Abstract
Diet influences host metabolism and intestinal microbiota; however, detailed understanding of this tripartite interaction is limited. To determine whether the nonfermentable fiber hydroxypropyl methylcellulose (HPMC) could alter the intestinal microbiota and whether such changes correlated with metabolic improvements, C57B/L6 mice were normalized to a high-fat diet (HFD), then either maintained on HFD (control), or switched to HFD supplemented with 10% HPMC, or a low-fat diet (LFD). Compared to control treatment, both LFD and HPMC reduced weight gain (11.8 and 5.7 g, respectively), plasma cholesterol (23.1 and 19.6%), and liver triglycerides (73.1 and 44.6%), and, as revealed by 454-pyrosequencing of the microbial 16S rRNA gene, decreased microbial α-diversity and differentially altered intestinal microbiota. Both LFD and HPMC increased intestinal Erysipelotrichaceae (7.3- and 12.4-fold) and decreased Lachnospiraceae (2.0- and 2.7-fold), while only HPMC increased Peptostreptococcaceae (3.4-fold) and decreased Ruminococcaceae (2.7-fold). Specific microorganisms were directly linked with weight change and metabolic parameters in HPMC and HFD mice, but not in LFD mice, indicating that the intestinal microbiota may play differing roles during the two dietary modulations. This work indicates that HPMC is a potential prebiotic fiber that influences intestinal microbiota and improves host metabolism.—Cox, L. M., Cho, I., Young, S. A., Kerr Anderson, W. H., Waters, B. J., Hung, S.-C., Gao, Z., Mahana, D., Bihan, M., Alekseyenko, A. V., Methé, B. A., Blaser, M. J. The nonfermentable dietary fiber hydroxypropyl methylcellulose modulates intestinal microbiota.
Keywords: prebiotics, microbial ecology, obesity, cholesterol, liver adiposity
The gastrointestinal microbiome is composed of trillions of organisms (1, 2) in any mammalian host, and is largely affected by a combination of host genetic factors and diet (3, 4). The intestinal microbiota is involved in many physiological processes (5), highlighting the need for methods to modulate the composition of the organisms present. Strategies to shape the microbiome include antibiotics to reduce specific populations, transfer of live organisms in either foods containing probiotics or via fecal transplant, or by delivering prebiotic compounds that stimulate the growth of specific microorganisms (6). Prebiotics have the advantage of being easy to transport and administer, and have few safety concerns. However, to date; the focus has been on compounds that are actively metabolized by members of the microbiota.
Obesity and the metabolic syndrome are increasing problems in the United States and worldwide (7, 8). The roots of these disorders often are multifactorial (9), and many potential treatments have emerged, including dietary and lifestyle modifications. Reducing dietary calorie and fat intake leads to weight loss and improved metabolic parameters by directly altering energy balance (10). Adding indigestible fiber to diets has long been associated with health benefits in both humans and animals, including effects on weight control (11, 12), metabolic syndrome (13, 14), blood cholesterol (15), and glucose tolerance (16–19); however, the mechanisms of action are not well understood.
Recently, much attention has focused on the role of intestinal microbiota in the regulation of host metabolism as a significant contributing factor to obesity (20–27). The microbiota perform a diverse range of metabolic functions, including short-chain fatty acid production from carbohydrate fermentation (28, 29), regulation of energy extraction from food (30, 31), and bile salt metabolism (32). Microbial interactions with dietary fiber, and consequently the mechanisms by which fiber-induced microbiome changes affect host metabolism, partially depends on the extent that the fiber can be fermented. However, fibers ranging from completely fermentable to nonfermentable (33), have been reported to improve host metabolism (34). Health improvements are usually attributed to an increase in short-chain fatty acid production for fermentable fibers (35, 36), but a similar conclusion cannot be drawn for nonfermentable fibers.
Hydroxypropyl methylcellulose (HPMC) is a nonfermentable dietary fiber with many beneficial health effects, making it an ideal substrate to probe the interaction between fiber, metabolism, and microbiota in a system in which the treatment was previously believed to be inert with respect to the microbiota (37). It is a semisynthetic cellulose derivative with hydroxypropyl and methyl side chains, used in the manufacturing of many foods (38). HPMC has a long safety record and is classified as having “generally recognized as safe (GRAS)” status, up to 20 g/d in the United States (39). HPMC supplementation lowers cholesterol and postprandial insulin levels, in both rodents (15, 40, 41) and humans (16, 42, 43), and decreases weight gain (44) and fat mass (45) in rodents fed a high-fat diet (HFD). HPMC is not absorbed by the host (46); however, it can modulate the intestinal nutrient environment by selectively increasing the excretion of fecal bile acids and fats (44) and increasing fecal water content (47), and thus have downstream effects on the intestinal microbiota.
In this study, we used high-throughput 16S rRNA taxonomic profiling to examine the effects of HPMC on the murine microbiome. We sought to determine whether HPMC and a low-fat diet (LFD), which yielded parallel metabolic effects, would impose similar changes on the intestinal microbiome. Our goal was to characterize overall changes in microbial community structure in response to dietary perturbation, and to identify specific taxa associated with particular diets or metabolic phenotypes.
MATERIALS AND METHODS
Animals and diets
Eighteen-week-old adult C57B6/L6J mice from Jackson Laboratories (Bar Harbor, ME, USA) were acclimated to an HFD (5.24 kcal/g, 60% kcal from fat; D12492; Research Diets, New Brunswick, NJ, USA) for 2 mo, then weighed and randomized into 3 groups of 10 mice: LFD (3.85 kcal/g, 10% kcal from fat; D12450B; Research Diets), continuing HFD, and HFD (60% kcal from fat) supplemented with 10% (weight percentage of the diet) of HPMC (4.71 kcal/g, K250M lot no. VJ241907R1; Dow Chemical, Midland, MI, USA). The mice were housed individually in polycarbonate cages maintained with a 12-h alternating light-dark cycle at normal temperature (22±4°C) with relative humidity 50 ± 15%. Total food intake (g) was measured for the duration of the 4-wk experiment, and total energy intake (kcal) was calculated by multiplying total grams of food consumed by the energy (kcal/g) in each diet. This study was performed according to an Institutional Animal Care and Use Committee-approved protocol (48) and conducted by In Vivo Services at the Jackson Laboratory West (Sacramento, CA, USA), an Office of Laboratory Animal Welfare–assured and Association for Assessment and Accreditation of Laboratory Animal Care–accredited organization.
Hepatic lipid analysis
Lyophilized liver samples were extracted using an accelerated solvent extractor (Dionex ASE; Dionex, Sunnyvale, CA, USA) at 100°C, ∼13.8 MPa with 75/25 hexane/2-propanol, dried, and weighed to determine the percentage of total hepatic lipids, and hepatic total cholesterol, free cholesterol, and triglyceride levels using colorimetric assays and a clinical analyzer (Roche Diagnostics/Hitachi 914; Roche Diagnostics, Indianapolis, IN, USA).
Fecal lipid analysis
Fecal lipids were extracted on a Dionex ASE system using a mixture of hexane and 2-propanol (3:2, v/v, 2% acetic acid) at 15 MPa and 60°C for 30 min. One aliquot was analyzed for saturated and unsaturated fatty acid composition by gas chromatography as described previously (44). The second aliquot was analyzed for total bile acids and sterols using a modified chromatographic method (49).
Bomb calorimetry
C57BL/6J mice were normalized to an HFD (60% kcal from fat) for 2 mo, then either maintained on an HFD or changed to an HFD supplemented with 8% HPMC. Fecal pellets were collected at baseline, prior to the change in diets, and 25 d after the dietary intervention. Samples were oven-dried at 55°C for 24 h. Total energy in the fecal pellets per gram was measured by bomb calorimetry using a Parr 6725 Semimicro calorimeter, 1109A semimicro oxygen bomb, and 6772 thermometer (Parr Instrument Co., Moline, IL, USA). The calorimetry energy equivalent factor was determined using benzoic acid standards, which showed 99.36–99.92% reproducibility.
Plasma biomarker analysis
Total cholesterol, free cholesterol, and triglycerides in plasma were determined by enzymatic colorimetric assays using a Roche Diagnostics/Hitachi 914 clinical analyzer as described previously (44). The VLDL-cholesterol levels were calculated by subtracting HDL-cholesterol and LDL-cholesterol from total cholesterol levels. Fasting plasma concentrations of adiponectin, leptin, and insulin of mice were determined after 12 h of food withdrawal using mouse adiponectin (B-Bridge International, Sunnyvale, CA, USA), leptin (Assay Designs, Ann Arbor, MI, USA), and insulin (Mercodia, Winston Salem, NC, USA) immunoassay kits, as described previously (41). Fasting glucose levels were measured from a drop of blood collected by tail vein puncture and analyzed using a OneTouch Ultra meter with FastDraw test strips (Johnson & Johnson, Milpitas, CA, USA).
Microbiome sample collection and processing
Fecal pellets were collected at baseline, 2 wk, and 4 wk, and cecal and ileal samples at euthanasia were frozen at −80°C until DNA extraction. DNA extraction was performed using the MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA).
Quantitative PCR
Quantitative PCR assays were performed on a Rotor Gene 3000 quantitative PCR cycler using the Power SYBR Green kit (Applied Biosystems, Carlsbad, CA, USA), according to the manufacturer's instructions, with the addition of 1 ng/μl of BSA. Total bacterial levels were determined by standardized quantitative PCR (qPCR) targeting universal regions of the 16S rRNA gene using primers Eub519F and Eub785R (50). Firmicutes and Bacteroidetes levels were determined using the primer pairs Firm934F/Firm1060R and Bact934F/Bact1060R, respectively (51), and were normalized on the basis of total bacteria 16S copies.
PCR amplification for 454-pyrosequencing
Samples were prepared for amplification and sequencing at the J. Craig Venter Institute (JCVI) Joint Technology Center (JTC). Genomic DNA sample concentrations were normalized to ∼2-6 ng/μl. The V3–V5 region of the 16S rRNA gene was amplified using forward primer 341F (5′-CCTACGGGAGGCAGCAG-3′) attached to the Roche B adapter for 454-library construction and reverse primer 926R (5′-CCGTCAATTCMTTTRAGT-3′) attached to the Roche A adapter and a 10-nt barcode [5′-A-adapter-N (10)+16S primer-3′]. A barcoded primer design was completed using a set of algorithms developed at the JCVI. Every effort was made to prevent contamination of PCR reactions with exogenous DNA, including a set of reactions in a laminar flow hood. PCR reactions were completed as follows (per reaction): 2 μl of gDNA, 1× final concentration of Accuprime PCR Buffer II (Invitrogen, Carlsbad, CA, USA), 200 nM forward and reverse primers, 0.75 U of Accuprime TaqDNA polymerase high fidelity (Invitrogen), and nuclease-free water to bring the final volume to 20 μl. PCR cycling conditions consisted of an initial denaturation of 2 min at 95°C, 30 cycles of 20 s at 95°C, 30 s at 50°C, and 5 min at 72°C. A negative control (water blank) reaction was examined after 35 cycles. Samples were then quantified, cleaned, and sequenced on the Roche 454-FLX (454 Life Sciences, Branford, CT, USA) as described previously (27), and a read processing pipeline consisting of a set of modular scripts designed at the JCVI were employed for deconvolution, trimming, and quality filtering, as described previously (27). The number of reads that passed the quality filter totaled 676,440 sequences (mean 4602±1625 reads/sample). The sequencing data were denoised with shhh.seqs, a denoising algorithm developed by Chris Quince (SeqNoise; http://dev.man-online.org/man1/SeqNoise/) as implemented in Mothur v 1.25.0 (52), with the following settings: minimum flow = 360, primer difference = 6, and barcode difference = 1.
Statistical and bioinformatics analysis
Quality-filtered sequences were further preprocessed through the Qiime pipeline (53) as described previously (27). The rarefactions for richness and Shannon diversity indices were calculated in the R statistical programming environment (54, 55) using Community Ecology Package vegan, on the denoised reads. Principal coordinates analysis (PCoA) plots from Qiime output were produced based on unweighted UniFrac (56) distances by Kinemage, Next Generation (KiNG 2.16; http://kinemage.biochem.duke.edu/software/king.php). Comparison of unweighted and weighted UniFrac distances was performed using 2-sided t tests. The operational taxonomic unit (OTU) relative abundances were calculated by dividing the absolute abundances by total sequence count per sample analyzed. The resulting relative abundance matrix was used to produce heat maps for major (relative abundance≥1%) taxa. Hierarchical clustering of major taxa from the resulting dense abundance matrices was performed using euclidean distances. Association of major clusters and treatment group was measured using Fisher's exact test at 0.05 significance threshold. Differences between dietary groups were evaluated using the Mann-Whitney U test. When multiple testing was performed, the P values were adjusted for false discovery using the Benjamini-Hochberg procedure (57).
RESULTS
Dietary interventions improve metabolic phenotypes
All C57BL/6J mice were fed an HFD for 2 mo prior to the study and then randomized to either continue the HFD, to receive the HFD with 10% HPMC supplementation (HPMC diet), or to receive an LFD. Over the course of the 4-wk study, the mice fed the HFD continued to gain weight, while HPMC mice showed reduced weight gain (Fig. 1A), despite isocaloric intake of food (Fig. 1B). Mice receiving the LFD lost weight and had significantly lower energy intake. Weight change and energy intake were correlated in both groups of HFD and LFD mice (Pearson correlation coefficient, r=0.84 and 0.65, P=0.003 and 0.042, respectively); when combined, the correlation strengthened (Pearson correlation coefficient, r=0.98, P<0.0001; Fig. 1C). No relationship was detected between energy intake and weight change in HPMC mice (P=0.39, for a nonzero slope).
Figure 1.
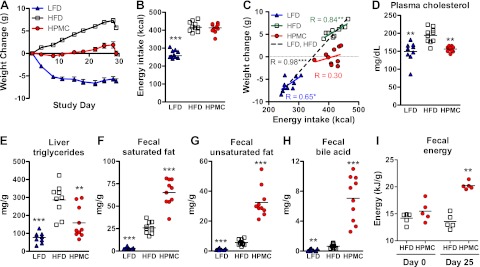
Effect of diet on host metabolism. Adult C57BL/6 mice were fed an HFD (60% kcal from fat) for 2 mo prior to the study, then continued on HFD, or switched to either an LFD (10% kcal from fat) or to HFD with 10% HPMC supplementation (HPMC group) (10 mice/group). A) Weight change over the 4-wk study. B) Total 4-wk energy intake (kcal; bar at median). C) Correlation by linear regression between 4-wk weight change and total energy intake. D–H) Biomarkers at 4 wk: fasting plasma cholesterol (D), liver triglycerides (E), fecal saturated fat (F), fecal unsaturated fat (G), fecal bile acids (H). I) Total energy content per gram in fecal pellets in C57BL/6J mice normalized to an HFD (60% kcal from fat) for 2 mo, then either maintained on HFD (n=5) or changed to HFD supplemented with 8% HPMC (n=5). Values are shown at baseline (d 0) before randomization into the two treatment groups and at 25 d after dietary intervention. *P < 0.05, **P < 0.01, ***P < 0.001; FDR-adjusted Mann-Whitney U (B, E–I); nonzero slope (C); Mann-Whitney U (D).
After 4 wk, mice fed the LFD and HPMC diets had significantly reduced total cholesterol, HDL, LDL, and VLDL cholesterol, leptin, liver triglycerides, and liver percentage adiposity, compared to the HFD mice (Fig. 1D, E and Supplemental Fig. S1A–C, H, J). The LFD mice had decreased fasting blood glucose, and free fatty acids (Supplemental Fig. S1D, F), while plasma insulin was decreased in the HPMC mice (Supplemental Fig. S1G). No changes were seen in serum triglycerides or adiponectin levels (Supplemental Fig. S1E, I). Compared to both the LFD and HFD, HPMC supplementation increased saturated, unsaturated, and trans-unsaturated fecal fat, as well as fecal monoacylglycerides/free fatty acids, and bile acids (Fig. 1F–H and Supplemental Fig. S1K, M). The levels of sterol excretion decreased in the HPMC group compared to the HFD group (Supplemental Fig. S1L), while diacylglycerides or triacylglycerides were unchanged (Supplemental Fig. S1M–O). HPMC-group mice had increased fecal energy, as measured by bomb calorimetry (Fig. 1I).
Microbial diversity and community structure
To assess changes in murine intestinal microbial community structure induced by the dietary changes, measures of richness and evenness were calculated for microbial 16S rRNA sequences from variable regions V3–V5 in fecal and cecal samples. At the OTU level, the LFD and HPMC mice had decreased fecal community richness compared to the HFD mice, and the HPMC mice had reduced cecal microbial richness (Fig. 2A). At the class level, richness in the fecal and cecal communities from the three groups was similarly maintained (Supplemental Fig. S2A). Community evenness, measured by the Shannon evenness metric, decreased at the OTU level in the fecal samples from LFD and HPMC mice, and similarly in cecal samples for the HPMC mice (Fig. 2B). However, Shannon evenness indices increased in the same groups at the class level (Supplemental Fig. S2B).
Figure 2.
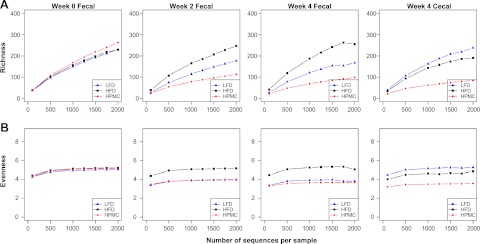
Assessment of microbial diversity in relation to treatments. Graphs depict α diversity at the OTU level, calculated on denoised sequences of C57B/L6J mouse fecal microbiota at wk 0, 2, and 4, and cecal microbiota at wk 4. Mice were normalized on HFD, then maintained on HFD, or switched to either LFD or HPMC; 10 mice/group. Rarefaction curves display the α diversity when subsampled at a lower depth. A) Population evenness calculated by the Shannon evenness metric. B) Taxonomic richness as measured by number of observed OTUs.
The phylogenetic differences within the intestinal microbial ecosystem between the three treatment groups also were assessed by PCoA of the unweighted UniFrac (58) distances, and 24.5% of the total variation was explained on the first 3 PCoA axes (Fig. 3). As expected, all samples cluster together at baseline (wk 0; Fig. 3A). At wk 2 (Fig. 3B) and wk 4 (Fig. 3C), the HFD samples were little different from baseline, whereas the LFD and HPMC communities had shifted in separate directions; HPMC samples formed a distinct cluster, while LFD samples were more variable. Pairwise unweighted UniFrac distances were calculated for all 10 mice within each dietary group, comparing baseline, wk 2, and wk 4 fecal microbiota to either baseline (Fig. 3D) or wk 2 (Fig. 3E) microbiota. There were no significant changes in the mean UniFrac distance over time for the HFD fecal microbiota, as might have been anticipated. However, for the LFD samples, pairwise distances significantly increased from wk 0 to 2, but not from wk 2 to 4. For the HPMC fecal microbiota, pairwise distances significantly increased from wk 0 to 2, and from wk 2 to 4. Analysis of the weighted UniFrac distances showed similar trends (Supplemental Fig. S2D, E). The community structure of the cecal samples was similar to the wk 4 fecal samples for HFD and HPMC samples; however, LFD cecal samples differed from the LFD fecal samples and were intermixed with HFD samples (Fig. 3F). The ileal specimens showed no distinctive clustering by treatment group (Fig. 3G).
Figure 3.
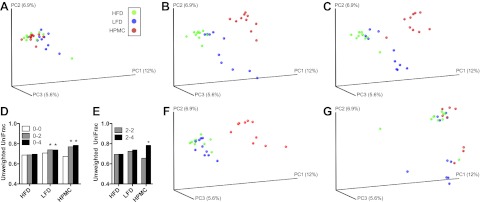
Effect of diet and fiber on microbial community structure. PCoA of the unweighted UniFrac distances of microbial 16S rRNA sequences from the V3–V5 region in fecal samples at wk 0 (baseline; A), wk 2 (B), and wk 4 (C), cecal samples at euthanasia (F), and ileal samples at euthanasia (G) in mice maintained on HFD, switched to LFD, or switched to HPMC, 10 mice/group. Panels D and E show change in β diversity over time. Average unweighted UniFrac distances from baseline microbiota (wk 0) to wk 0, 2, and 4 microbiota, within a dietary group (D) and distances from wk 2 microbiota to 2- or 4 wk microbiota (E). Each sample is represented as a colored circle. *P < 0.001.
Hierarchical clustering of the most abundant (≥1%) families in fecal specimens was visualized on heat map plots (Supplemental Fig. S2C). The samples did not cluster according to treatment at baseline, as expected. As the study progressed, the fecal samples gradually shifted toward 3 clusters based on diet. The cecal specimens showed a clear separation (P<0.001, Fisher's exact test; Supplemental Fig. S2) of the HPMC mice from the HFD and LFD groups, which were not distinguishable, consistent with the PCoA results (Fig. 3). In the ileal specimens, the LFD group clustered distinctly from the HFD and HPMC groups (P<0.05, Fisher's exact test; Supplemental Fig. S2).
Microbial population changes induced by dietary intervention
Quantitative PCR of the 16S rRNA gene showed that prior to randomization to the three treatment groups, there were no significant differences in total fecal bacteria or Bacteroidetes/Firmicutes (B/F) ratio between the mice (Fig. 4A). Over time, total bacteria levels were significantly decreased in fecal, cecal, and ileal samples from HPMC mice compared to the LFD and HFD groups. In addition, the LFD mice showed lower ileal bacteria compared to the HFD mice. As expected, the B/F ratio in fecal samples was unchanged in the HFD mice but increased in both the LFD and HPMC mice and in cecal samples from HPMC mice. In contrast, in ileal samples, the B/F ratios were decreased in LFD and HPMC mice. These data indicate that the dietary changes, especially HPMC supplementation, affected the overall size and composition of the intestinal microbiota.
Figure 4.
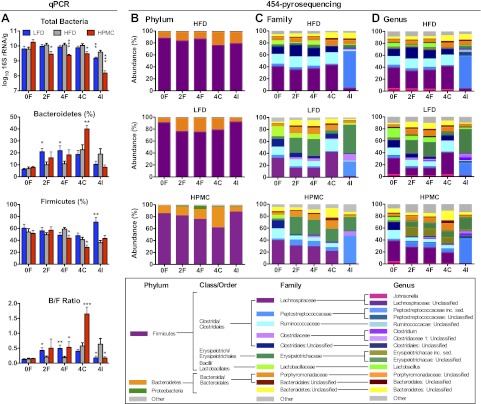
Changes in intestinal microbiota composition with dietary treatments. A) Total bacteria per gram of sample, and Bacteroidetes and Firmicutes, normalized by total bacteria, as determined by quantitative PCR using primers that target 16S rRNA (see Materials and Methods), and calculated B/F ratio for the following samples: wk 0 fecal (0F), wk 2 fecal (2F), wk 4 fecal (4F), wk 4 cecal (4C), wk 4 ileal (4I). *P < 0.05, **P < 0.01, ***P < 0.001; Mann-Whitney U test, 10 mice/group. B, C, D) Relative abundance of the predominant intestinal microbiota (≥1%) at the phylum (B), family (C), and the genus (D) level, determined by 454-pyrosequencing of the V3–V5 region of the 16S rRNA gene.
To assess specific changes in intestinal microbiota, we compared the relative abundance of predominant (≥1%) taxa identified from 454-pyrosequencing in HFD vs. LFD mice (to assess for the effects of dietary fat) and HFD vs. HPMC mice (to assess for the effects of fiber supplementation) (Supplemental Table S1 and Fig. 4B–D). No significant differences were seen in wk 0 fecal samples, as expected. Over the course of the experiment, changes at the phylum level were not significant after false discovery rate (FDR) adjustment; however, the trends observed from the pyrosequencing results were similar to those detected by qPCR (Fig. 4A). All significant differences at lower taxonomic levels were within the phylum Firmicutes, including members of class Bacilli, Clostridia, and Erysipelotrichi, and of order Lactobacillales, Clostridiales, and Erysipelotrichales.
At the family level, populations of the predominant microbiota in fecal samples from mice fed HFD were consistent over the course of the 4-wk study (Fig. 4C), with major stable populations of Lachnospiraceae, Peptostreptococcaceae, Ruminococcaceae, Erysipelotrichaceae, and Porphyromonadaceae, while a major Lactobacillaceae population is transiently present only in baseline and 4-wk fecal samples. The cecal microbiota of HFD mice is similar to fecal samples, but with near absence of Erysipelotrichaceae and Lactobacillaceae. The ileal microbiota of HFD mice is dominated by Peptostreptococcaceae and contains minor populations of families seen in the fecal specimens. In both treatment groups, changes in family level microbiota was observed in the 2-wk fecal sample and maintained in the 4-wk fecal sample, including a significant decrease in Lachnospiraceae and an increase in Erysipelotrichaceae. HPMC mice also had significantly increased Peptostreptococcaceae and decreased Ruminococcaceae in fecal and cecal samples, while LFD mice had reduced ileal Peptostreptococcaceae. While both dietary treatments altered fecal microbiota, differences in cecal and ileal populations were limited to HPMC and LFD mice, respectively. At the genus level, there were significant changes in the community representation (Fig. 4D and Supplemental Table S1); HPMC-supplementation led to significant decreases in Johnsonella and Lactobacillus, and significant increases in Erysipelotrichaceae incertae sedis (inc. sed.) and Peptostreptococcus inc. sed. LFD led to decreases in Johnsonella and Erysipelotrichaceae inc. sed., and increases in Clostridium. Thus, the dietary changes from HFD or to LFD and HPMC induced significantly different compositions within Firmicutes, with reproducible compositional effects at the family and genus levels.
Specific associations of weight change and metabolic variables via partial correlation analysis
Since dietary intervention affects both host metabolism and intestinal microbiome, we attempted to factor out treatment-mediated changes and directly capture interactions of the host and microbiome. In standard correlation analysis, significant correlations can result from large treatment-mediated changes, rather than a direct interaction of the two variables. We performed partial correlation analysis (59–61) to remove the effect of dietary variables by conditioning on either presence of fiber or percentage of fat in the diet. Significant FDR-adjusted P values for each pairwise comparison of taxa, in particular, samples with each metabolic variable are indicated (Supplemental Table S2); regression analysis was used to avoid reporting correlations influenced by outlier samples (Supplemental Fig. S3). Weight changes in HFD mice were positively correlated with cecal Firmicutes and cecal Erysipelotrichaceae inc. sed. and negatively associated with cecal Bacteroidetes (Supplemental Fig. S3A–C). For the HPMC mice, weight changes and fecal saturated fat were positively associated with cecal Erysipelotrichaceae inc. sed. and cecal Erysipelotrichales, respectively (Supplemental Fig. S3C, SF). The 4-wk fecal abundances of Lachnospiraceae were negatively correlated with energy intake in the HFD and HPMC mice (Supplemental Fig. S3D), and cecal Porphyromonadaceae correlated positively with liver free cholesterol in HFD and HPMC mice (Supplemental Fig. S3E). However, the only significant correlation verified by linear regression analysis in the LFD group was between baseline fecal Clostridiales and insulin (Supplemental Fig. S3O) and was not observed in the other diet groups.
DISCUSSION
The current study confirms and extends prior work, indicating that reducing dietary fat or adding dietary fiber improves markers of metabolic health in mice receiving a high-calorie HFD (12, 62–65). In particular, the addition of HPMC disrupts the relationship between energy intake and weight change (66), consistent with a prior study (44), and is accompanied with increased excretion of bile salts and fats in the feces, resulting in an increased loss in calories. LFD mice consumed 36.6% fewer calories and weighed 29.2% less than the HFD mice, whereas HPMC mice did not differ in caloric intake, but excreted 49.1% more energy and weighed 12.6% less than HFD mice. Thus, in mice with diet-induced obesity, HPMC supplementation improved metabolic biomarkers to an extent comparable to that of caloric reduction, which was associated with increased fecal loss of specific metabolites and energy content.
We have shown that changing from HFD to LFD or 10% HPMC supplementation resulted in marked shifts in the intestinal microbiota over a 4-wk period by PCoA representations of the UniFrac analyses (67) and by heat map analysis. The induction of intestinal microbial community shifts by dietary changes is no longer surprising (3, 68–70); however, we provide the first evidence that HPMC alters the intestinal microbiota, challenging the previously held notion that HPMC is inert with respect to the microbiota. The results are highly consistent within the experimental groups and show progressive changes in the microbiota sampled in the feces, with similar shifts in the cecal and ileal samples for HPMC and LFD, respectively. Although ecological richness showed no changes at higher taxonomic orders, richness of the LFD- or HPMC-affected intestinal community and evenness at the OTU (species) level declined. Such dynamics are consistent with HPMC selection for a small group of specialist organisms that become over-represented at the expense of the preexisting community members.
Because all of the major HPMC-induced shifts involve families within the phylum Firmicutes, differences in their functional or anatomic niches merit further exploration. Members of the microbial communities interact cooperatively by mutualistic cross-feeding (71), or compete for nutrients and physical space, or by secreting antimicrobial peptides (72). The populations of Lachnospiraceae, Ruminococcaceae (both members of the order Clostridiales), and unclassified Clostridiales diminished after both dietary changes, suggesting that the HFD diet specifically provides a competitive advantage to Clostridiales. This observation provides a confirmation and further characterization of the altered Bacteroidetes/Firmicutes ratio described by Ley et al. (20).
HPMC and LFD both improve metabolic health and shape the microbiome; however, correlations between specific microbial taxa and host metabolic phenotypes using partial correlation analysis, which minimizes the effect of the diet, could only be identified in HPMC mice and not in LFD mice. This dichotomy suggests that specific microbes may play a lesser role in the beneficial effects of LFD, than in the health improvements induced by HPMC. We now identify specific taxa that are associated with weight gain (e.g., Erysipelotrichaceae inc. sed.), and with other metabolic phenotypes. Although these associations do not indicate the direction of causality between microbiome and host metabolic change, they provide candidate taxa to be explored in future studies involving dietary manipulations. Therefore, HPMC supplementation may be used as a probe for better understanding of intestinal microbiome population structure due to dietary fiber intervention.
Although both LFD and HPMC increase levels of Erysipelotrichaceae, HPMC increases organisms within Erysipelotrichaceae inc. sed., while LFD increases members of the Erysipelotrichaceae that cannot be classified at the genus level. The taxonomy of bacteria is constantly evolving as our knowledge expands (73). Incertae sedis means “uncertain seat” in Latin, and is typically used when a new genus is justified, but a name has not yet been assigned. The genus Erysipelotrichaceae inc. sed. contains species previously classified in Clostridiaceae, Eubacteriaceae, and Lactobacillaceae (Table 1), and several members, including Eubacterium bioforme, E. cylindroides, E. dolichum, and Clostridium spiroforme increase in response to an HFD (68). The high levels of fecal fat in HPMC mice could be driving this specific increase. In contrast, LFD is likely increasing other genera within Erysipelotrichaceae, potentially Allobaculum, which has been reported to be higher in animals fed LFD compared to HFD and increases in response to weight loss (74). LFD also shows the greatest decrease in Lachnospiraceae, which have been observed to be lower in animals on LFD compared to HFD, and lowest in animals on LFD after undergoing weight loss (74).
Table 1.
Classification within inc. sed. genera and related families in RDP
| Erysipelotrichaceae |
Peptostreptococcaceae |
||
|---|---|---|---|
| Genera | Inc. sed. | Genera | Inc. sed. |
| Erysipelothrix | Clostridium catenaformis | Peptostreptococcus | Eubacterium yurii |
| Allobaculum | Clostridium cocleatum | Filifactor | |
| Bulleidia | Clostridium innocuum | Sporacetigenium | |
| Catenibacterium | Clostridium ramosum | Tepidobacter | |
| Coprobacillus | Clostridium spiroforme | Anaerobium | |
| Holdemania | Eubacterium bioforme | Proteocatella | |
| Solobacterium Erysipelotrichaceae inc. sed. Turicibacter | Eubacterium cylindroides Eubacterium dolichum Eubacterium tortuosum | Peptostreptococcaceae inc. sed. Clostridium XI | |
| Lactobacillus catenaformis | |||
| Lactobacillus vitulinus | |||
| Streptococcus pleomorphus | |||
RDP, Ribosomal Database Project (Michigan State University, Ann Arbor, MI, USA; http://rdp.cme.msu.edu/).
Fermentable dietary fibers, such as inulin (35), arabinoxylan (12, 64), and chitin-glucan (65), improve metabolic health and alter the microbiota, partially by increasing short-chain fatty acid production (33, 75). Similar to HPMC, arabinoxylan and chitin-glucan lowered total bacteria in the cecum. Arabinoxylan increased Bacteroides/Prevotella, which is consistent with increased Bacteroidetes in HPMC-fed mice. However, unlike HPMC, members of the family Lachnospiraceae as classified by the Ribosomal Database Project (RDP; Michigan State University, Ann Arbor, MI, USA; Roseburia and Eubacterium rectale/Clostridium coccoides group) were increased by arabinoxylan and chitin-glucan supplementation, whereas Lachnospiraceae was decreased by HPMC. In a study of adult men, polydextrose supplementation decreased populations of Lachnospiraceae, similarly to HPMC, and specifically decreased Roseburia (76). Bifidobacterium was increased by arabinoxylan as measured by qPCR, but not by chitin-glucan, and not detected as a predominant phylotype (>1%) in the HPMC-fed mice. These fermentable dietary fibers and nonfermentable HPMC similarly improve metabolism; however, the subsequent shifts in microbiota differ by type of fiber, highlighting the need for further characterization of the effects of specific fibers on the microbiota and host metabolism. The ability of HPMC to modulate the intestinal microbiota and maintain stable changes, whether due to a primary effect or to a secondary increase in fecal fats, indicates that it may be used as a prebiotic agent, able to reduce populations of specific microbes, including members of Lachnospiraceae (which contains common intestinal genera Blautia, Butyrivibrio, Coprococcus, Dorea, Johnsonella and Roseburia) and Ruminococcaceae (containing Faecalibacterium, Oscillospira, Ruminococcus, and Subdoligranulum), and induce the growth of specific microbes, such as members of Erysipelotrichaceae (containing Allobaculum, Coprobacillus, Holdemania, and Turicibacter) and Peptostreptococcaceae (containing Peptostreptococcus, Sporacetigenum, and clostridial cluster XI; ref. 77).
HPMC is considered to be not fermentable by gut microbiota in vitro (78) and consequently, is not absorbed. In a study administering 14C-radiolabeled HPMC to rats, 99% of the radioactivity was detected in the feces, 1% detected in the urine, and none detected in the tissues of the animal, demonstrating that HPMC does not undergo substantial degradation in the intestinal tract (46); thus, the way in which it alters the intestinal microbiota and improves health must differ from other commonly used prebiotic dietary fibers. The increases in fecal fats may be a primary source of microbiome perturbation; examination of similarly structured nonfermentable fibers may provide further insights. The similar fiber methylcellulose (MC) is not absorbed on passage through the rat digestive tract (79) or in humans (80) but is subject to partial glycosidic hydrolysis (81); in vitro, MC is subject to weak fermentation by fecal bacteria (78, 80, 82). Fermentation of hydroxyethylcellulose by human fecal microbiota in vitro yields 10-fold higher short-chain fatty acid concentrations than MC (82), providing evidence that substitution of modifying groups on cellulose affects fermentative potential, and raising the question whether side groups of HPMC could be acted on by the intestinal microbiota.
Although the results obtained in the current study do not provide evidence for direct interaction of HPMC with fecal microbiota, specific experiments for delineation of the mechanisms by which HPMC mediates changes in the microbial ecosystem can be designed in future investigations. Nevertheless, including baseline samples has allowed the assessment of early microbiologic changes and the identification of taxa that respond to HPMC treatment and that may interact with host metabolism. In addition, individual animals differed in phenotypic responses to dietary constituents; thus, such variation was used to identify candidate taxa associated with particular stressors. The current study provides the first observations of variation in genus-level taxa, in broad analyses without a priori candidates.
Supplementary Material
Acknowledgments
This study was supported by the U.S. National Institutes of Health (1UL1RR029893 and R01DK098989), the Dow Chemical Company, and the Diane Belfer Program for Human Microbial Ecology.
The authors declare no conflicts of interest.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- B/F
- Bacteroidetes/Firmicutes
- HFD
- high-fat diet
- HPMC
- hydroxypropyl methylcellulose
- inc. sed.
- incertae sedis
- LFD
- low-fat diet
- MC
- methyl cellulose
- OTU
- operational taxonomic unit
- PCoA
- principal coordinates analysis
- qPCR
- quantitative polymerase chain reaction
REFERENCES
- 1. Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.-M., Hansen T., Le Paslier D., Linneberg A., Nielsen H. B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Antolin M., Artiguenave F., Blottiere H., Borruel N., Bruls T., Casellas F., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Forte M., Friss C., Van De Guchte M., Guedon E., Haimet F., Jamet A., Juste C., Kaci G., Kleerebezem M., Knol J., Kristensen M., Layec S., Le Roux K., Leclerc M., Maguin E., Melo Minardi R., Oozeer R., Rescigno M., Sanchez N., Tims S., Torrejon T., Varela E., De Vos W., Winogradsky Y., Zoetendal E., Bork P., Ehrlich S. D., Wang J. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ley R. E., Peterson D. A., Gordon J. I. (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 [DOI] [PubMed] [Google Scholar]
- 3. Walker A. W., Ince J., Duncan S. H., Webster L. M., Holtrop G., Ze X., Brown D., Stares M. D., Scott P., Bergerat A., Louis P., McIntosh F., Johnstone A. M., Lobley G. E., Parkhill J., Flint H. J. (2010) Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muegge B. D., Kuczynski J., Knights D., Clemente J. C., Gonz·lez A., Fontana L., Henrissat B., Knight R., Gordon J. I. (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Hara A., Shanahan F. (2006) The gut flora as a forgotten organ. EMBO Rep. 7, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kootte R. S., Vrieze A., Holleman F., Dallinga-Thie G. M., Zoetendal E. G., de Vos W. M., Groen A. K., Hoekstra J. B. L., Stroes E. S., Nieuwdorp M. (2012) The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes. Metab. 14, 112–120 [DOI] [PubMed] [Google Scholar]
- 7. Flegal K. M., Carroll M. D., Ogden C. L., Curtin L. R. (2010) Prevalence and tends in obesity among US adults, 1999-2008. JAMA 303, 235–241 [DOI] [PubMed] [Google Scholar]
- 8. Gortmaker S., Swinburn B., Levy D., Carter R., Mabry P., Finegood D., Huang T., Marsh T., Moodie M. (2011) Changing the future of obesity: science, policy, and action. Lancet 378, 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pataky Z., Bobbioni-Harsch E., Golay A. (2010) Obesity: a complex growing challenge. Exp. Clin. Endocrinol. Diabetes 118, 427–433 [DOI] [PubMed] [Google Scholar]
- 10. Hill J. O. (1998) Environmental contributions to the obesity epidemic. Science 280, 1371–1374 [DOI] [PubMed] [Google Scholar]
- 11. Slavin J. L. (2005) Dietary fiber and body weight. Nutrition 21, 411–418 [DOI] [PubMed] [Google Scholar]
- 12. Neyrinck A., Van Hee V., Piront N., De Backer F., Toussaint O., Cani P., Delzenne N. (2012) Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutr. Diabetes 2, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weickert M., Roden M., Isken F., Hoffmann D., Nowotny P., Osterhoff M., Blaut M., Alpert C., Ggebakan O., Bumke Vogt C., Mueller F., Machann J., Barber T., Petzke K., Hierholzer J., Hornemann S., Kruse M., Illner A.-K., Kohl A., Loeffelholz C., Arafat A., Mhlig M., Pfeiffer A. (2011) Effects of supplemented isoenergetic diets differing in cereal fiber and protein content on insulin sensitivity in overweight humans. Am. J. Clin. Nutr. 94, 459–471 [DOI] [PubMed] [Google Scholar]
- 14. Psaltopoulou T., Ilias I., Alevizaki M. (2010) The role of diet and lifestyle in primary, secondary, and tertiary diabetes prevention: a review of meta-analyses. Rev. Diabet. Stud. 7, 26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carr T., Gallaher D., Yang C., Hassel C. (1996) Increased intestinal contents viscosity reduces cholesterol absorption efficiency in hamsters fed hydroxypropyl methylcellulose. J. Nutr. 126, 1463–1469 [DOI] [PubMed] [Google Scholar]
- 16. Maki K. C., Carson M. L., Miller M. P., Turowski M., Bell M., Wilder D. M., Reeves M. S. (2007) High-viscosity hydroxypropylmethylcellulose blunts postprandial glucose and insulin responses. Diabetes Care 30, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 17. Topping D. L., Oakenfull D., Trimble R. P., Illman R. J. (1988) A viscous fibre (methylcellulose) lowers blood glucose and plasma triacylglycerols and increases liver glycogen independently of volatile fatty acid production in the rat. Br. J. Nutr. 59, 21–30 [DOI] [PubMed] [Google Scholar]
- 18. Ban S., Rico C., Um I., Kang M. (2012) Antihyperglycemic and antioxidative effects of hydroxyethyl methylcellulose (HEMC) and hydroxypropyl methylcellulose (HPMC) in mice fed with a high-fat diet. Int. J. Mol. Sci. 13, 3738–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maki K. C., Reeves M. S., Carson M. L., Miller M. P., Turowski M., Rains T. M., Anderson K., Papanikolaou Y., Wilder D. M. (2009) Dose-response characteristics of high-viscosity hydroxypropylmethylcellulose in subjects at risk for the development of type 2 diabetes mellitus. Diabetes Technol. Ther. 11, 119–125 [DOI] [PubMed] [Google Scholar]
- 20. Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005) Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102, 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bäckhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., Semenkovich C. F., Gordon J. I. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 101, 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Turnbaugh P. J., Gordon J. I. (2009) The core gut microbiome, energy balance and obesity. J. Physiol. 587, 4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., Hu C., Wong F. S., Szot G. L., Bluestone J. A., Gordon J. I., Chervonsky A. V. (2008) Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cani P. D., Delzenne N. M. (2009) The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 15, 1546–1558 [DOI] [PubMed] [Google Scholar]
- 25. Flier J. S., Mekalanos J. J. (2009) Gut check: testing a role for the intestinal microbiome in human obesity. Sci. Transl. Med. 1, 6ps7. [DOI] [PubMed] [Google Scholar]
- 26. Sonnenburg J. L., Fischbach M. A. (2011) Community health care: therapeutic opportunities in the human microbiome. Sci. Transl. Med. 3, 78ps12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cho I., Yamanishi S., Cox L., Methe B. A., Zavadil J., Li K., Gao Z., Mahana D., Raju K., Teitler I., Li H., Alekseyenko A. V., Blaser M. J. (2012) Antibiotics in early life alter murine colonic microbiome and adiposity. Nature 488, 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong J. M. W., de Souza R., Kendall C. W. C., Emam A., Jenkins D. J. A. (2006) Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40, 235–243 [DOI] [PubMed] [Google Scholar]
- 29. Louis P., Duncan S. H., McCrae S. I., Millar J., Jackson M. S., Flint H. J. (2004) Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186, 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Savage D. C. (1977) Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133 [DOI] [PubMed] [Google Scholar]
- 31. Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1131 [DOI] [PubMed] [Google Scholar]
- 32. Savage D. C. (1986) Gastrointestinal microflora in mammalian nutrition. Annu. Rev. Nutr. 6, 155–178 [DOI] [PubMed] [Google Scholar]
- 33. Cook S. I., Sellin J. H. (1998) Review article: short chain fatty acids in health and disease. Aliment. Pharmacol. Ther. 12, 499–507 [DOI] [PubMed] [Google Scholar]
- 34. Delzenne N. M., Neyrinck A. M., Backhed F., Cani P. D. (2011) Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 7, 639–646 [DOI] [PubMed] [Google Scholar]
- 35. Ramirez-Farias C., Slezak K., Fuller Z., Duncan A., Holtrop G., Louis P. (2009) Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 101, 541–550 [DOI] [PubMed] [Google Scholar]
- 36. Macfarlane G. T., Macfarlane S. (2011) Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 45, S120. [DOI] [PubMed] [Google Scholar]
- 37. Santos E. (1986) Expert panel. Final report on the safety assessment of hydroxymethylcellulose, hydroxypropylcellulose, methylcellulose, hydroxypropylmethylcellulose, and cellulose gum. J. Am. Coll. Toxicol. 5, 1–59 [Google Scholar]
- 38. BeMiller J. N., Whistler R. L. (1996) Food Chemistry, Marcel Dekker, New York, NY [Google Scholar]
- 39. Burdock G. A. (2007) Safety assessment of hydroxypropyl methylcellulose as a food ingredient. Food Chem. Toxicol. 45, 2341–2351 [DOI] [PubMed] [Google Scholar]
- 40. Gallaher D. D., Hassel C. A., Lee K. J., Gallaher C. M. (1993) Viscosity and fermentability as attributes of dietary fiber responsible for the hypocholesterolemic effect in hamsters. J. Nutr. 123, 244–252 [DOI] [PubMed] [Google Scholar]
- 41. Hung S.-C., Bartley G., Young S. A., Albers D. R., Dielman D. R., Anderson W. H. K., Yokoyama W. (2009) Dietary fiber improves lipid homeostasis and modulates adipocytokines in hamsters. J. Diabetes 1, 194–206 [DOI] [PubMed] [Google Scholar]
- 42. Reppas C., Swidan S. Z., Tobey S. W., Turowski M., Dressman J. B. (2009) Hydroxypropylmethylcellulose significantly lowers blood cholesterol in mildly hypercholesterolemic human subjects. Eur. J. Clin. Nutr. 63, 71–77 [DOI] [PubMed] [Google Scholar]
- 43. Maki K. C., Carson M. L., Miller M. P., Turowski M., Bell M., Wilder D. M., Rains T. M., Reeves M. S. (2008) Hydroxypropylmethylcellulose and methylcellulose consumption reduce postprandial insulinemia in overweight and obese men and women. J. Nutr. 138, 292–296 [DOI] [PubMed] [Google Scholar]
- 44. Hung S. C., Anderson W. H. K., Albers D. R., Langhorst M. L., Young S. A. (2011) Effect of hydroxypropyl methylcellulose on obesity and glucose metabolism in a diet induced obesity mouse model. J. Diabetes 3, 158–167 [DOI] [PubMed] [Google Scholar]
- 45. Islam A., Civitarese A. E., Hesslink R. L., Gallaher D. D. (2012) Viscous dietary fiber reduces adiposity and plasma leptin and increases muscle expression of fat oxidation genes in rats. Nature 20, 349–355 [DOI] [PubMed] [Google Scholar]
- 46. Gorzinski S. J., Takahashi I. T., Hurst G. H. (1986) The fate of ultra-low viscosity 14C-hydroxypropyl methylcellulose in rats following gavage administration. Drug Chem. Toxicol. 9, 83–100 [DOI] [PubMed] [Google Scholar]
- 47. Wyatt G. M., Horn N., Gee J. M., Johnson I. T. (1988) Intestinal microflora and gastrointestinal adaptation in the rat in response to non-digestible dietary polysaccharides. Br. J. Nutr. 60, 197–207 [DOI] [PubMed] [Google Scholar]
- 48. Clark J., Baldwin R., Bayne K., Brown M., Gebhart G., Gonder J., Gwathmey J., Keeling M., Kohn D., Robb J. (1996) Guide for the Care and Use of Laboratory Animals. Institute of Laboratory Animal Resources, National Research Council, Washington, DC [Google Scholar]
- 49. Hong Y. J., Turowski M., Lin J. T., Yokoyama W. H. (2007) Simultaneous characterization of bile acid, sterols, and determination of acylglycerides in feces from soluble cellulose-fed hamsters using HPLC with evaporative light-scattering detection and APCI-MS. J. Agric. Food. Chem. 55, 9750–9757 [DOI] [PubMed] [Google Scholar]
- 50. Gao Z., Perez-Perez G. I., Chen Y., Blaser M. J. (2010) Quantitation of major human cutaneous bacterial and fungal populations. J. Clin. Microbiol. 48, 3575–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo X., Xia X., Tang R., Wang K. (2008) Real-time PCR quantification of the predominant bacterial divisions in the distal gut of Meishan and Landrace pigs. Anaerobe 14, 224–228 [DOI] [PubMed] [Google Scholar]
- 52. Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. R Development Core Team (2009) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org [Google Scholar]
- 55. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R. (2010) UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 58. Lozupone C. A., Knight R. (2008) Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 32, 557–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fisher R. A. (1924) The distribution of the partial correlation coefficient. Metron 3, 329–332 [Google Scholar]
- 60. Prokhorov A. V. (2002) Partial correlation coefficient. In Encyclopaedia of Mathematics (Hazewinkel M., ed), Springer, New York [Google Scholar]
- 61. Kim S. H. (2006) Correlated asymmetry of sequence and functional divergence between duplicate proteins of Saccharomyces cerevisiae. Mol. Biol. Evol. 23, 1068–1075 [DOI] [PubMed] [Google Scholar]
- 62. Huang X.-F., Yu Y., Beck E., South T., Li Y., Batterham M., Tapsell L., Chen J. (2011) Diet high in oat β-glucan activates the gut-hypothalamic (PYY3-36-NPY) axis and increases satiety in diet-induced obesity in mice. Mol. Nutr. Food. Res. 55, 1118–1121 [DOI] [PubMed] [Google Scholar]
- 63. Wang Q., Perrard X. D., Perrard J. L., Mansoori A., Raya J. L., Hoogeveen R., Smith C. W., Ballantyne C. M., Wu H. (2011) Differential effect of weight loss with low-fat diet or high-fat diet restriction on inflammation in the liver and adipose tissue of mice with diet-induced obesity. Atherosclerosis 219, 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neyrinck A. M., Possemiers S., Druart C., Van de Wiele T., De Backer F., Cani P. D., Larondelle Y., Delzenne N. M. (2011) Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS One 6, e20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Neyrinck A. M., Possemiers S., Verstraete W., De Backer F., Cani P. D., Delzenne N. M. (2011) Dietary modulation of clostridial cluster XIVa gut bacteria (Roseburia spp.) by chitin-glucan fiber improves host metabolic alterations induced by high-fat diet in mice. J. Nutr. Biochem. 23, 51-59 [DOI] [PubMed] [Google Scholar]
- 66. Mayer J. (1955) Regulation of energy intake and the body weight: the glucostatic theory and the lipostatic hypothesis. Ann. N. Y. Acad. Sci. 63, 15. [DOI] [PubMed] [Google Scholar]
- 67. Lozupone C., Knight R. (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Turnbaugh P., Backhed F., Fulton L., Gordon J. (2008) Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Faith J. J., McNulty N. P., Rey F. E., Gordon J. I. (2011) Predicting a human gut microbiotaís response to diet in gnotobiotic mice. Science 333, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., Gordon J. I. (2009) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Willing B. P., Russell S. L., Finlay B. B. (2011) Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 9, 233–243 [DOI] [PubMed] [Google Scholar]
- 72. Stecher B., Hardt W. D. (2011) Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 14, 82–91 [DOI] [PubMed] [Google Scholar]
- 73. Ludwig W., Schleifer K.-H., Whitman W. (2009) Revised road map to the phylum Firmicutes. In Bergey's Manual of Systematic Bacteriology (Vos P., Garrity G., Jones D., Krieg N., Ludwig W., Rainey F., Schleifer K.-H., Whitman W., eds) pp. 1–13, Springer, New York [Google Scholar]
- 74. Ravussin Y., Koren O., Spor A., LeDuc C., Gutman R., Stombaugh J., Knight R., Ley R. E., Leibel R. L. (2012) Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity 20, 738–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Martìnez I., Wallace G., Zhang C., Legge R., Benson A. K., Carr T. P., Moriyama E. N., Walter J. (2009) Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl. Environ. Microbiol. 75, 4175–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hooda S., Boler B. M. V., Serao M. C. R., Brulc J. M., Staeger M. A., Boileau T. W., Dowd S. E., Fahey G. C., Swanson K. S. (2012) 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J. Nutr. 142, 1259–1265 [DOI] [PubMed] [Google Scholar]
- 77. Collins M. D., Lawson P. A., Willems A., Cordoba J. J., Fernandez-Garayzabal J., Garcia P., Cai J., Hippe H., Farrow J. A. (1994) The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44, 812–826 [DOI] [PubMed] [Google Scholar]
- 78. Ferguson M., Jones G. (2000) Production of short chain fatty acids following in vitro fermentation of saccharides, saccharide esters, fructo oligosaccharides, starches, modified starches and non starch polysaccharides. J. Sci. Food Agric. 80, 166–170 [Google Scholar]
- 79. Braun W. H., Ramsey J. C., Gehring P. J. (1974) The lack of significant absorption of methylcellulose, viscosity 3300 CP, from the gastrointestinal tract following single and multiple oral doses to the rat. Food Cosmet. Toxicol. 12, 373–376 [DOI] [PubMed] [Google Scholar]
- 80. Machle W., Heyroth F. F., Witherup S. (1944) The fate of methylcellulose in the human digestive tract. J. Biol. Chem. 153, 551 [Google Scholar]
- 81. Yokoyama W., Knuckles B., Davis P., Daggy B. (2002) Stability of ingested methylcellulose in the rat determined by polymer molar mass measurements by light scattering. J. Agric. Food. Chem. 50, 7726–7730 [DOI] [PubMed] [Google Scholar]
- 82. Cai X., Yang L., Zhang L.-M., Wu Q. (2009) Synthesis and anaerobic biodegradation of indomethacin-conjugated cellulose ethers used for colon-specific drug delivery. Bioresour. Technol. 100, 4164–4170 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


