were selected, which in combination produced the best prediction in distinguishing lung adenocarcinoma patients from normal subjects with 80.6% sensitivity and 91.7% specificity. Validation of the marker panel in the independent populations confirmed the sensitivity and specificity that provided a significant improvement over any single one alone. The sputum markers demonstrated the potential of translation to laboratory settings for improving the early detection of lung adenocarcinoma.
Keywords: MicroRNA, sputum, lung adenocarcinoma, real-time RT-qPCR, diagnosis
Introduction
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer death in the United States. NSCLC is histologically subdivided into four major subtypes with distinct pathological characteristics: adenocarcinoma, squamous cell carcinoma, large cell carcinoma and “other” (neuroendocrine cancers, carcinoids etc.). The disease is usually diagnosed at advanced stages when the prognosis is poor, resulting in an overall 5-year survival rate of approximately 14% 1. However, the 5-year survival rate in patients with stage I NSCLC that has been resected can be as high as 83% 1. Therefore, finding early stage NSCLC may reduce the mortality 1. In particular, early identification of lung adenocarcinoma is clinically important, because it is now the most common type of lung cancer 1, accounting for 40% of all NSCLCs. Furthermore, the incidence of lung adenocarcinoma is on the rise in many countries, mainly, in women and nonsmokers 2,3. In addition, because adenocarcinoma arises in peripheral lung tissue and originates from the smaller airways, it is more difficult to be detected by bronchoscopy or sputum cytology 3. Moreover, computed tomography (CT) provides excellent anatomic information and can detect lung tumor at small size, however the improved sensitivity is associated with over-diagnosis 1-3. Thus, the major obstacle in management of lung adenocarcinoma is the lack of adequate method for its early detection.
MicroRNAs (miRNAs) are a new class of small noncoding RNAs that regulate gene expression and are involved in a variety of biologic and pathologic processes 4. The differential expression of miRNAs in human cancers and its potential diagnostic values have been previously investigated 4-7. For instance, by analyzing changes of a large-scale miRNAs on 540 human cancer specimens including lung, breast, stomach, prostate, colon, and pancreatic tumors, Volinia et al, identified a solid cancer miRNA signature composed by a large portion of overexpressed miRNAs that provides potential diagnostic targets for the tumors 4. Our recent proof of principle study 8 showed that endogenous miRNAs were present in sputum in a remarkably stable form and could reliably be detected by real-time reverse transcription (RT)-quantitative (q)PCR. Furthermore, detecting elevated expression of a single miRNA, miR-21, produced a higher sensitivity in diagnosis of lung cancer compared with sputum cytology. Our data suggested that the measurement of altered miRNA expressions in sputum sample could be a useful noninvasive approach for lung cancer diagnosis. However, the sensitivity reached by a single miRNA is low for clinical application 8.
It has been widely accepted that lung tumor is a heterogeneous disease and develops from complex and multistep processes 2, 9. We therefore hypothesized that simultaneous assessment of a panel of tumor-specific miRNAs that, used in combination in sputum, could provide a highly sensitive and specific diagnostic test for early stage lung adenocarcinoma. To verify the hypothesis, we first identified miRNA signatures of stage I lung adenocarcinoma using miRNA profiling on primary tumor tissues. From these signatures, we then optimized and validated a panel of miRNAs that could be detected in sputum for the early detection of lung adenocarcinoma.
Materials and Methods
Patients and clinical specimens
To define miRNA signatures for lung adenocarcinoma, surgical specimens were obtained from 20 lung cancer patients who had either a lobectomy or a pneumonectomy. All cases were diagnosed with histologically confirmed stage I lung adenocarcinoma (Table 1). None of the patients had received preoperative adjuvant chemotherapy or radiotherapy. Tumor tissues were intraoperatively dissected from the surrounding lung parenchyma; paired normal lung tissues were also obtained from the same patients at an area distant from their tumors. Serial cryostat sections from the specimens were stained with hematoxylin and eosin to confirm the diagnosis based on the most recent WHO classification of tumors of the lung 10.
Table 1. Demographics of 20 patients diagnosed with lung adenocarcinoma.
| Age* | 65 (55-78) |
| Sex | |
| Female | 8 |
| Male | 12 |
| Smoking status | 17 smokers |
| Pack-years | 35 ± 22 |
| Location of tumor † | All are peripheral tumors |
| Stage | All are stage I |
| Histology | All are lung adenocarcinoma |
Data are presented as median (range).
Peripheral tumors were located at or within 1 cm of the visceral pleura.
To optimize a panel of miRNAs that could be detected in sputum, 36 stage I lung adenocarcinoma patients and an equal number of normal subjects were recruited. The case and control were matched in the ratio of 1:1 by age, gender, and smoking history as a nested case-control cohort (Supplement Table 1). Sputum was collected from the participants as described in our recent reports 8, 11-2. To further validate the identified sputum markers, we collected sputum specimens from a total of 64 NSCLC patients and 58 healthy controls. The demographic and clinical characteristics of the NSCLC patients are summarized in Table 2. Tumors were classified as peripheral or central on the basis of radiographic studies, bronchoscopic or operative findings, and pathologic analysis. The study was approved by Institutional Review Board.
Table 2. Demographic and clinical characteristics of NSCLC patients and healthy subjects in an independent corhot.
| NSCLC (n = 64) | Controls (n = 58) | P-value | |
|---|---|---|---|
| Age | 67 (SD 11.5) | 65 (SD 10.6) | >0.05 |
| Sex | |||
| Female | 25 | 23 | >0.05 |
| Male | 39 | 35 | >0.05 |
| Race | |||
| White | 41 | 37 | >0.05 |
| African American | 23 | 21 | >0.05 |
| Smoking status | |||
| Pack-years | 30.9 (SD 24.8) | 27.7 (SD 28.4) | >0.05 |
| Location of tumor * | >0.05 | ||
| Central | 30 | ||
| Peripheral | 34 | ||
| Stage | >0.05 | ||
| Stage I | 16 | ||
| Stage II | 15 | ||
| Stage III | 17 | ||
| Stage IV | 16 | ||
| Histology | >0.05 | ||
| AC of lung | 33 | ||
| SC of lung | 31 |
Abbreviations: NSCLC: non–small-cell lung cancer; SD, standard deviation. AC, adenocarcinoma; SC, squamous cell carcinoma.
Peripheral tumors were located at or within 1 cm of the visceral pleura.
RNA isolation
Total RNA containing small RNA was extracted from the tissue and sputum specimens as described in our previous study 8 by using a mirVana miRNA Isolation Kit (Ambion, Austin, TX). The purity and concentration of RNA were determined from OD260/280 readings using a dual beam UV spectrophotometer (Eppendorf AG, Hamburg, Germany). RNA integrity was determined by capillary electrophoresis using the RNA 6000 Nano Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Only RNA extracts with RNA integrity number values >6 underwent in further analysis.
MiRNA profiling of surgical resected lung adenocarcinoma tissues
MiRNA profiling was performed using Taqman human microRNA array A (System Biosciences, Mountain View, CA), which is a PCR-based array and contains 377 most commonly found human mature miRNAs and 4 small RNA endogenous controls. Briefly, 100 ng of total RNA was polyadenylated by poly(A) polymerase and then reverse transcribed to cDNA. RT-qPCR was performed using miRNA specific primers provided by the manufacturer in ABI PRISM 7500 Real-time PCR system (Applied Biosystems, Foster City, CA). The cycle threshold (Ct) was defined as the number of cycles required for the fluorescent signal to cross the threshold in PCR. ΔCt was calculated by subtracting the Ct values of the small control RNAs from the Ct values of the miRNA of interest. ΔΔCt was then computed by subtracting ΔCt of the normal control tissue from ΔCt of the tumor specimen, and fold-change of miRNA gene was determined by the equation 2– ΔΔCt.
Analysis of miRNA expression in sputum samples
Expression of the identified miRNA signatures was evaluated in sputum by using real-time RT-qPCR with Taqman miRNA assays (Applied Biosystems) as previously described 8. Expression of target miRNAs was normalized in relation to expression of small nuclear U6 RNA. U6 RNA was proven as an internal control for miRNA quantification in sputum in our previous study 8. All assays were performed in triplicates, and one no-template control and two interplate controls were carried along in each experiment. Expression levels of the mRNAs were calculated using comparative Ct method as previously described 8.
To determine the sensitivity and dynamic range of miRNA quantification in sputum, RNA was extracted from ten sputum specimens and then diluted at different orders of magnitude in diethylpyrocarbonate (DEPC) water (Sigma Chemical Co.. St. Louis, MO). Expressions of the miRNAs were then assessed by using RT-qPCR in the samples as described above. All tests were performed in triplicates.
Statistical analysis
To find miRNA genes that were statistically differentially expressed between lung adenocarcinoma specimens and the corresponding normal tissues, we expected the acceptable number of false positives to be 1.0, fold difference between normal and tumor of samples at 2.0, standard deviation of the gene measurements on the base-two logarithmic scale at 0.7, and desired power at 0.8. Given 377 miRNAs included in the array, at least 15 specimens for each tissue type were required to achieve the statistical criteria 13. To define an optimal miRNA marker panel that can be detected in sputum for distinguishing cancer patients from normal controls, a case-control study was designed that consisted of lung cancer cases and cancer-free individuals. We used receiver-operator characteristic (ROC) curve and the area under ROC curve (AUC) to determine sample size in the case-control study. The ROC curve is a plot of diagnostic test's sensitivity, or true positive rate versus 1-specifcity, or the false positive rate at various discrimination cutoffs depicting the trade-offs between the true positives and the false positives in diagnostic accuracy 14. The AUC represents an overall summary of diagnostic accuracy. ROC analysis is considered as a powerful tool to evaluate diagnostic tests and predictive models by assessing accuracy quantitatively or comparing accuracy between tests or predictive models 14,15. Furthermore, ROC analysis can be used to select optimal threshold under a variety of clinical circumstances, balancing the inherent tradeoffs that exist between sensitivity and sensitivity. In addition, ROC analysis is one of the most important approaches that are commonly used to determine sample size 16. In the case-control cohort study, the AUC of H0 (the null hypothesis) was set at 0.5. H1 represented the alternative hypothesis; accordingly, at least 28 subjects were required in each category to show a minimum difference of interest between an AUC of 0.75 versus an AUC of 0.5 with 80% power at the 5% significance level 14-6.
Statistical analysis of RT-qPCR data was done using Statistical Analysis System software version 6.12 (SAS Institute, Cary, NC). All P values shown were two sided, and a P value of <0.05 was considered statistically significant. ROC curve analysis was undertaken using expression level for each miRNA in sputum from cancer patients and cancer-free controls by Analyse-it software (Analyse-it Software, Leeds, UK) 8. Briefly, for each miRNA, we constructed the ROC curve and computed the AUC value by numerical integration of the ROC curve. Using this approach, the AUC identified maximum sensitivity and specificity levels at which to distinguish cancer patients from healthy subjects, yielding corresponding optimal thresholds defining expression levels of the tested genes. Logistic regression was used to generate prediction model building. Validated biomarkers were fitted into logistic regression models, and the stepwise backward model selection was performed to determine the best discriminating combinations of miRNAs. Furthermore, contingency table and logistic regression analysis were applied to determine the associations between the expression levels of the miRNAs and both clinicopathologic and demographic characteristics of the cases and controls.
Results
Identifying miRNA signatures whose aberrant expression levels were associated with lung adenocarcinoma
We used a TaqMan-based miRNA array to profile mature miRNAs in the matched lung adenocarcinoma and normal lung tissues. To determine if expression of these 377 miRNAs was readily detectable, wee prepared two RNA pools that contain equal amounts of RNA from 20 tumor tissues and 20 normal lung tissues, respectively. We then performed the miRNA array analysis on the pooled RNAs. 346 (92%) of the miRNAs had ≤ 30 Ct value, however only 32 (8%) of the genes displayed >30 Ct value (Supplement Fig. 1). Furthermore, four replicate sets of raw threshold data obtained by two research staff at two different times on the same specimens are directly compared. The results demonstrated a high degree of correlation (R2 > 0.992), suggesting that assay format yielded excellent reproducibility on the surgical resected specimens. Therefore, the miRNAs could be accurately and reliably measured in the clinical samples by the PCR-based miRNA array.
When P value <0.01 was used as a cutoff, of the 377 miRNA targets, 9 miRNAs were downregulated and 11 miRNAs were up-regulated with ≥ 1.5 fold-change in cancer group (Fig. 1 and Supplement Tables 2-3). Using a predefined criterion of a fold-change ≥2, we identified seven miRNAs that statistically differently expressed between the paired tumor and normal samples. These included three miRNAs (miR-486, miR-126, and miR-145) that were underexpressed, and four miRNAs (miR-21, miR-182, miR-375, and miR-200b) that were overexpressed in tumor specimens. It should be noted that altered expressions of the seven miRNAs existed in all 20 lung adenocarcinoma tissues compared with the paired normal specimens. We therefore assigned the seven miRNAs for further analysis.
Fig. 1.
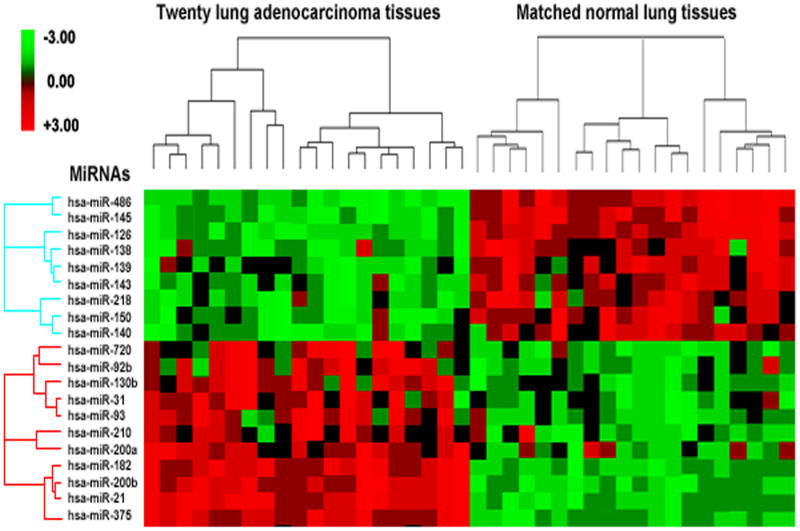
MiRNAs differentially express in lung adenocarcinomas versus normal lung tissues. Hierarchical clustering of 20 miRNA genes with a significantly different expression (p < 0.01) in tumor tissues. Rows represent individual genes; columns represent individual tissue samples. The scale represents the intensity of gene expression (log2 scale ranges between −3.0 and 3.0).
Optimizing a panel of highly specific and sensitive sputum miRNA markers for lung adenocarcinoma
We have previously demonstrated feasibility of measuring expressions of human endogenous miRNAs, miR-21 and miR-155, in sputum by RT-qPCR 8. To determine if the seven newly identified miRNAs could be reliably detected in the specimens, we prepared two RNA pools containing equal amounts of RNA from sputum samples of 10 cancer patients and 10 cancer-free individuals, respectively. All tested miRNAs had ≤30 Ct values in both pools, indicating that the miRNAs could easily be measured in sputum (data not shown). To further determine the sensitivity of detecting the miRNAs by RT-qPCR in sputum, the total RNA was diluted in DEPC water at different concentrations. The serially diluted RNAs served as experimental samples for measuring expression of each miRNA. There was an excellent linearity between the RNA input and the Ct values for the miRNA tested (Supplement Fig.2). In addition, the assay had a dynamic range of at least six orders of magnitude (R2= 0.998), and was capable of detecting as little as 0.86 pg of RNA and 10 copies of the target genes. Altogether, the miRNAs identified from the primary tumor tissues were readily detectable in sputum. The seven miRNAs were therefore continually tested in all individual sputum samples collected from a case-control of 36 patients diagnosed with stage I lung adenocarcinoma and 36 health subjects.
MiR-486, miR-126, and miR-145 showed lower expression levels, whereas miR-21, miR-182, miR-375, and miR-200b displayed higher expression levels in cancer patients' sputum compared with sputum of cancer-free individuals (Table 3) (All p < 0.01). The data was in agreement with the results obtained from the tissue specimens. ROC analyses were performed to evaluate the capability of using the miRNAs in sputum to discriminate between cancer patients and cancer-free individuals. As depicted in Table 3, the seven miRNAs showed 0.807-0.846 AUC values. When optimum cutoffs were selected, the miRNAs yielded 59.5-72.6% sensitivity and 73.8-82.9% specificity, respectively, implying that the miRNAs held promise as cancer-specific markers in sputum.
Table 3. Expression levles of the seven miRNAs and their diagnostic significace in sputum of 36 lung adenocarcinoma patients and 36 healthy controls.
| MiRNA | Mean (SD) in cancer patients | Mean (SD) in healthy controls | P* | AUC* (SE) | Cutoffs | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|
| MiR-486 | 0.051 (0.034) | 0.182 (0.167) | <0.001 | 0.834 (0.034) | 0.108 | 66.9 | 79.4 |
| MiR-126 | 0.083 (0.087) | 0.258 (0.198) | <0.001 | 0.824 (0.032) | 0.156 | 67.2 | 73.8 |
| MiR-145 | 0.132 (0.073) | 0.351 (0.297) | <0.001 | 0.807 (0.031) | 0.226 | 59.5 | 82.9 |
| MiR-21 | 8.217 (5.680) | 2.730 (2.354) | <0.001 | 0.846 (0.038) | 5.664 | 72.6 | 79.2 |
| MiR-182 | 7.463 (5.822) | 2.212 (1.164) | <0.001 | 0.825 (0.033) | 4.973 | 64.3 | 79.5 |
| MiR-200b | 6.623 (5.367) | 2.931 (1.285) | <0.001 | 0.823 (0.036) | 4.838 | 62.9 | 78.5 |
| MiR-375 | 7.207 (6.012) | 2.894 (1.766) | <0.001 | 0.822 (0.035) | 5.267 | 63.9 | 80.6 |
Abbreviations: AUC, the area under receiver operating characteristic curve; SD, standard deviation; SE, standard error.
Both the P value and AUC were obtained using the U6-normalized values
To optimize a small panel of miRNA markers for the early detection of lung adenocarcinoma with high sensitivity and specificity, logistic regression of all seven miRNAs using a backward elimination approach was performed. One of the logistic regression models was built based on four miRNAs, miR-486, miR-21, miR-200b, and miR-375, which in combination provided the best prediction. Combing the four miRNAs produced 0.896 AUC, being considerably higher than 0.807-0.846 AUC values of each individual gene in distinguishing cancer patients from normal subjects (All P<0.05) (Fig. 2). Accordingly, the ROC curves revealed that the sensitivity and specificity for the combination of the four miRNAs were 80.6% and 91.7%, which were significantly higher than 59.5-72.6% sensitivity and 73.8-82.9% specificity of the individual miRNAs (All p<0.05) (Supplement Fig.3).
Fig. 2.
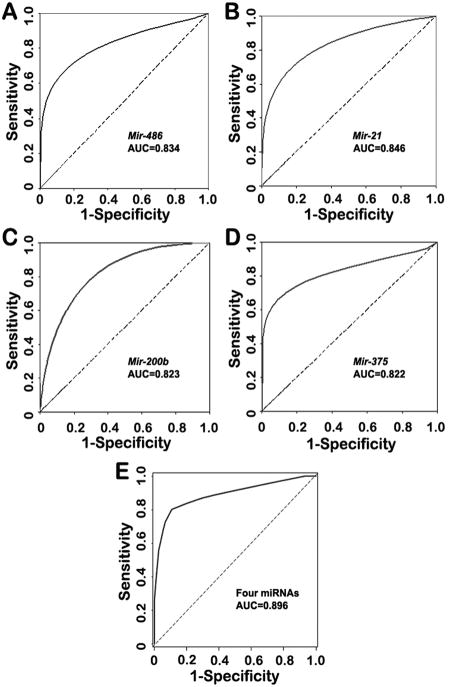
Receiver-operator characteristic (ROC) curve analysis of expression levels of the four miRNAs in sputum of 36 patients diagnosed with stage I lung adenocarcinoma and 36 health subjects. The area under the ROC curve (AUC) for each miRNA conveys its accuracy for differentiation of lung adenocarcinoma patients and healthy subjects in terms of sensitivity and specificity. The individual genes produce 0.822-0.846 AUC values (A-D), being significantly lower than 0.896 AUC by the four genes combined as a marker panel (E) (All P<0.05).
Validating the sputum miRNA markers in an independent set of NSCLC patients
To further evaluate the diagnostic performance of the optimal markers, the four miRNAs were assessed on sputum samples of 64 patients with different stages and histological types of NSCLC and 58 healthy controls. The miRNAs had significantly different expression levels in sputum between NSCLC and cancer-free controls (All p<0.001) (Supplement Table 4). The ROC curve analysis showed that the individual miRNAs displayed 0.713-0.789 AUC values in identification of NSCLC patients. When optimal cutoffs were selected, the individual miRNAs produced 55.1-62.6% sensitivity and 69.4-73.8% specificity (Supplement Table 4). The four miRNAs in combination could differentiate the NSCLC patients from healthy controls with 0.839 AUC, producing 70.3% sensitivity and 80.0% that were statistically higher than those by any single one used alone (all p<0.05) (Supplement Table 4). Furthermore, the panel of markers had different diagnostic efficiency for different histological types of NSCLC (Table 4): the sensitivity and specificity for lung adenocarcinoma were 80.6% and 92.5%, being similar to those (80.6% and 91.7%) in the above case-control cohort that only consisted of the patients diagnosed with stage I adenocarcinoma and healthy controls (All p>0.05). The parameters were statistically higher than those in diagnosis of squamous cell carcinoma (64.1 % and 71.3%, all p<0.05), suggesting that the miRNA markers had higher diagnostic efficiency for adenocarcinoma compared with squamous cell carcinoma of the lung. In addition, the sensitivity and specificity of the four miRNAs combined were 78.3% and 93.8% for peripheral cancer, whereas 65.8% and 70.9% for central tumor, respectively (Table 4), indicating that the miRNAs had better efficiency in detecting peripherally located cancers than central tumors (All p<0.05). However, no statistically significant difference was found in the sensitivity and specificity of the markers for stage I, II, III, and IV lung adenocarcinomas (p>0.05) (Table 4). Moreover, in a univariate analysis, histological type and location of the tumors were associated with expression levels of the miRNAs in sputum samples (Supplement Table 5) (All p > 0.05). There was no association of expressions of the miRNAs with the age, gender, ethnic group, tumor stage, or histories of smoking of the lung cancer patients and normal individuals (Supplement Tables 5-6) (All p > 0.05). Taken together, the results confirm that the optimal set of miRNAs could be used as specific biomarkers for the early detection of lung adenocarcinoma.
Table 4. Diagnostic efficiency of the miRNA marker panel on sputum of 64 NSCLC patients and 58 healthy subjects*.
| Sensitivity | Specificity | P | |
|---|---|---|---|
| Histological types | All <0.05 | ||
| AC | 80.60% | 92.50% | |
| SC | 64.10% | 71.30% | |
| Tumor location | All <0.05 | ||
| Peripheral tumor † | 78.26% | 93.80% | |
| Central tumor | 65.80% | 70.92% | |
| Stage of NSCLC | All >0.05 | ||
| I | 69.22% | 81.70% | |
| II | 69.90% | 82.50% | |
| III | 71.50% | 79.10% | |
| IV | 70.90% | 82.70% |
The results were determined by using ROC curve analysis with optimum cutoffs.
Abbreviations: NSCLC, non–small-cell lung cancer; AC, adenocarcinoma; SC, squamous cell carcinoma.
Peripheral tumors were located at or within 1 cm of the visceral pleura.
Discussion
The development of highly accurate biomarkers that can be detected in easily accessible body fluids is a major research effort in the field of lung cancer early detection 17. Sputum, particularly, has been considered as potential surrogate material for noninvasive diagnosis of lung cancer, because it is a mirror to lung disease 17. Conventional cytologic analysis of sputum has been used clinically to diagnose lung cancer; however, it was no more effective than chest radiographs in detecting lung cancer in several large prospective randomized trials 18. The molecular genetic alterations could occur before morphological changes that can be found by a cytological test 8, 11,12, 19-22. Furthermore, the molecular genetic changes seen in sputum may reflect the same abnormalities found in lung tumors 11, 21. Therefore, there is a long history of identifying and developing molecule genetic changes that can be tested in sputum as biomarkers 8, 11, 12, 17, 19-22. For instance, mutations of oncogene (e.g., K-ras) or tumor suppressor gene (e.g., P53) were detected in sputum of patients with primary adenocarcinoma of the lung 19,20. Hypermethylation of p16 gene was found in sputum collected from patients with lung cancer, 5-35 months before sputum cytological and clinical diagnoses 21. However, to date there is no molecular genetic marker accepted in clinical settings.
In our recent proof of principle study 8, we demonstrated that measuring elevated expression level of a single miRNA in sputum produced higher sensitivity in identification of lung cancer than did conventional sputum cytology. To enhance the diagnostic power of miRNAs in sputum for lung adenocarcinoma, here we developed and characterized a sputum-based miRNA marker panel with a sensitivity of 80.6% and a specificity of 91.7%. This study further extends our previous research efforts to develop sputum-based diagnostic tool for lung cancer 8, 11,12, 22. Given the expenses associated with quantitative molecular analyses, a marker panel with the smallest number of miRNAs and highest diagnostic accuracy would provide a cost-effective diagnostic assay for lung adenocarcinoma.
Among the four miRNAs identified, up-regulation of miR-21 has been found in many human cancer specimens 23. Therefore, extensive efforts have been taken to identify the downstream genes and gene networks regulated by miR-21 and the upstream factors that can regulate dysfunction of miR-2123-25. For example, elevated miR-21 expression might be associated with apoptosis inhibition and acquisition of invasive properties, likely mediated by its downregulating effects on the expression of target tumor suppressors PTEN, TPM, and PDCD423-25. More importantly, miR-21 itself displays oncogenic activity and can be classed as an oncomir, whose overexpression lead to tumor development and progression 24. This current study confirmed our previous finding 8 that the assessment of miR-21 overexpression in sputum had higher sensitivity compared with cytologic examination. Therefore, miR-21 can serve as an important biomarker for the early detection of lung cancer. MiR-200b locates on chromosome 1p36.33, one of the most common regions with genomic amplicons in solid tumors including lung cancer 27-30. Although biological mechanism of miR-200b dysfunction in lung tumorigenesis is unclear, miR-200b was recently identified as one of a set of miRNAs whose aberrant expressions were related to recurrence of stage I NSCLC after surgical resection 31. Consistently, we herein found that miR-200b overexpression existed in lung adenocarcinomas, one of the major histological types of NSCLC. Altogether, the observations suggest that miR-200b could be a potential target of the genomic amplification in 1p36.33, and its activation might be involved in lung carcinogenesis. MiR-375 down-regulation was found in some human malignancies 32,33. However, miR-375 was consistently up-regulated in adenocarcinoma rather than squamous cell carcinoma of lungs 34. Furthermore, when comparing cancerous tissue expression between adenocarcinoma and squamous cell carcinoma patients with esophagus cancer, Mathé et al. 33 found that miR-375 was elevated in adenocarcinoma patients. In good agreement with the findings, our present data showed that miR-375 was overexpressed in lung adenocarcinoma, and Moreover, measuring its expression in sputum displayed higher accuracy in diagnosis of lung adenocarcinoma compared with squamous cell lung cancer. On the other hand, miR-486 is located on one of the most frequent genomic rearrangement regions, chromosome 8p11.21 that contain potential tumor suppressor genes in lung tumorigenesis 27,28. Navon et al. recently found that miR-486 was underexpressed in eight types of human tumors, including lung cancer 35. The observation from our present research is consistent with the previous findings, suggesting that miR-486 might be a potential tumor suppressor in carcinogenesis. Although miR-182 was not eventually included in the panel of the four miRNAs, its over-expression was found in primary NSCLC tissues and sputum from the patients. MiR-182 is a member of a miRNA cluster in a chromosomal locus (7q31-34) frequently amplified in solid tumors 36. Segura, et al recently found that miR-182 was commonly up-regulated in human melanoma cell lines and tissue samples, and this up-regulation correlated with gene copy number in a subset of melanoma cell lines 36. Furthermore, the aberrant miR-182 expression could promote tumorigenesis by repressing FOXO3 and microphthalmia-associated transcription factor in several types of human cancers 37-9. Our primary goal of the current study is marker development. We showed for the first time that measuring altered expressions of a small panel of miRNAs in sputum might be a potential noninvasive test for the early detection of lung adenocarcinoma. The biological relevance of the miRNA dysfunctions in lung tumorigenesis are currently being investigated at our laboratory.
The panel of sputum markers could also identify squamous cell lung cancer with 64.1 % sensitivity and 71.3% specificity. However, the markers are more accurate to lung adenocarcinoma with 80.6% sensitivity and 92.5% specificity. The main reason might be that the miRNAs are identified from surgically resected primary adenocarcinomas, thus should be more specific for the type of NSCLC. It is also not surprising to find that the abnormal miRNAs expression levels are related to the tumors located in peripheral airways, because most peripheral tumors are adenocarcinomas. These observations would be clinically important, because the majority of NSCLCs detected by cytologic analysis and visible by bronchoscopy are squamous cell carcinoma predominantly locating in central areas of the lungs, rather than adenocarcinoma that is the most common type in NSCLC. Once confirmed, the sputum miRNA panel might improve the detection rate for lung adenocarcinomas that are more difficult to be found by these conventional techniques.
Most of the previously identified lung cancer associated molecular genetic changes were related to the smoking status. Some of the changes can be found in healthy smokers who never develop lung cancer 2,3, 8, 17. The use of such molecular genetic alterations as biomarkers might produce high false positive diagnostic rate or over-diagnosis, thus impeding their application in clinical settings in screening or early detection of lung cancer. The four miRNA markers identified from the present research is encouraging, because expression levels of the miRNAs appear to be independent of subject age, gender, ethnic subgroup, and tobacco smoking. The identified miRNAs could dysregulate in a cancer-specific manner. In addition, no significant differences of the miRNA expression levels were observed for the cancerous samples at different stages of the disease, implying that the potential markers were not stage-specific. The results further provide evidence that this miRNA marker panel might be useful in the early detection of lung adenocarcinoma, although whether the expression levels of the four miRNAs are affected in non-cancer associated lung pathologies remains to be investigated.
This panel of miRNA markers detected by RT-qPCR platform provides a significant improvement over any single one, and hence shows promise as a lung adenocarcinoma-specific test on sputum. In the future, developing and using an independent methodology, e.g., solution hybridization 40, to evaluate the expression levels of the miRNAs may continue to improve efficiency of the sputum-based biomarkers. Furthermore, comparing the miRNAs on sputum to CT imaging for the early detection of lung adenocarcinoma will lead to more understanding the diagnostic value of the biomarkers. In addition, integration of the sputum-based markers with the current conventional modalities, especially CT, could facilitate noninvasive diagnostic efficiency and accuracy for early lung adenocarcinoma or screening of high-risk patients for the cancer.
In conclusion, we have developed a panel of miRNAs that can be reliably measured in sputum. Detection of the miRNAs could be used as a noninvasive and cost-effective diagnostic tool for early lung adenocarcinoma. Nonetheless, a large multi-center clinical project to further validate the full utility is warranted before it could potentially be adopted in routine clinical settings.
Supplementary Material
Brief statements describing the novelty and impact of the paper
MicroRNAs (miRNAs) are emerging as highly tissue-specific biomarkers with potential for clinical applicability in indentifying and defining cancer type. Here we aimed to develop a panel of miRNAs that can be used as highly sensitive and specific sputum markers for early detection of lung adenocarcinoma, the most common type of lung cancer. This study was divided into three phases: (1) marker discovery using reverse transcription (RT) PCR-based miRNA profiling on 20 stage I lung adenocarcinoma tissues; (2) marker optimization by real-time RT-quantitative (q)PCR on sputum of 36 cancer patients and 36 health individuals; and (3) validation on an independent set of 64 lung cancer patients and 58 cancer-free subjects. We successfully developed a small panel of sputum markers consisting of four miRNAs, which in combination yielded 80.6% sensitivity and 91.7% specificity for the diagnosis of early stage lung adenocarcinoma. This is the first report that measuring a miRNA panel in sputum for early detection of lung adenocarcinoma. Future integration of the sputum-based markers with the current conventional modalities, especially computed tomography, could provide a noninvasive and cost-effective diagnostic tool for early stage lung adenocarcinoma or screening of high-risk patients for the disease.
Acknowledgments
Grant support: This work was supported in part by National Cancer Institute (NCI) grants CA-135382, CA-137742, and CA-133956, American Cancer Society Research Scholar Grant, an clinical innovator award from Flight Attendant Medical Research Institute, a scholar career development award from NIH K12RR023250-University of Maryland Multidisciplinary Research Career Development Program, and an exploratory research grant from Maryland Stem Cell Fund (F. J.).
Abbreviations used
- NSCLC
non-small-cell lung cancer
- MiRNAs
microRNAs
- CT
computed tomography
- ROC
receiver-operator characteristic
- AUC
the area under receiver operating characteristic curve
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- Ct
cycle threshold
- DEPC
diethylpyrocarbonate
References
- 1.American Cancer Society: Cancer Facts & Figures. Atlanta: American Cancer Society; 2009. [Google Scholar]
- 2.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 3.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7:778–90. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 4.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9:572–94. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- 8.Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–6. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colby TV, Wistuba II, Gazdar A. Precursors to pulmonary neoplasia. Adv Anat Pathol. 1998;5:205–9. doi: 10.1097/00125480-199807000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Colby TV, Noguchi M, Henschke C. Adenocarcinoma. In: Travis WD, Brambilla E, Muller-Hermelink HK, et al., editors. Pathology and Genetics, Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; pp. 2005pp. 35–44. [Google Scholar]
- 11.Li R, Todd NW, Qiu Q, Fan T, Zhao RY, Rodgers WH, Fang HB, Katz RL, Stass SA, Jiang F. Genetic deletions in sputum as diagnostic markers for early detection of stage I non-small cell lung cancer. Clin Cancer Res. 2007;13:482–7. doi: 10.1158/1078-0432.CCR-06-1593. [DOI] [PubMed] [Google Scholar]
- 12.Qiu Q, Todd NW, Li R, Peng H, Liu Z, Yfantis HG, Katz RL, Stass SA, Jiang F. Magnetic enrichment of bronchial epithelial cells from sputum for lung cancer diagnosis. Cancer. 2008;114:275–83. doi: 10.1002/cncr.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahai H, Kurshid A. Formulae and tables for the determination of sample sizes and power in clinical trials for testing differences in proportions for the two-sample design: A review. Statistics in Medicine. 1996;15:1–21. doi: 10.1002/(SICI)1097-0258(19960115)15:1<1::AID-SIM134>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 14.Dodd LE, Pepe MS. Partial AUC estimation and regression. Biometrics. 2003;59:614–23. doi: 10.1111/1541-0420.00071. [DOI] [PubMed] [Google Scholar]
- 15.Zou KH, O′Malley AJ, Mauri L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation. 2007;115:654–7. doi: 10.1161/CIRCULATIONAHA.105.594929. [DOI] [PubMed] [Google Scholar]
- 16.Obuchowski NA, McClish DK. Sample size determination for diagnostic accuracy studies involving binormal ROC curve indices. Stat Med. 1997;16:1529–42. doi: 10.1002/(sici)1097-0258(19970715)16:13<1529::aid-sim565>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 17.Thunnissen FB. Sputum examination for early detection of lung cancer. J Clin Pathol. 2003;11:805–10. doi: 10.1136/jcp.56.11.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flehinger BJ, Melamed MR, Zaman MB, Heelan RT, Perchick WB, Martini N. Early lung cancer detection: results of the initial (prevalence) radiologic and cytologic screening in the Memorial Sloan-Kettering study. Am Rev Respir Dis. 1984;4:555–60. doi: 10.1164/arrd.1984.130.4.555. [DOI] [PubMed] [Google Scholar]
- 19.Yakubovskaya MS, Spiegelman V, Luo FC, Malaev S, Salnev A, Zborovskaya I, Gasparyan A, Polotsky B, Machaladze Z, Trachtenberg AC. High frequency of K-ras mutations in normal appearing lung tissues and sputum of patients with lung cancer. Int J Cancer. 1995;63:810–4. doi: 10.1002/ijc.2910630611. [DOI] [PubMed] [Google Scholar]
- 20.Mao L, Hruban RH, Boyle JO, Tockman M, Sidransky D. Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res. 1994;1:1634–7. [PubMed] [Google Scholar]
- 21.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, Franklin WA, Herman JG, Baylin SB, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 22.Jiang F, Todd NW, Qiu Q, Liu Z, Katz RL, Stass SA. Combined genetic analysis of sputum and computed tomography for noninvasive diagnosis of non-small-cell lung cancer. Lung Cancer. 2009;66:58–63. doi: 10.1016/j.lungcan.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 3007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 24.Pezzolesi MG, Platzer P, Waite KA, Eng C. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet. 2008;82:1141–9. doi: 10.1016/j.ajhg.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 3007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 26.Allgayer H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol. 2010;73:185–91. doi: 10.1016/j.critrevonc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Jiang F, Yin Z, Caraway NP, Li R, Katz RL. Genomic profiles in stage I primary non small cell lung cancer using comparative genomic hybridization analysis of cDNA microarrays. Neoplasia. 2004;6:623–35. doi: 10.1593/neo.04142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, Wang H, Bekele BN, Yin Z, Caraway NP, Katz RL, Stass SA, Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2006;18:2628–35. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- 29.Jeon JP, Shim SM, Nam HY, Baik SY, Kim JW, Han BG. Copy number increase of 1p36.33 and mitochondrial genome amplification in Epstein-Barr virus-transformed lymphoblastoid cell lines. Cancer Genet Cytogenet. 2007;2:122–30. doi: 10.1016/j.cancergencyto.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Scaruffi P, Parodi S, Mazzocco K, Defferrari R, Fontana V, Bonassi S, Tonini GP. Detection of MYCN amplification and chromosome 1p36 loss in neuroblastoma by cDNA microarray comparative genomic hybridization. Mol Diagn. 2004;8:93–100. doi: 10.1007/BF03260051. [DOI] [PubMed] [Google Scholar]
- 31.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 32.Avissar M, McClean MD, Kelsey KT, Marsit CJ. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. 2009;12:2059–63. doi: 10.1093/carcin/bgp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathé EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, Altorki NK, Casson AG, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, Nonaka D, Li J, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–7. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 35.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;25:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;6:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;28:23204–16. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM, Anderson ML, Matzuk MM. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 2010;22:447–63. doi: 10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;1:367–77. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coutlee F, Rubalcaba EA, Viscidi RP, Gern JE, Murphy PA, Lederman HM. Quantitative detection of messenger RNA by solution hybridization and enzyme immunoassay. J Biol Chem. 1990;265:11601–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


