Abstract
Protein and nucleic acid methylating enzymes are implicated in myriad cellular processes. These enzymes utilize diverse chemical mechanisms ranging from nucleophilic substitution-displacement to a novel radical-based reaction found in bacterial iron–sulfur cluster proteins. Within the cell, methylation activity is governed by interactions with endogenous molecular machinery. Of particular interest are the observations that methylating enzyme activity can be allosterically controlled by regulatory binding partners. Recent advances and emerging trends in the study of methylating enzyme mechanisms and regulation highlight the importance of protein and nucleic acid methylation in cellular physiology and disease.
Introduction
Protein and nucleic acid methylation play diverse roles in cellular signaling and regulation of macromolecular function [1–4]. Protein arginine methyltransferases (PRMTs) and protein lysine methyltransferases (PKMTs) are the predominant enzymes that catalyze S-adenosylmethionine (SAM)-dependent methylation of protein substrates. Broadly, theses enzymes promote a nucleophilic substitution-displacement reaction: methyl addition to an arginine or lysine side chain nitrogen atom concomitant with displacement of S-adenosylhomocysteine (SAH). Additional protein methyltransferases — those that target various other peptidyl side chains (glutamate, glutamine, and histidine) or N-termini and C-termini — use a similar polar mechanism [5–9]. In particular, the recent discovery of NRMT, responsible for eukaryotic N-terminal methylation has renewed attention on the biochemical role of this modification [8,10]. Unlike protein methyltransferases, nucleic acid methylating enzymes employ various chemical mechanisms to install methyl groups on their DNA and RNA substrates. The activities of RNA methylating enzymes are especially notable given their diversity of substrates (mRNA, tRNA, and rRNA) and modification sites (N-methylation, O-methylation, and C-methylation) [11].
Together, protein and nucleic acid methylation expands the repertoire of functions available to their modified substrates. This review focuses on chemical, regulatory, and physiological mechanisms of protein arginine and lysine methyltransferases as well as nucleic acid methylating enzymes.
Signature features of PRMT catalysis
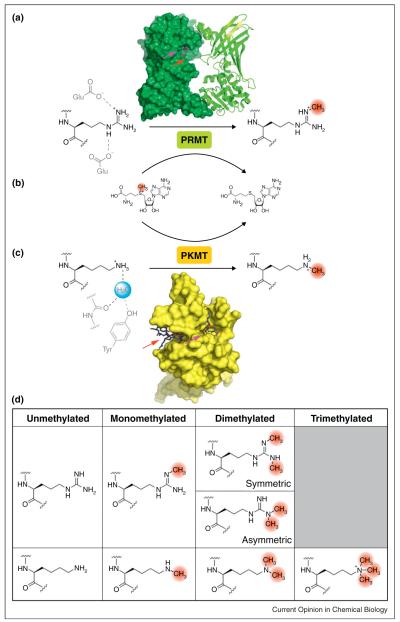
In humans, there are nine canonical protein arginine methyltransferase paralogs (PRMT1-9) [1,12]. Beyond the commonly accepted PRMTs, 34 genes share sequence homology [13]. Structural evidence and mutational analysis from representative members of the PRMT family have informed a spatial model of the active site, in which arginine is juxtaposed with the S-methylsulfonium of SAM along one face of the enzyme cavity (Figure 1a,b) [14]. By structural inference, a pair of conserved glutamate residues opposite the interface between SAM and the methyl acceptor is thought to polarize the guanidino group of arginine, thus priming nucleophilic attack [14]. Another proposed feature is a proton relay, consisting of one or more amphoteric residues thought to facilitate arginine deprotonation [15].
Figure 1.
Substitution-displacement reaction mechanism of PRMTs and PKMTs. (a) PRMT1 dimer model (1OR8; colored green) with reaction scheme. The substrate residue (colored black) and catalytic residues (colored gray) are shown. The SAM and methyl acceptor binding sites are identified with magenta and red arrows, respectively. (b) PRMTs and PKMTs utilize S-adenosylmethionine as a methyl group donor. The methyl group (colored red) is transferred to the substrate residue concomitant with S-adenosylhomocysteine formation. (c) SET7/9 (2F69; colored yellow) with substrate, catalytic residues, and catalytic water (colored blue). (d) Substrates and methylated products of protein methyltransferases.
PRMTs are classified by product selectivity
In vivo, methylarginine exists in three major forms: mono N-methylarginine, symmetric N,N’-dimethylarginine, and asymmetric N,N-dimethylarginine (Figure 1d) [1]. The PRMT paralogs are defined as type I or type II based on the capacity to form either asymmetric or symmetric dimethylated arginine, respectively. Both PRMT types are able to form monomethylarginine as a reaction intermediate en route to the dimethylated states. Structural and sequence-based comparison between members of the two PRMT types has identified a conserved, type-specific residue that controls product selectivity [16]. For the type II enzyme PRMT5, substitution at Phe379 to a methionine residue, characteristic of type I PRMTs, produces a mutant that catalyzes both symmetric and asymmetric dimethylation. Proximity of SAH to the determinant residue Phe379, revealed in a co-crystal structure, suggests that product selectivity may be partly controlled by restricted methyl donor conformations relative to the substrate nucleophile. Two minor PRMT subclasses, type III and type IV, have also been reported to exclusively mono-methylate arginine on the terminal or internal nitrogen atom, respectively [1].
Allosteric modulation of PRMT activity via multimerization
Seminal structural investigations revealed that PRMTs form multimeric complexes composed of repeating homo-dimer units [14,15,17]. It was proposed that the proximity of active sites in PRMT dimers promotes dimethylation through a quasi-processive mechanism, in which the paired enzymes each add a methyl group to a single arginine residue. However, for at least a subset of enzymes and their substrates, biochemical data support distributive or partially processive mechanisms [18–20]. While the role of dimerization in processivity remains inconclusive, PRMT homo-dimers are essential to catalysis. Mutations that abrogate homo-dimerization concomitantly eliminate methylation activity, which has been attributed to disruption of the SAM-binding surface proximal to the dimer interface [14,15,17]. Moreover, a study of PRMT1 catalytic activity as a function of multimerization showed that apparent turnover rate correlates with oligomer formation, with both metrics reaching a maximum at enzyme concentrations in the range of 0.5–1.0 μM [19]. These observations support a model in which catalytic activity is dependent upon multimerization of a minimal dimer unit.
Studies of PRMT hetero-multimers suggest a regulatory function. For example, both PRMT1 and PRMT2 function as transcriptional co-activators with the ability to methylate histone H4 [21]. Through co-immunoprecipitation and fluorescence microscopy experiments, it was shown that these paralogs physically interact [22•]. Furthermore, this association increases PRMT1 activity independent of the catalytic capacity of PRMT2, suggesting stimulation through allostery. In another case, PRMT5 and regulatory accessory proteins coexist in a large heterogeneous complex responsible for depositing methyl marks necessary during proper spliceosome assembly [23]. Among these complex partners is pICLn, a protein that restricts the inherently promiscuous substrate specificity of PRMT5 [23]. Together, these examples demonstrate that hetero-multimers — whether among PRMTs or with other proteins — can regulate catalytic behavior.
Signature features of PKMT catalysis
The SET domain-containing enzyme family, named after DrosophilaSu(var), E(z) and Trithorax histone lysine methyltransferases, shares sequence homology with 51 human genes [13]. The ~130-residue SET domain comprises non-contiguous, conserved regions known as nSET/SET-N and cSET/SET-C, denoting proximity to respective primary sequence termini [24,25]. Separating these regions is a variable iSET/SET-I region, which is thought to influence substrate specificity and catalytic activity. Together, these regions comprise the active site, which organizes SAM and the peptidyl lysine substrate at opposing ends of a narrow channel that accommodates the substrate side chain (Figure 1b,c) [26]. Along with the canonical lysine methyltransferases, there are at least two non-SET domain PKMTs: the histone methyltransferase DOT1L that possesses seve-n-β-strand architecture and a multi-subunit methyltransferase (WDR5, RbBp5, Ash2L, and DPY-30) derived from the MLL1 core complex that targets histone H3 [27,28].
Transition state modeling of the SET7/9 homolog suggests a collinear arrangement of the lysyl ε-nitrogen and the S-methylsulfonium bond of SAM, consistent with a substitution-displacement mechanism [29]. At physiological pH, the side chain of lysine exists primarily as an ammonium species, which requires deprotonation for nucleophilic addition into SAM. Dynamical simulations of a composite active site, derived from LSMT, vSET, and SET7/9 structures, indicate the existence of a water channel that relays protons to bulk solvent [30]. Therefore, it is expected that at high pH, facile deprotonation drives formation of the nucleophilic amine species. Accordingly, PKMT catalysis is pH-dependent, reaching near-maximal activity at pH ~9 [31,32]. In agreement with a nucleophilic substitution-displacement mechanism, deprotonation is a critical step that activates the lysyl amine for methyl transfer.
Substrate binding site influences degree of product methylation by PKMTs
In vivo, protein lysine methylation results in monomethylated, dimethylated, or trimethylated species (Figure 1d) [33]. Because specific physiological functions are dependent upon methylation status, product selectivity is precisely regulated to preclude spurious signaling [34,35]. The methyl acceptor binding site architecture is a major determinant that governs the extent of product methylation. Analysis of the DIM-5 methyltransferase indicated that an acceptor site mutation at Phe281 influences methylation multiplicity. In particular, the F281Y mutation causes a product distribution shift towards lower-order methylated states [35,36]. The reciprocal mutation in SET7/9 (Y305F) imparts an additional capacity to form trimethylated product. From this work, a Phe/Tyr switch model was developed to explain PKMT product selectivity. For enzymes containing a phenylalanine determinant residue, higher-order methylation products are expected. A corresponding tyrosine residue, conversely, is predicted to limit methylation extent. Consistent with this model, available structural evidence from methyltransferases with a phenylalanine determinant residue shows sterically permissive methyl acceptor binding sites capable of accommodating partially methylated substrates [37]. With tyrosine as the determinant residue, imposition of a phenolic hydroxyl group precludes binding of intermediate methylated species; thus, these enzymes are restricted to production of lower-order methylated products [37].
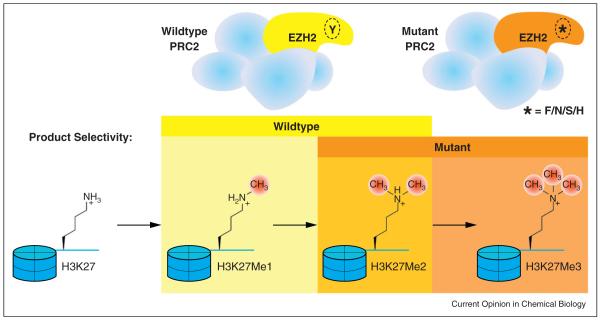
The Phe/Tyr switch phenomenon had been ascribed to a single structurally homologous site until the identification of several disease-associated mutations in a gene encoding EZH2, a histone H3 methyltransferase [38,39••]. Discovered in a screen of non-Hodgkin’s lymphoma samples, substitutions at Tyr641 (Y641F/N/S/H) were originally thought to abrogate methyltransferase activity on non-methylated substrates. However, subsequent work using heterogeneous extracted nucleosomes indicates catalytic competence, attributed to the preference of mutant EZH2 for partially methylated substrates (i.e. monomethylated and dimethylated) [39••]. Because of their complementary substrate specificities, these enzymes coordinate to produce H3K27Me3 (histone H3, lysine 27, trimethyl) through sequential methyl transfers — initially by wild-type EZH2 then by a Y641/F/N/S/H mutant (Figure 2) [39••]. Similar to the structural alterations caused by canonical determinant residue substitution, homology modeling of the EZH2 active site suggests that mutations at Tyr641 exhibit an analogous reduction of steric hindrance [40].
Figure 2.
Complementary product selectivities of wildtype and mutant EZH2 underlie the hyper-trimethylation phenotype among heterozygotes. Wildtype EZH2, a subunit of PRC2, monomethylates and dimethylates H3K27. Mutant EZH2 (Y641F/N/S/H), with its product selectivity shifted to higher-order methylation products, has the capacity to form H3K27Me3.
Allosteric modulation of PKMT activity
In an example of allosteric regulation, H3K27 trimethylation by EZH2 is modulated by protein–protein interactions. EZH2 is the enzymatic subunit of the polycomb repressive complex 2 (PRC2) responsible for gene silencing [41]. Another subunit of PRC2 is EED, which was shown to preferentially bind H3K9Me3 and H3K27Me3 using a peptide binding screen and coprecipitation with methyllysine analog-modified nucleosomes (Figure 3) [41,42]. On the basis of the respective subunit functions, PRC2 has the dual-capacity to generate and bind H3K27Me3. Steady-state kinetic analysis of PRC2 in the presence of H3K27Me3 peptide revealed a ~7-fold increase in apparent maximal reaction rate, while the substrate concentration necessary for half-maximal velocity remained unchanged. These findings support a model of allosteric feed-forward regulation, whereby H3K27Me3 interaction with EED stimulates EZH2 to methylate nearby unmodified H3K27. This mechanism is consistent with a proposed spreading process that explains the contiguous distribution of repressive histone marks along chromatin [43].
Figure 3.
Allosteric activation of PRC2 methyltransferase activity via EED interaction with H3K27Me3. PRC2 is a multi-subunit complex that establishes the repressive H3K27Me3 mark. (a) PRC2 binds non-methylated chromatin to install H3K27Me3 via EZH2 activity. (b) EED binds H3K27Me3, which causes allosteric activation of EZH2. With its improved catalytic efficiency, EZH2 methylates chromatin regions adjacent to the founding H3K27Me3 mark. (c) Nucleosomes modified with methyllysine analogs were used to assay EED substrate recognition and allostery.
DNA methyltransferases
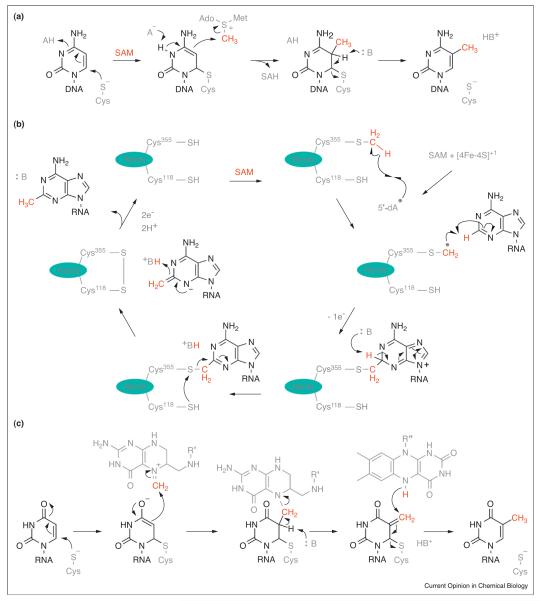
Enzymatic DNA methylation in prokaryotes yields several species: N6-methyladenosine, N4-methylcytosine and 5-methylcytosine. Among eukaryotes, DNA is exclusively methylated on C5 of cytosine [44]. This epigenetic modification is commonly associated with loci-specific repressive chromatin states. Deposition of this modification is performed by DNA methyltransferases (DNMTs), which utilize SAM as the methyl donor [45]. Common among these enzymes is the ability to flip their target cytosine out of helical DNA before nucleophilic addition of a catalytic cysteine at C6 [46]. The resultant localization of negative charge on C5 promotes reactivity with SAM. Subsequent pyrimidine re-aromatization drives β-elimination of the appended cysteine, thus regenerating the resting-state enzyme and releasing the methylated cytosine (Figure 4a). Investigations into the regulatory mechanisms that control DNMT catalytic activity have revealed that some DNMTs are sensitive to the post-translational modification status of their chromatin substrates. For example, the ADD domain of murine Dnmt3a is necessary for efficient DNA methylation on in vitro reconstituted oligo-nucleosomes lacking post-translational modification; while, DNMT activity on the corresponding substrate harboring H3K4 methylation is reduced by three to fivefold — consistent with the physiological distribution of these mutually exclusive epigenetic marks [47]. In another example of DNMT regulation, human DNMT3A forms homomultimers and heteromultimers that are necessary for processive methylation of multiple cytosine bases along a DNA strand [48].
Figure 4.
Chemical mechanisms of nucleic acid methylation. (a) DNA methylation at C5 of cytosine. (b) Proposed mechanism of rRNA methylation by the radical SAM methyl synthase RlmN. (c) Proposed mechanism of the folate/FAD-dependent tRNA methylating enzyme TrmFO. The transferred methyl group or its component parts are colored red. R’ = (p-aminobenzoyl)-glutamate and R” = adenosine-5′-pyrophosphate-ribityl.
RNA methylating enzymes
Methylation of RNA is a widespread post-transcriptional modification that expands structural and functional diversity [11]. The RNA substrates contain nucleophilic sites, which methylating enzymes target for reaction with the methyl group of SAM. These sites include various nucleoside heteroatoms as well as an enolate of uracil produced via conjugate-addition by an enzyme-derived cysteine residue [49–51]. However, these reactions are not compatible with C2 and C8 methylation of adenosine [52••,53••,54••,55••]. Methylation of these electrophilic sites is achieved by the bacterial radical SAM enzymes RlmN and Cfr, which employ a unique methyl synthase mechanism. These enzymes assemble methyl groups on their substrate from a methylene fragment and a hydrogen atom at the site of methylation [53••]. Essential to catalysis is the utilization of SAM in two distinct ways: First, the methyl group of SAM is transferred to a cysteine residue within the enzyme that serves as a methylene precursor [54••,55••]. Secondly, another molecule of SAM is homolytically cleaved to produce a 5′-deoxyadenosyl radical, which abstracts a hydrogen atom from the methylated cysteine. The resultant thiomethylene radical subsequently adds into the substrate (Figure 4b) [52••,55••]. A proposed series of acid-base steps resolves the enzyme-substrate adduct to yield methyladenosine. Because 2-methyladenosine formed by RlmN resides in the peptidyltransferase center (PTC) of the ribosome, it is probable that this modification fine-tunes translation activity [56]. The Cfr paralog evolved an additional capacity to methylate C8, which decreases the effectiveness of many PTC-targeting antibiotics [57]. The folate-dependent and flavin-dependent RNA methylating enzyme TrmFO utilizes an atypical mechanism to install methyl groups [58,59]. Although the catalytic mechanism is yet to be fully elucidated, it is proposed that the enzyme-activated uridine substrate reacts with the electrophilic carbon of methylene-tetrahydrofolate. The resulting adduct is then reduced by flavin to yield 5-methyluridine (Figure 4c).
Conclusion
While significant advancements have been made to elucidate the underlying chemistry of methylating enzyme catalysis, there remains much to be revealed about their regulation. Many methylating enzymes participate in large protein complexes, suggesting the possibility of allosteric regulation. Moreover, the factors that govern substrate recognition are largely unknown. For example, both protein and nucleic acid methylating enzymes target chromatin, which harbors myriad chemical modifications that may affect enzyme–substrate interactions. Furthermore, pharmacological inhibition of methylation activity offers the potential to dissect methylation-independent functions of multi-protein methylating complexes, while maintaining the quaternary structure — often perturbed by genetic knockdown/knockout methods. As demonstrated by the body of work compiled in this review and elsewhere, chemical biological techniques have proven to be powerful tools for the investigation of mechanisms underlying protein and nucleic acid methylating enzymes.
Acknowledgements
We would like to thank Megan Riel-Mehan (UCSF) for producing the graphical abstract and consulting on artwork, and NIAID (R01AI095393), NSF (NSF graduate fellowship to DDL and CAREER award to DGF), the Sidney Kimmel Foundation for Cancer Research, the V Foundation and Searle Scholars Program for funding. We apologize to those authors whose important work in the field of enzymatic methylation was not included in this review due to space restrictions.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 3.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 4.Grosjean H. Fine-tuning of RNA functions by modification and editing.Topics in Current Genetics. Springer; 2005. pp. 1–22. [Google Scholar]
- 5.Oleksiuk O, Jakovljevic V, Vladimirov N, Carvalho R, Paster E, Ryu WS, Meir Y, Wingreen NS, Kollmann M, Sourjik V. Thermal robustness of signaling in bacterial chemotaxis. Cell. 2011;145:312–321. doi: 10.1016/j.cell.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liger D, Mora L, Lazar N, Figaro S, Henri J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgue-Hamard V, Graille M. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011;39:6249–6259. doi: 10.1093/nar/gkr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb KJ, Zurita-Lopez CI, Al-Hadid Q, Laganowsky A, Young BD, Lipson RS, Souda P, Faull KF, Whitelegge JP, Clarke SG. A novel 3-methylhistidine modification of yeast ribosomal protein Rpl3 is dependent upon the YIL110W methyltransferase. J Biol Chem. 2010;285:37598–37606. doi: 10.1074/jbc.M110.170787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M, Minor W, Hunt DF, Macara IG. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature. 2010;466:1125–1128. doi: 10.1038/nature09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Kulkarni K, Manolaridis I, Zhang Z, Dodd RB, Mas-Droux C, Barford D. Mechanism of isoprenylcysteine carboxyl methylation from the crystal structure of the integral membrane methyltransferase ICMT. Mol Cell. 2011;44:997–1004. doi: 10.1016/j.molcel.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Webb KJ, Lipson RS, Al-Hadid Q, Whitelegge JP, Clarke SG. Identification of protein N-terminal methyltransferases in yeast and humans. Biochemistry. 2010;49:5225–5235. doi: 10.1021/bi100428x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, Jerva LF, Scott MP, Copeland RA. Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011;78:199–210. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S, Gong W, Xu RM. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc Natl Acad Sci U S A. 2011;108:20538–20543. doi: 10.1073/pnas.1106946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat Struct Biol. 2000;7:1165–1171. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 18.Kolbel K, Ihling C, Bellmann-Sickert K, Neundorf I, Beck-Sickinger AG, Sinz A, Kuhn U, Wahle E. Type I arginine methyltransferases PRMT1 and PRMT-3 act distributively. J Biol Chem. 2009;284:8274–8282. doi: 10.1074/jbc.M809547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Xie N, Jin M, Stahley MR, Stivers JT, Zheng YG. A transient kinetic analysis of PRMT1 catalysis. Biochemistry. 2011;50:7033–7044. doi: 10.1021/bi200456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obianyo O, Thompson PR. Kinetic mechanism of protein arginine methyltransferase 6 (PRMT6) J Biol Chem. 2012;287:6062–6071. doi: 10.1074/jbc.M111.333609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakowski TM, Frankel A. Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem J. 2009;421:253–261. doi: 10.1042/BJ20090268. [DOI] [PubMed] [Google Scholar]
- 22 •.Pak ML, Lakowski TM, Thomas D, Vhuiyan MI, Husecken K, Frankel A. A protein arginine N-methyltransferase 1 (PRMT1) and 2 heteromeric interaction increases PRMT1 enzymatic activity. Biochemistry. 2011;50:8226–8240. doi: 10.1021/bi200644c. [DOI] [PubMed] [Google Scholar]
- 23.Pesiridis GS, Diamond E, Van Duyne GD. Role of pICLn in methylation of Sm proteins by PRMT5. J Biol Chem. 2009;284:21347–21359. doi: 10.1074/jbc.M109.015578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao B, Wilson JR, Gamblin SJ. SET domains and histone methylation. Curr Opin Struct Biol. 2003;13:699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 2005;19:1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith BC, Denu JM. Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 28.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2011;286:3359–3369. doi: 10.1074/jbc.M110.174524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu P, Zhang Y. Catalytic mechanism and product specificity of the histone lysine methyltransferase SET7/9: an ab initio QM/MM-FE study with multiple initial structures. J Am Chem Soc. 2006;128:1272–1278. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Bruice TC. Enzymatic mechanism and product specificity of SET-domain protein lysine methyltransferases. Proc Natl Acad Sci U S A. 2008;105:5728–5732. doi: 10.1073/pnas.0801788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Cheung T, Grande C, Ferguson AD, Zhu X, Theriault K, Code E, Birr C, Keen N, Chen H. Biochemical characterization of human SET and MYND domain-containing protein 2 methyltransferase. Biochemistry. 2011;50:6488–6497. doi: 10.1021/bi200725p. [DOI] [PubMed] [Google Scholar]
- 32.Chin HG, Patnaik D, Esteve PO, Jacobsen SE, Pradhan S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45:3272–3284. doi: 10.1021/bi051997r. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couture JF, Dirk LM, Brunzelle JS, Houtz RL, Trievel RC. Structural origins for the product specificity of SET domain protein methyltransferases. Proc Natl Acad Sci U S A. 2008;105:20659–20664. doi: 10.1073/pnas.0806712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39 ••.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 44.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 45.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 46.Gerasimaite R, Merkiene E, Klimasauskas S. Direct observation of cytosine flipping and covalent catalysis in a DNA methyltransferase. Nucleic Acids Res. 2011;39:3771–3780. doi: 10.1093/nar/gkq1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holz-Schietinger C, Matje DM, Harrison MF, Reich NO. Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. J Biol Chem. 2011;286:41479–41488. doi: 10.1074/jbc.M111.284687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou YM, Perona JJ. Stereochemical mechanisms of tRNA methyltransferases. FEBS Lett. 2010;584:278–286. doi: 10.1016/j.febslet.2009.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benitez-Paez A, Villarroya M, Armengod ME. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA. 2012 doi: 10.1261/rna.033266.112. [Ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52 ••.Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53 ••.Yan F, Fujimori DG. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc Natl Acad Sci U S A. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54 ••.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55 ••.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 56.Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS. Insights into the structure, function and evolution of the radical-SAM 23S rRNA methyltransferase Cfr that confers antibiotic resistance in bacteria. Nucleic Acids Res. 2010;38:1652–1663. doi: 10.1093/nar/gkp1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamdane D, Argentini M, Cornu D, Myllykallio H, Skouloubris S, Hui-Bon-Hoa G, Golinelli-Pimpaneau B. Insights into folate/FAD-dependent tRNA methyltransferase mechanism: role of two highly conserved cysteines in catalysis. J Biol Chem. 2011;286:36268–36280. doi: 10.1074/jbc.M111.256966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimasu H, Ishitani R, Yamashita K, Iwashita C, Hirata A, Hori H, Nureki O. Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase. Proc Natl Acad Sci USA. 2009;106:8180–8185. doi: 10.1073/pnas.0901330106. [DOI] [PMC free article] [PubMed] [Google Scholar]