Abstract
Horizontal gene transfer (HGT) between bacteria occurs in the intestinal tract of their animal hosts and facilitates both virulence and antibiotic resistance. A model in which both the pathogen and the host are genetically tractable facilitates developing insight into mechanistic processes enabling or restricting the transfer of antibiotic resistance genes. Here we develop an in vivo experimental system to study HGT in bacteria using Caenorhabditis elegans as a model host. Using a thermosensitive conjugative system, we provide evidence that conjugation between two Escherichia coli strains can take place in the intestinal lumen of N2 wild-type worms at a rate of 10−3 and 10−2 per donor. We also show that C. elegans age and genotype are important determinants of the frequency of conjugation. Whereas ∼1 transconjugant for every 100 donor cells could be recovered from the intestine of N2 C. elegans, for the age-1 and tol-1 mutants, the detected rate of transconjugation (10−3 and 10−4 per donor cell, respectively) was significantly lower. This work demonstrates that increased recombination among lumenal microbial populations is a phenotype associated with host aging, and the model provides a framework to study the dynamics of bacterial horizontal gene transfer within the intestinal environment.—Portal-Celhay, C., Nehrke, K., Blaser, M. J. Effect of Caenorhabditis elegans age and genotype on horizontal gene transfer in intestinal bacteria.
Keywords: conjugation, mating, in vivo
Horizontal gene transfer (HGT) is an ancient phenomenon that affects the entire biosphere (1). Of particular importance is microbial HGT, which has a continuing strong influence on evolution (2–4) and is a model to understand evolution of more complex organisms. For microbes that interact with host species, HGT plays important roles shaping these interactions (5–8). However, although pioneering work began in the 1960s (9–11), the effect of animal host environments on HGT between bacteria has been little-studied over the years (12). Gene exchange that may be limited under in vitro conditions may readily occur in vivo in environments to which the microbes have evolved, and in response to host signals. The gut in different animal species, including humans, may be an important milieu for gene transfer, since large and diverse microbial communities reside there (13, 14).
Residential bacteria not only exchange antibiotic-resistance genes among themselves (15), but also interact with transient bacteria, which leads to the bidirectional transfer of resistance genes (16). Bacteria colonizing the intestine of laboratory animals have been observed to transfer antibiotic resistance genes under particular circumstances (17–20). Antibiotic resistance genes can be disseminated among bacterial populations by transduction, transformation, or by conjugation. A model in which both the pathogen and the host are genetically tractable facilitates developing insight into mechanistic processes enabling or restricting the transfer of antibiotic resistance genes. As such, we developed an in vivo experimental system to study horizontal gene transfer in bacteria, using the nematode Caenorhabditis elegans as a model host.
When C. elegans ingest bacteria as their food source, a proportion remains viable and can colonize and proliferate in its intestinal tract (21, 22). Specific C. elegans innate immune pathways regulate intestinal bacterial proliferation, via expression of antimicrobial peptides (23, 24), and differences in pH (25) as well as differing concentrations of antimicrobial peptides in the intestinal milieu (26) also could affect conjugation rates. In this study, we assess the frequency of conjugation of Escherichia coli in the intestinal lumen of C. elegans wild-type and mutant worms. Since both E. coli and C. elegans are genetically defined and tractable, this approach permits identifying factors that regulate HGT between bacteria in vivo.
MATERIALS AND METHODS
C. elegans strains and growth conditions
All strains were provided by the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN, USA)and maintained on modified (0.35% peptone) nematode growth mediuma (mNGM), using standard procedures (27). E. coli DY330 [TetR (efflux pump Tn10), StrS], which contains a derepressed form of plasmid R27 (the prototypical IncHI1 conjugative plasmid), and E. coli OP50 (StrR chromosomal marker, TetS), were kindly provided by Diane E. Taylor (University of Alberta, Edmonton, AB, Canada; ref. 28) and Fred M. Ausubel (Harvard Medical School, Boston, MA, USA), respectively. Bacterial cultures were grown in Luria-Bertani (LB) broth at 37°C, supplemented with either tetracycline (20 μg/ml) or streptomycin (60 μg/ml). Table 1 indicates the C. elegans and bacterial strains used in this study.
Table 1.
C. elegans and E. coli strains used in this study
| Strain | Relevant phenotype | Source and references |
|---|---|---|
| C. elegans | ||
| N2 | Wild type, reference C. elegans strain | Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN, USA) |
| daf-16 (mu86)I | Decreased lifespan; decreased resistance to heat, oxidative stress, and pathogens | 40 |
| age-1 (hx546)II | Extended lifespan; increased resistance to heat, oxidative stress, and pathogens | 40 |
| tol-1 (nr2033)I | Unable to avoid pathogenic bacteria; susceptible to killing by gram-negative bacteria | 41,42 |
| phm-2 (ad597)I | Defective terminal bulb; allows greater numbers of intact bacteria to enter the intestinal tract | 43 |
| E. coli | ||
| OP50mgh | Uracil auxotroph, StrR | Fred M. Ausubel (Harvard Medical School, Boston, MA, USA) |
| DY330 with plasmid drR27 | RifR, TetR, KmR | Diane E. Taylor (University of Alberta, Edmonton, AB, Canada) |
In vitro conjugation
Bacterial conjugation was achieved in vitro by mixing E. coli strains DY330/(donor)/(OP50)/(recipient). After incubation at different temperatures, the bacterial mixture was plated onto antibiotic-containing selective medium [LB agar supplemented with either streptomycin (60 μg/ml), tetracycline (20 μg/ml), or both] that allowed identification of donor, recipient, and potential transconjugant strains.
In vivo conjugation assay
To examine in vivo bacterial conjugation, C. elegans embryos were grown on mNGM agar plates seeded with E. coli DY330 (donor strain; see Fig. 1A). Based on our preliminary studies (29, 30), at d 4 of adulthood, worms were washed to remove surface bacteria and transferred to mNGM agar plates seeded with E. coli OP50 (recipient strain). After 24, 48, or 72 h, 10 worms were washed, homogenized by grinding, and plated on selective agars, as above, to quantify the three distinct bacterial populations. Conjugation frequency was estimated by dividing the number of transconjugants per worm by the number of donor bacteria per worm. All mating experiments were conducted at 25°C.
Figure 1.

Bacterial conjugation can occur in the intestine of older C. elegans. A) C. elegans embryos were grown on NGM agar plates seeded with donor E. coli DY330 strain. At various days of adulthood [e.g., d 4 (L4+4)], worms were washed and transferred to plates seeded with lawns of recipient E. coli OP50 strain. After 24, 48, or 72 h, worms were washed and homogenized by grinding, and the lysates were plated on selective agar to quantify bacterial populations with 3 patterns of antibiotic resistance. B) Worms were initially grown on lawns of donor E. coli DY330 (solid bars) and then moved at different points in worm maturation (L4 stage +2, +3, +4, or +7 d) to plates with lawns of E. coli OP50 (open bars; recipient strain). Graph shows intestinal densities of donor E. coli DY330, recipient E. coli OP50, and transconjugant E. coli (shaded bars) recovered from N2 C. elegans 24 h after shifting the worms to lawns of recipient strain. Data represent means ± sd. C) Worms were grown on donor DY330 (solid circles) and moved on d 4 (L4 stage+4) to lawns of recipient OP50 (open squares). Intestinal densities of the donor E. coli DY330, recipient E. coli OP50, and transconjugant E. coli (shaded triangles) isolated from N2 C. elegans after following the same worm population over the 3 d following transfer are shown. Data represent means ± sd.
Confirmation of in vitro conjugation by random amplified polymorphic DNA (RAPD)-PCR
To determine whether conjugation actually occurred, and to rule out spontaneous mutations conferring StrR in the donor strain, transconjugants were characterized by RAPD-PCR fingerprinting (31). Colonies were isolated from agar plates selective for the resistance phenotype (TetR StrR) of putative transconjugants, and genomic DNA was extracted and purified, followed by PCRs with RAPD primers 1254 (5′-CCGCAGCCAA-3′) or 1290 (5′-GCGGAAATAG-3′), using conditions as described previously (32). Template DNA from the donor and recipient strains was included in the PCR as positive controls to indicate strain origin of the putative transconjugants.
Measurement of C. elegans intestinal pH
The vital dye Oregon Green 488 Dextran (1 μM) was fed to N2 and mutant C. elegans in S medium supplemented with E. coli OP50 for 1 h at room temperature. The worms then were pipetted onto an NGM-agarose plate for imaging. Image acquisition, analysis, and calibration were performed as described previously (33), with dual excitation wavelengths of 490 and 440 nm.
Statistical analysis
Transconjugant populations and conjugation frequencies in wild-type C. elegans and mutants were compared using 2-sample t tests assuming equal variances; values of P < 0.05 were considered significantly different from control.
RESULTS
Temperature-sensitive transfer of H incompatibility group plasmid in vitro
The wild-type C. elegans N2 strain could be cultivated at temperatures between 15 and 25°C (34). To determine whether C. elegans can be used as a model host to study HGT, a conjugation system that can transfer DNA at these temperatures was required. The ability to transfer DNA between 15 and 25°C was first studied in vitro to provide an indication of the likelihood of genetic transfer occurring within worms raised at that temperature. We tested a self-transmissible resistance plasmid (drR27) belonging to the IncHI1 plasmid group (35), which has temperature-sensitive conjugative transfer. These plasmids have predominantly been identified in Enterobacteriaceae (36) and play a central role in the emergence and reemergence of bacterial pathogens, since they encode resistance to multiple antibiotics (37, 38).
After 24 h of mating at 20°C and 25°C, donor E. coli strain DY330, which carries pdrR27, showed rates of transfer of 10−3 and 10−2, respectively, to recipient E. coli strain OP50 (data not shown). In contrast, transfer at 37°C was negligible, confirming previous observations that showed that IncHI1 plasmids transfer optimally between 22 and 30°C (39). From these results, we conclude that pdrR27 transfer can be examined in vivo in C. elegans, since its occurrence would be predicted to be a relatively frequent event at temperatures at which C. elegans can be cultivated.
Bacterial gene transfer in the intestine of C. elegans
Next, we asked whether exchange of genetic material among E. coli strains could occur within the C. elegans intestinal tract. The C. elegans wild-type strain N2 was maintained and propagated on donor strain E. coli DY330 using standard techniques. Then worms were transferred to lawns composed of recipient strain E. coli OP50 and then sampled for transconjugants on several days after transfer to determine which day of worm life was optimal to perform the conjugation assay (Fig. 1A). As increasingly older worms were exposed to the introduced recipient E. coli strain (OP50), progressively greater intestinal persistence of the donor E. coli strain DY330 was found. When worms were transferred on d 2 of adulthood, only recipient E. coli OP50 could be isolated from the intestine. However, when transfer was done later, bacterial densities of persisting donor strains within the C. elegans intestine increased significantly (P<0.001), with smaller concomitant increases in the introduced recipient strain (Fig. 1B). These results are consistent with previous observations, indicating that worm age determines the persistence of founder bacteria (29). Under the conditions tested, putative bacterial conjugation was detected and increased significantly as the worms aged. As a control, we harvested and plated all bacteria growing on the NGM plates. Finding only cells of the recipient strain (E. coli OP50) ruled out the possibility of gene transfer outside the worms and of worms feeding on transconjugants. Based on the results from these experiments, we selected transfer at d 4 as a condition for further study.
To further test the ability of conjugation to proceed in vivo, we next investigated the persistence of donor, recipient, and putative transconjugant cells in the C. elegans intestine. In these experiments, C. elegans were grown on lawns of E. coli donor DY330 and transferred to the recipient E. coli strain OP50 on adult d 4 (Fig. 1C, arrow), and then groups of worms were sampled 1 to 3 d after the transfer. As shown in Fig. 1C, the recipient strain introduced on d 4 rapidly colonized the intestine with 104 to 105 cfu/worm, as expected (29). Although no further introduction of the donor strain was starting on d 4, it also persisted in the intestine of the worms, at stable levels of ∼103 cfu/worm. Strains with both antibiotic resistances could be detected in the intestinal contents 24 h after the recipient strain was introduced, at stable concentrations of ∼101 cfu/worm.
Confirmation of in vivo conjugation by RAPD-PCR
To confirm that conjugation had occurred and to rule out spontaneous mutations conferring streptomycin resistance in the donor strain, putative transconjugants isolated from the plates containing both of the antibiotics (Tet and Str) on which selection was based were characterized by RAPD-PCR fingerprinting. DNA from the donor and recipient strains was included in the PCR as positive controls to indicate strain origin of the putative transconjugants. RAPD-PCR assays revealed band profiles that clearly distinguished between the donor and recipient strains (Fig. 2); 2–3 major polymorphic bands were amplified with each of the primers. In the experiment shown, four putative transconjugant isolates with the expected phenotype were examined, and the results show that all had RAPD-PCR profiles identical to those of the recipient strain, as expected, since spontaneous mutation conferring resistance to streptomycin are rare. Based on these findings, we conclude that conjugation had been achieved and that the double-resistant phenotype of transconjugants results from DNA transfer from the expected donor to recipient strain.
Figure 2.

RAPD profiles of donor, recipient, and putative transconjugant strains. The donor E. coli DY330 (letter D) recipient E. coli OP50 (R) and 4 putative transconjugants (T1–T4) were studied by RAPD. Amplifications were performed using primer 1290 (left panel) or primer 1254 (right panel). Lane at left contains a 1-kb molecular size marker (M); lanes C represent the negative controls. Labels at bottom indicate the antibiotic-resistance phenotype of the strains (S, susceptible; R, resistant).
Role of the host intestinal milieu in bacterial gene transfer
We next examined the role of variation within the intestinal milieu within C. elegans on the rate of bacterial gene transfer. We studied age-1 and daf-16 worms that have essentially opposite phenotypes, restricting and enhancing bacterial colonization in the intestinal lumen, respectively (30, 40). We also examined mutants in tol-1, the sole Toll-like receptor (TLR) in C. elegans, which are required for worms to avoid pathogenic bacteria (41) and for the expression of ABF-2, which is a defensin-like molecule expressed in the pharynx of the worms, and of HSP-16, which is involved in stress resistance (42). Finally, we examined phm-2 mutants, which have an abnormal pharyngeal grinder (43), and therefore increased delivery of living bacterial cells to the intestine. In all four C. elegans mutants, sufficient members of both donor and recipient E. coli cells could persist in the intestine after the introduction of the recipient strain on d 4 to allow transfer (Fig. 3); however, each worm genotype showed different bacterial densities and kinetics of colonization (Fig. 3). In the age-1 mutants, donor E. coli DY330 intestinal counts were low (∼10 cfu/worm) at d 4, but after the introduction of the recipient strain increased to concentrations of 103 on each subsequent day (Fig. 3A). This increase represents intraintestinal proliferation, since the worms no longer had DY330 cells in their external environment. In contrast, phm-2 mutants had ∼105 viable donor E. coli DY330 cells/worm on d 4, but concentrations decreased ∼1 log after transfer to lawns with recipient strain E. coli OP50 (Fig. 3D); this decrease represents the effects of competition between the founder (donor) and the introduced (recipient) bacteria. Intestinal concentrations of the donor strain in the daf-16 and tol-1 mutants were maintained similar to N2 wild-type worms, despite no further introduction (Fig. 3B, C). The introduced recipient strain colonized the intestines of all four mutants to similar extents as with N2 (104 to 105 cfu/worm; Fig. 3). Thus, in both the wild-type and mutant worms, cocolonization of the intestinal lumen by both donor and recipient strains was achieved. In each worm background, transconjugation had occurred (Fig. 3).
Figure 3.
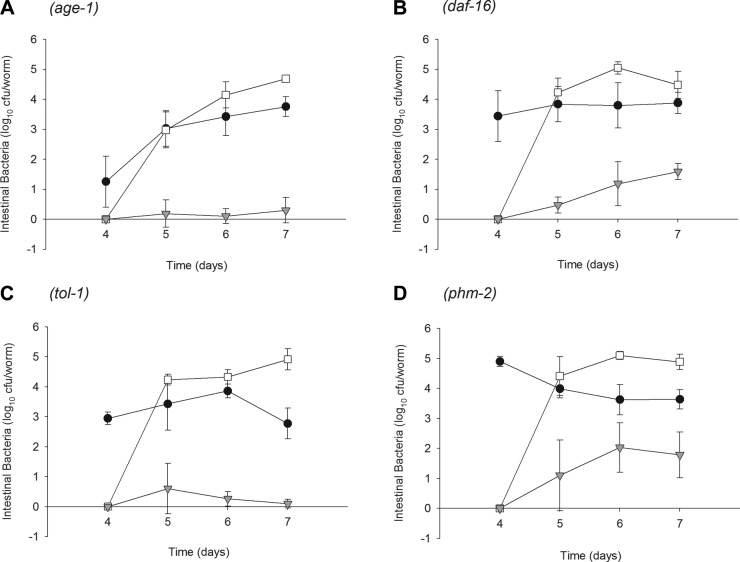
Bacterial conjugation in the intestinal lumen of C. elegans mutants. Intestinal densities of donor E. coli DY330 (solid circles), recipient-E. coli OP50 (open circles), and transconjugant E. coli (shaded triangles) isolated from: age-1 (A); daf-16 (B); tol-1 (C); or phm-2 (D) mutants. Worms were grown on E. coli DY330, and then on d 4 shifted to lawns of E. coli OP50.
Role of host genotype on conjugation frequencies
Although transconjugants were detected in all four mutants by the day after the introduction of the recipient strain and colonized the intestine throughout the experiment, patterns differed by host genotype (Fig. 4A, top panel). The trends became most apparent by d 6. A progressive increase was found in the number of transconjugants until d 6 in the N2, phm-2, and daf-16 mutants. However, the number of intestinal transconjugants in the age-1 and tol-1 mutants did not rise and remained significantly (P<0.005) lower than in the N2 worms (Fig. 4A, bottom panel).
Figure 4.

Host genotype determines the size of the transconjugant E. coli population and the conjugation rate. A) Top panel: intestinal densities of transconjugant E. coli within N2 and defined C. elegans mutants throughout the conjugation assay are highlighted. Bottom panel: transconjugant populations within the intestine of N2 and mutant C. elegans on d 6 of conjugation assay. *P < 0.005 vs. N2 worms. B) Top panel: densities of the donor E. coli strain DY330 within the C. elegans intestine throughout the conjugation assays. Bottom panel: donor populations within the intestine on d 6 of conjugation assay in N2 and mutant C. elegans worms. C) Top panel: conjugation rates within N2 C. elegans and defined mutants throughout the conjugation assay. Bottom panel: bacterial gene exchange in N2 and mutant C. elegans on d 6 of conjugation assay. *P = 0.01, **P = 0.005 vs. N2 worms.
As expected, each worm genotype showed different donor bacterial densities at the day of transfer (d 4); however, after the introduction of the recipient strain, the donor concentrations (on d 6) were nearly the same in all five worm genotypes (Fig. 4B). We next examined the frequency of conjugation in relation to the number of donor cells. At 24 h after transfer (d 5), the calculated conjugation rates of the bacteria within the intestine of N2 and mutants were very similar (Fig. 4C, top panel); however, by 48 h (d 6) after the introduction of the recipient strain, significant differences in frequencies were observed (Fig. 4C, bottom panel). Whereas ∼1 transconjugant for every 100 donor cells could be recovered from the intestine of N2 (wild-type) C. elegans, and from the phm-2 and daf-16 mutants, for the age-1 and tol-1 mutants, the detected rate of transconjugation (10−3 and 10−4 per donor cell), respectively, was significantly lower than for the N2 wild-type worms (Fig. 4C, bottom panel). Thus, C. elegans genotype clearly affects genetic exchange in the intestinal lumen, with lowest rates in age-1 and tol-1 worms.
Effect of intestinal pH on conjugation rates
Among factors known to affect the rate of plasmid transfer in bacterial cells are pH (44–46), temperature (39, 44, 47), nutrients (48, 49), and bacterial density (50, 51). The intestinal pH of C. elegans might vary according to genetic background and, consequently, could affect conjugation rates in the lumen. To test this hypothesis, we measured the intestinal pH of the mutants by feeding them dextran conjugated to the pH-sensitive vital dye Oregon Green-488 in S-basal medium for 1 h, followed by live fluorescent imaging (25, 33). Although the pH in the intestinal lumen of C. elegans oscillates during defecation (25, 52), each of the tested mutants had similar luminal pH levels (Supplemental Fig. S1), which were consistent with the intestinal pH of wild-type worms described previously (25). Thus, intestinal pH is well-conserved in these mutants, and therefore, luminal pH variation cannot explain the different transconjugation frequencies observed.
DISCUSSION
HGT has had an enormous effect on bacterial biodiversity and evolution (53). HGT in the intestinal environment of animals is of particular interest, due to its contribution to the stability of the microbiota, emergence of new pathogens and to antimicrobial-resistant strains (54–56). Using a temperature-sensitive conjugative plasmid (39), we now show that C. elegans can be used as a model system to study HGT in vivo in the intestinal tract, and that host genotype affects the phenomenon.
The high numbers of both donor and recipient bacteria cocolonizing the intestine of 4-d-old adult C. elegans allowed mating to occur at detectable levels. Higher ratios of transconjugants per donor in vivo compared to in vitro assays has been reported (48, 57), suggesting that in vitro systems underestimate the transfer potential; however, the opposite relationship also has been observed (58). In our study, conjugation frequencies obtained in the N2 worms ranged between 10−3 and 10−2 per donor, similar or higher than the in vitro transfer rates of 1 × 10−3 transconjugants/donor. The ability of the donor strain to persist in the C. elegans intestine is necessary for conjugation to occur; high donor density increases encounters of donors and recipients, increasing mating events (59). In previous studies, we found a strong correlation between bacterial counts and life span (30), and we created a model system to distinguish between continuing accumulation vs. bacterial proliferation (29). We found that the host's age, as well as bacterial strain, determines the nature of bacterial persistence in the C. elegans intestine. Using microscopy and strains labeled with fluorescence, we confirmed dual colonization. Our work also provided evidence for active competition in vivo for colonization sites, as well as evidence for in vivo bacterial adaptation (29). Thus, aging in C. elegans, which permits intestinal persistence of founding colonizing strains (Fig. 3 and ref. 29), facilitates intraluminal genetic transfer. This is a new phenotype associated with aging in C. elegans.
However, transfer frequency events do not solely depend on bacterial density. The nematode intestine is dynamic, and bacterial mating in vivo may be more complex than mating in vitro. Since environmental factors have the potential to affect the frequency and outcome of conjugative gene transfer, by studying C. elegans mutants, we could evaluate roles of particular host genotypes in gene transfer. We found no significant differences in conjugation rates between the phm-2 mutants and the daf-16 mutants compared to wild-type (Fig. 4B) despite differences in numbers of bacteria delivered to the intestine (phm-2) and in their ability to colonize the intestine (daf-16) (30). The significantly lower rates of conjugation in age-1 and tol-1 mutants also do not reflect intestinal levels of bacterial colonization (Fig. 4B) but must reflect variation in the host intestinal milieu. It is possible that increased levels of antimicrobial peptides and antioxidant enzymes are responsible of the decreased HGT observed in age-1 mutants. However, lower conjugation rate phenotype was surprising for tol-1 mutants, which have decreased expression of abf-2 (42), an insect like-defensin with proven antimicrobial properties, and of hsp-16.41, which is highly expressed in the worms pharynx in response to stress and belongs to a family of heat-shock proteins recently found to be required for C. elegans immunity (60). Since higher levels of HGT had been expected, other compensating defense molecules may have been up-regulated.
There is no single optimum pH for conjugation. However, conjugal transfer is affected by pH. In general, extremes of pH reduce the rate of transfer; this inhibition is more marked under acidic conditions than under alkaline conditions and is more significant as temperature decreases (44). Although differences in intestinal luminal pH do not appear to play a role in this phenomenon (Supplemental Fig. S1), other luminal factors, such as the content of reactive oxygen species (ROS), antimicrobial peptides, or intestinal physical defenses, such as peristaltic movements, may be significant. In the future, studies using animals deficient in antioxidant enzymes such as ctl-2 and sod-3, antimicrobial peptides such as lysozymes and caenopores (61), or defecation mutants (62) should be done.
Increases in the numbers of transconjugants, such as observed between d 4 and 6 in the N2, phm-2, and daf-16 worms (Fig. 4A), may represent either ongoing conjugation due to the continuing contact of mating strains or could represent proliferation of transconjugants in the intestinal lumen. While the studies we performed were not designed to differentiate between these two possibilities, the altered kinetics in the age-1 and tol-1 worms argues against the latter hypothesis. Similarly, although it is possible that the transconjugants are more fit in the intestinal milieu than their parents, this is unlikely since the introduced marker was not selected for in vivo. That the density of the transconjugants reached a relative plateau is consistent with that notion. However, the clear evidence of proliferation of the donor strain inside the C. elegans lumen may serve as a model for the dynamics (proliferation) of the transconjugants. The increase in the level of transconjugants over time observed in the daf-16 mutants could result from the lack of antioxidant enzymes and/or antimicrobial peptides in the intestinal milieu (59), factors that might inhibit the process of transfer. Experiments designed to confirm this hypothesis, such as testing a daf-16;age-1 double mutant and transformation rescue should be performed, as well as the development of a conditional GFP reporter to indicate in vivo mating.
Overall, our results indicate that we can reproducibly achieve bacterial conjugation in the intestinal lumen of a living organism (C. elegans) that is both genetically defined and experimentally tractable. We also provide evidence that both the age of the host and its genotype play roles in the in vivo regulation of bacterial gene transfer. The C. elegans intestinal lumen model system of microbial colonization, competition, and persistence provides a powerful tool that should lead to a better understanding of both host and microbial factors that affect horizontal gene transfer in vivo.
Supplementary Material
Acknowledgments
This work was supported in part by U.S. National Institutes of Health (NIH) grant RO1 GM63270 and National Science Foundation grant IOS0919848 (K.N.), the Michael Saperstein Medical Scholars Program, the Ellison Medical Foundation, and the Diane Belfer Program for Human Microbial Ecology. C. elegans strains were provided by the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN, USA), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- HGT
- horizontal gene transfer
- LB
- Luria-Bertani
- mNGM
- modified nematode growth medium
- RAPD
- random amplified polymorphic DNA
REFERENCES
- 1. Arber W. (2009) Systemic aspects of biological evolution. J. Biotechnol. 144, 242–244 [DOI] [PubMed] [Google Scholar]
- 2. Levin B. R., Cornejo O. E. (2009) The population and evolutionary dynamics of homologous gene recombination in bacterial populations. PLoS Genet. 5, e1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boto L. Horizontal gene transfer in evolution: facts and challenges. Proc. Biol. Sci. 277, 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barkay T., Kritee K., Boyd E., Geesey G. (2010) A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ. Microbiol. 12, 2904–2917 [DOI] [PubMed] [Google Scholar]
- 5. Blaser M. J. (1997) The versatility of Helicobacter pylori in the adaptation to the human stomach. J. Physiol. Pharmacol. 48, 307–314 [PubMed] [Google Scholar]
- 6. Zaneveld J., Turnbaugh P. J., Lozupone C., Ley R. E., Hamady M., Gordon J. I., Knight R. (2008) Host-bacterial coevolution and the search for new drug targets. Curr. Opin. Chem. Biol. 12, 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kassen R., Rainey P. B. (2004) The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58, 207–231 [DOI] [PubMed] [Google Scholar]
- 8. Atherton J. C., Blaser M. J. (2009) Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 119, 2475–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jarolmen H., Kemp G. (1969) R factor transmission in vivo. J. Bacteriol. 99, 487–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walton J. R. (1966) Infectious drug resistance in Escherichia coli isolated from healthy farm animals. Lancet 2, 1300–1302 [DOI] [PubMed] [Google Scholar]
- 11. Walton J. R., Fulton F. (1967) Segregation during transfer of infectious drug resistance in Enterobacteriaceae. Nature 215, 179–180 [DOI] [PubMed] [Google Scholar]
- 12. Sundin G. W., Bender C. L. (1996) Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 5, 133–143 [DOI] [PubMed] [Google Scholar]
- 13. Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., Gordon J. I. (2008) Evolution of mammals and their gut microbes. Science 320, 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ley R. E., Lozupone C. A., Hamady M., Knight R., Gordon J. I. (2008) Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shoemaker N. B., Vlamakis H., Hayes K., Salyers A. A. (2001) Evidence for extensive resistance gene transfer among Bacteroides spp., and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salyers A. A., Gupta A., Wang Y. (2004) Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416 [DOI] [PubMed] [Google Scholar]
- 17. Boer P., Wagenaar J. A., Achterberg R. P., Putten J. P., Schouls L. M., Duim B. (2002) Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44, 351–359 [DOI] [PubMed] [Google Scholar]
- 18. Lester C. H., Frimodt-Moller N., Hammerum A. M. (2004) Conjugal transfer of aminoglycoside and macrolide resistance between Enterococcus faecium isolates in the intestine of streptomycin-treated mice. FEMS Microbiol. Lett. 235, 385–391 [DOI] [PubMed] [Google Scholar]
- 19. Bourgeois-Nicolaos N., Moubareck C., Mangeney N., Butel M. J., Doucet-Populaire F. (2006) Comparative study of vanA gene transfer from Enterococcus faecium to Enterococcus faecalis and to Enterococcus faecium in the intestine of mice. FEMS Microbiol. Lett. 254, 27–33 [DOI] [PubMed] [Google Scholar]
- 20. Schjorring S., Struve C., Krogfelt K. A. (2008) Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J. Antimicrob. Chemother. 62, 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aballay A., Yorgey P., Ausubel F. M. (2000) Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10, 1539–1542 [DOI] [PubMed] [Google Scholar]
- 22. Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., Murray B. E., Calderwood S. B., Ausubel F. M. (2001) A simple model host for identifying gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98, 10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mallo G. V., Kurz C. L., Couillault C., Pujol N., Granjeaud S., Kohara Y., Ewbank J. J. (2002) Inducible antibacterial defense system in C. elegans. Curr. Biol. 12, 1209–1214 [DOI] [PubMed] [Google Scholar]
- 24. Alegado R. A., Tan M. W. (2008) Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell. Microbiol. 10, 1259–1273 [DOI] [PubMed] [Google Scholar]
- 25. Nehrke K. (2003) A reduction in intestinal cell pHi due to loss of the Caenorhabditis elegans Na+/H+ exchanger NHX-2 increases life span. J. Biol. Chem. 278, 44657–44666 [DOI] [PubMed] [Google Scholar]
- 26. Alper S., McBride S. J., Lackford B., Freedman J. H., Schwartz D. A. (2007) Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol. Cell. Biol. 27, 5544–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stiernagle T. (2006) Maintenance of C. elegans. WormBook doi: 10.1895/wormbook.1.101.1, http:/www.wormbook.org [DOI] [PMC free article] [PubMed]
- 28. Lawley T. D., Gilmour M. W., Gunton J. E., Standeven L. J., Taylor D. E. (2002) Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184, 2173–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Portal-Celhay C., Blaser M. J. (2012) Competition and resilience between founder and introduced bacteria in the Caenorhabditis elegans gut. Infect. Immun. 80, 1288–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Portal-Celhay C., Bradley E. R., Blaser M. J. (2012) Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Power E. G. (1996) RAPD typing in microbiology–a technical review. J. Hosp. Infect. 34, 247–265 [DOI] [PubMed] [Google Scholar]
- 32. Pacheco A. B., Guth B. E., Soares K. C., Nishimura L., de Almeida D. F., Ferreira L. C. (1997) Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J. Clin. Microbiol. 35, 1521–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pfeiffer J., Johnson D., Nehrke K. (2008) Oscillatory transepithelial H(+) flux regulates a rhythmic behavior in C. elegans. Curr. Biol. 18, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riddle D. L., Blumenthal T., Meyre B. J., Priess J. R., eds. (1997) C. elegans II, 2nd Ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA: [PubMed] [Google Scholar]
- 35. Rooker M. M., Sherburne C., Lawley T. D., Taylor D. E. (1999) Characterization of the Tra2 region of the IncHI1 plasmid R27. Plasmid 41, 226–239 [DOI] [PubMed] [Google Scholar]
- 36. Sherburne C., Taylor D. E. (1997) Effect of lipopolysaccharide mutations on recipient ability of Salmonella typhimurium for incompatibility group H plasmids. J. Bacteriol. 179, 952–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taylor D. E. (1989) General properties of resistance plasmids. In Handbook of Experimental Pharmacology, Vol. 91, Springer-Verlag KG, Berlin, Germany [Google Scholar]
- 38. Pang T., Levine M. M., Ivanoff B., Wain J., Finlay B. B. (1998) Typhoid fever–important issues still remain. Trends Microbiol. 6, 131–133 [DOI] [PubMed] [Google Scholar]
- 39. Taylor D. E., Levine J. G. (1980) Studies of temperature-sensitive transfer and maintenance of H incompatibility group plasmids. J. Gen. Microbiol. 116, 475–484 [DOI] [PubMed] [Google Scholar]
- 40. Garsin D. A., Villanueva J. M., Begun J., Kim D. H., Sifri C. D., Calderwood S. B., Ruvkun G., Ausubel F. M. (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300, 1921. [DOI] [PubMed] [Google Scholar]
- 41. Pujol N., Link E. M., Liu L. X., Kurz C. L., Alloing G., Tan M. W., Ray K. P., Solari R., Johnson C. D., Ewbank J. J. (2001) A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11, 809–821 [DOI] [PubMed] [Google Scholar]
- 42. Tenor J. L., Aballay A. (2008) A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Reports 9, 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Avery L. (1993) The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singleton P., Anson A. E. (1983) Effect of pH on conjugal transfer at low temperatures. Appl. Environ. Microbiol. 46, 291–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagachinta S., Chen J. (2008) Transfer of class 1 integron-mediated antibiotic resistance genes from shiga toxin-producing Escherichia coli to a susceptible E. coli K-12 strain in storm water and bovine feces. Appl. Environ. Microbiol. 74, 5063–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fisher K. W. (1957) The nature of the endergonic processes in conjugation in Escherichia coli K-12. J. Gen. Microbiol. 16, 136–145 [DOI] [PubMed] [Google Scholar]
- 47. Singleton P., Anson A. E. (1981) Conjugal transfer of R-plasmid R1drd-19 in Escherichia coli below 22 degrees C. Appl. Environ. Microbiol. 42, 789–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feld L., Schjorring S., Hammer K., Licht T. R., Danielsen M., Krogfelt K., Wilcks A. (2008) Selective pressure affects transfer and establishment of a Lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J. Antimicrob. Chemother. 61, 845–852 [DOI] [PubMed] [Google Scholar]
- 49. Freter R., Brickner H., Botney M., Cleven D., Aranki A. (1983) Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect. Immun. 39, 676–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lampkowska J., Feld L., Monaghan A., Toomey N., Schjorring S., Jacobsen B., van der Voet H., Andersen S. R., Bolton D., Aarts H., Krogfelt K. A., Wilcks A., Bardowski J. (2008) A standardized conjugation protocol to asses antibiotic resistance transfer between lactococcal species. Int. J. Food. Microbiol. 127, 172–175 [DOI] [PubMed] [Google Scholar]
- 51. Kingsman A., Willetts N. (1978) The requirements for conjugal DNA synthesis in the donor strain during flac transfer. J. Mol. Biol. 122, 287–300 [DOI] [PubMed] [Google Scholar]
- 52. Allman E., Johnson D., Nehrke K. (2009) Loss of the apical V-ATPase a-subunit VHA-6 prevents acidification of the intestinal lumen during a rhythmic behavior in C. elegans. Am. J. Physiol. Cell. Physiol. 297, C1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thomas C. M., Nielsen K. M. (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 [DOI] [PubMed] [Google Scholar]
- 54. Marteau P., Pochart P., Bouhnik Y., Rambaud J. C. (1993) The fate and effects of transiting, nonpathogenic microorganisms in the human intestine. World Rev. Nutr. Diet. 74, 1–21 [DOI] [PubMed] [Google Scholar]
- 55. Vedantam G., Hecht D. W. (2003) Antibiotics and anaerobes of gut origin. Curr. Opin. Microbiol. 6, 457–461 [DOI] [PubMed] [Google Scholar]
- 56. Witte W. (2000) Ecological impact of antibiotic use in animals on different complex microflora: environment. Int. J. Antimicrob. Agents 14, 321–325 [DOI] [PubMed] [Google Scholar]
- 57. Dahl K. H., Mater D. D., Flores M. J., Johnsen P. J., Midtvedt T., Corthier G., Sundsfjord A. (2007) Transfer of plasmid and chromosomal glycopeptide resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist stably in vivo in the absence of glycopeptide selection. J. Antimicrob. Chemother. 59, 478–486 [DOI] [PubMed] [Google Scholar]
- 58. Jacobsen L., Wilcks A., Hammer K., Huys G., Gevers D., Andersen S. R. (2007) Horizontal transfer of tet(M) and erm(B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol. Ecol. 59, 158–166 [DOI] [PubMed] [Google Scholar]
- 59. Licht T. R., Christensen B. B., Krogfelt K. A., Molin S. (1999) Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145 (Pt 9), 2615–2622 [DOI] [PubMed] [Google Scholar]
- 60. Singh V., Aballay A. (2006) Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. U. S. A. 103, 13092–13097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roeder T., Stanisak M., Gelhaus C., Bruchhaus I., Grotzinger J., Leippe M. (2010) Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev. Compara. Immunol. 34, 203–209 [DOI] [PubMed] [Google Scholar]
- 62. Rae R., Witte H., Rodelsperger C., Sommer R. J. (2012) The importance of being regular: Caenorhabditis elegans and Pristionchus pacificus defecation mutants are hypersusceptible to bacterial pathogens. Inter. J. Parasitol. 42, 747–753 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


