Abstract
Byonic™ is the name of a software package for peptide and protein identification by tandem mass spectrometry. This software, which has only recently become commercially available, facilitates a much wider range of search possibilities than previous search software such as SEQUEST and Mascot. Byonic allows the user to define an essentially unlimited number of variable modification types. Byonic also allows the user to set a separate limit on the number of occurrences of each modification type, so that a search may consider only one or two chance modifications such as oxidations and deamidations per peptide, yet allow three or four biological modifications such as phosphorylations, which tend to cluster together. Hence Byonic can search for 10s or even 100s of modification types simultaneously without a prohibitively large combinatorial explosion. Byonic’s Wildcard Search™ allows the user to search for unanticipated or even unknown modifications alongside known modifications. Finally, Byonic’s Glycopeptide Search allows the user to identify glycopeptides without prior knowledge of glycan masses or glycosylation sites.
Keywords: Proteomics, mass spectrometry, post-translational modifications, glycosylation, glycopeptide, phosphopeptide
INTRODUCTION
Byonic™ is a software package for identifying peptides and proteins by tandem mass spectrometry. Byonic has existed as research software at the Palo Alto Research Center for about 6 years (Bern et al., 2007), and has been used in biological studies at several research centers (Zhu et al., 2009) (Charvatova et al., 2008) (Bern et al., 2009) (Bhatia et al., 2012). Byonic is now available as a commercial product from Protein Metrics Inc. (www.proteinmetrics.com) with a 30-day free trial. Byonic plays the same role as Mascot (Perkins et al., 1999), SEQUEST (Eng et al., 1994; UNIT 13.3), and X!Tandem (Craig and Beavis, 2004), but offers a wider range of capabilities. In particular, Byonic provides three major features not found in these other search engines: Modification Fine Control™, Wildcard Search™, and Glycopeptide Search.
Modification Fine Control enables the user to search for 10s or even 100s of modification types at a time without a combinatorial explosion. For example, a user might allow up to three phosphoserines, S[+80], per peptide, but allow at most one beta elimination, S[[−18], and at most one deamidated asparagine, N[+1]. To further reduce the search, the user might allow at most one of either S[−18] or N[+1], that is, disallowing peptides containing one of each. Modification fine control empowers the user to tailor the search to the sample, and thus avoid overly narrow searches that miss interesting peptides and overly broad searches that run for hours or days and produce “noisy” results with many false positives. Byonic provides a simple language for specifying the limits and scope of modifications. This unit provides sufficient examples for the reader to start writing customized modification rules for Byonic searches.
Wildcard Search enables the user to search for unanticipated modifications. A wildcard can modify any residue by any mass delta within a user-settable range, for example, −50 to +150 Daltons. Wildcard masses occur at roughly one Dalton spacing, just like molecular masses. Because wildcard searches are already very broad, there is a limit of one wildcard per peptide. Wildcard search can find known but unanticipated modifications, and also novel modifications not represented in the proteomics literature or in databases such as Unimod (www.unimod.org). For example, a modification with a mass shift of 34 Daltons was discovered by Byonic’s wildcard search and later identified as homocysteic acid (Bern et al., 2010).
Glycopeptide Search enables the user to search for glycosylated peptides without prior knowledge of either glycosylation sites or glycan masses. Fully automatic glycan search allows one glycosylation per peptide and uses internal tables of N-linked and O-linked glycans that cover the most likely possibilities. Alternatively, the user can perform glycopeptide searches by specifying glycan masses using Byonic’s usual modification fine control mechanism. This option enables the user to customize the list of glycans and/or allow more than one glycan per peptide.
We now explain the basic protocols: how to set up a Byonic search, and how to view and interpret Byonic results. A Support Protocol describing how to install Byonic is also included.
BASIC PROTOCOL 1 SETTING UP A BYONIC SEARCH
Like other proteomics search engines, Byonic requires two essential inputs: a set of tandem mass spectra (MS/MS spectra) and a database of protein sequences. Byonic currently requires centroided mass spectra in MGF format and a protein database in FASTA format (APPENDIX 1B), but the software will translate vendor formats in the future. Byonic also asks the user to set a number of parameters: the digestion specificity, the type of MS/MS fragmentation, m/z measurement tolerances, and modification types and prevalences.
The optimal settings of these parameters are not usually known in advance, because the digestion specificity may vary with endogenous proteases, the m/z accuracy may vary with the instrument calibration, and so forth. To address this problem, Protein Metrics offers a program called Preview ™ (Kil et al., 2011) as a companion tool for Byonic. Preview runs a large number of fast initial searches to measure mass errors, digestion specificity, and a wide range of protein modifications. Optionally, Preview recalibrates m/z measurements based on confident identifications and outputs an MGF file with improved accuracy. Byonic and other search engines generally give more accurate and sensitive identifications on the recalibrated MGF.
Necessary Resources
Hardware
Windows PC or workstation, preferably with 64-bit architecture and a minimum of 8 GB RAM. Editions for Mac and Unix will be released later.
Software
Byonic and Java 6 or 7 (See Support Protocol 1)
-
1
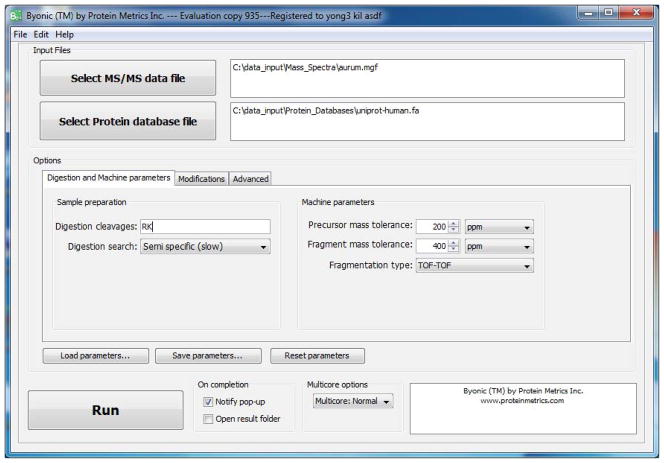
Launch Byonic by double-clicking on the green Byonic icon. A splash screen will be shown for a few seconds followed by the input GUI shown in Figure 1.
-
2
Input the MS/MS data file in MGF format. The input GUI supports both browsing and direct text entry.
-
3
Input the protein database file in FASTA format. The protein database should include decoy proteins, denoted by protein names beginning with >Reverse or >Decoy, so that Byonic can compute confidences and False Discovery Rate (FDR). The conventional way to configure directories is to keep MGF files in C:\data_input\Mass_Spectra and FASTA files in C:\data_input\Protein_Databases, and to create a folder called C:\data_results for holding Byonic’s output as subfolders.
-
4
Input information on the Digestion and Machine parameters tab. This tab asks for amino acid residues as one-letter abbreviations in the box labeled Digestion cleavages. Digestion is assumed to occur C-terminal to these residues; support for digestion enzymes that cut on the N-terminal side will be added soon. In Figure 1, the residues are RK for trypsin. The next box asks for the specificity of the search: fully specific, nonspecific digestion at the peptide N-terminus, nonspecific digestion at the C-terminus, nonspecific at either but not termini, and fully nonspecific. In Figure 1, the choice is Semispecific, meaning that either terminus, but not both, may be nonspecific. The top two boxes on the right ask for m/z tolerances for precursor and fragment ion measurements; these tolerances may be given in either ppm or Daltons. The third box on the right asks the user to choose the fragmentation type. Byonic currently supports four options: low-energy CID, beam-type CID (meaning QTOF or HCD), TOF-TOF, and ETD/ECD. Unsupported fragmentation types should be searched as the most similar supported type, for example, IRMPD should be searched with the low-energy CID setting.
Figure 1.
The input GUI with the Digestion and Machine Parameters tab open.
Modifications Tab
-
5
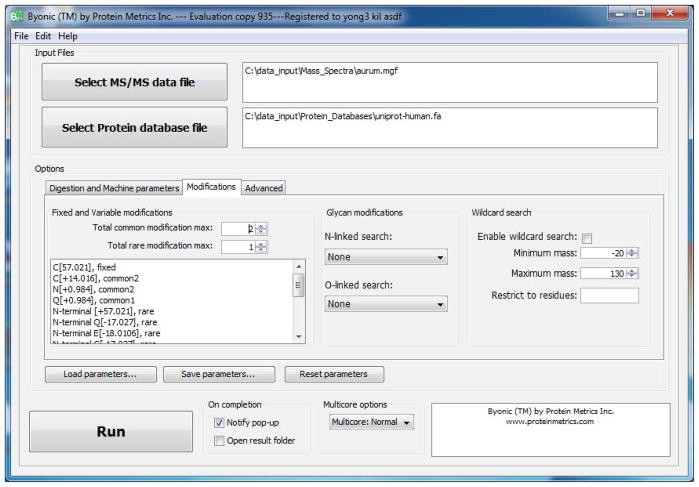
Input fixed and variable modifications. Byonic’s modification box, on the left in Figure 2, recognizes a number of keywords (fixed, common, rare, N-terminal, C-terminal, Nglycan, …) that control the placement and prevalence of modifications. In Figure 2, the user specified C[+57.021], fixed, meaning carbamidomethylated cysteine (camC). The user also specified C[+14.016], common2, directing the program to consider each cysteine residue with and without this modification, up to a limit of two such modifications per peptide. The keyword common1 would allow only one occurrence of this modification per peptide. Variable modifications are added on top of fixed modifications, so the total mass added to C will be 57.021+14.016 = 71.037, which represents cysteine propionamide. Modification masses should be specified to 3 or 4 decimal digits, even for low-resolution MS instruments, because certain modification masses cue Byonic to expect neutral losses. As a convenience, Byonic maps certain integers to exact masses, for example 57 is shorthand for 57.021464; consult the User’s Manual before using this feature.
A typical search allows a total of at most two common modifications and a total of at most one rare modification per peptide. To search for, say, three phosphoserines per peptide, the user can change Total common modification max to 3 or split phosphorylated serine between two rules: S[+79.966], common2 and S[+79.966], rare1. Depending upon the other modification rules, the latter approach will generally give a faster search. By specifying most modifications as rare it is quite feasible to search for 10 – 20 modification types at once with Byonic. Even larger searches are possible with focused protein databases, for example, mutation searches that allow 200+ possible amino acid residue substitutions or oxidative footprinting searches that allow 50+ types of oxidations.
In Figure 2, the two rules, N[+0.984], common2 and Q[+0.984], common1, represent deamidation; here the user chose to allow up to two deamidated asparagines (the more common deamidation) but only one deamidated glutamine per peptide. (Byonic allows the user to bundle amino acid residues into one modification rule, so that [NQ][+0.984], common2 is equivalent to the two rules N[+0.984], common2 and Q[+0.984], common2.) The next rule N-terminal [+57.021], rare1 specifies a common artifact (over-alkylation) that occurs on the peptide N-terminus. The rule N-terminal Q[−17.027], rare1 specifies a modification that occurs only on peptides with N-terminal glutamine. Conceptually, Byonic has one modification “slot” for each residue, along with slots for the peptide’s N- and C-termini. A variable modification such as S[+79.966] uses up the residue slot; a nonspecific terminal modification such as N-terminal [+57.021] uses up the terminal slot; but residue-specific N-terminal modifications, such as N-terminal Q[−17.027], use up both the residue and the N-terminal slots.
-
6
Input special-case modifications. The keyword NGlycan as in NGlycan[+2204.772], rare1 applies a mass shift only to asparagine in the NX{S/T} motif for N-linked glycosylation.
To enable a modification only on protein termini, rather than all peptide termini, add a vertical slash and the keyword ProteinTerminalMod, as in this modification rule: N-terminal [42.011], rare | ProteinTerminalMod. The keywords common and rare without numbers are equivalent to common1 and rare1. The + is unnecessary to define a positive mass addition.
To enable a modification only on certain proteins, for example hydroxyproline on collagens, write a rule like this: P[15.995], common3 | ProteinLabel[icase]{collagen}. The keyword [icase] specifies case-insensitive match, so that the string collagen will also match proteins that include Collagen, procollagen, or collagenase in their protein names. The keyword [case] specifies case-sensitive match. To match a single protein and avoid inadvertent matches like collagenase, use a unique identifier such as the accession number.
-
7
Input Glycopeptide search options. The box labeled Glycan modifications contains two pull-down menus for fully automatic glycopeptide searches. The menu labeled N-linked search currently includes two tables, common human glycans and mammalian glycans. The menu labeled O-linked search also includes two tables, simply labeled small and large. Fully automatic glycopeptide searches represent a convenient alternative to writing long lists of modification rules. These searches employ tables of known N- and O-linked glycan masses, and are equivalent to writing a long list of rules such as NGlycan[+2204.772], rare1 and [ST][+203.079], rare1. These searches allow only one glycan per peptide, which is adequate for N-linked glycosylation but quite restrictive in the case of O-linked glycosylation, because O-glycosylation sites tend to cluster together. Thus an intensive O-glycosylation search will employ both the automatic option as well as handcrafted modification rules. (The automatic glycosylation searches can be combined with each other, and with handcrafted modification rules.) In the example shown in Figure 2, the user did not choose glycosylation search, which is best used only for data from samples enriched for glycopeptides.
-
8
Input Wildcard search options. The rightmost box of the Modification tab allows the user to turn on wildcard search, set the range for the wildcard mass, and if desired restrict the wildcard to certain residues. If the precursor mass tolerance is low (less than 100 ppm) Byonic obtains the exact mass of the wildcard from the difference between the observed precursor mass and the calculated mass of the candidate peptide. If the precursor mass tolerance is high (at least 100 ppm), Byonic uses a mass defect (fractional part of the modification mass) characteristic of an organic molecule. In the example in Figure 2, the wildcard range is set to −20 to +130 Da, but the user did not check the box to use a wildcard.
The glycopeptide searches, especially the O-glycosylation search, and the wildcard search enlarge the search space by 2 – 3 orders of magnitude, even more if the wildcard range is very wide, so these options should be used with care, and in conjunction with only the most common variable modifications such as oxidized methionine.
Figure 2.
GUI view with the Modifications tab open.
Advanced Tab
-
9
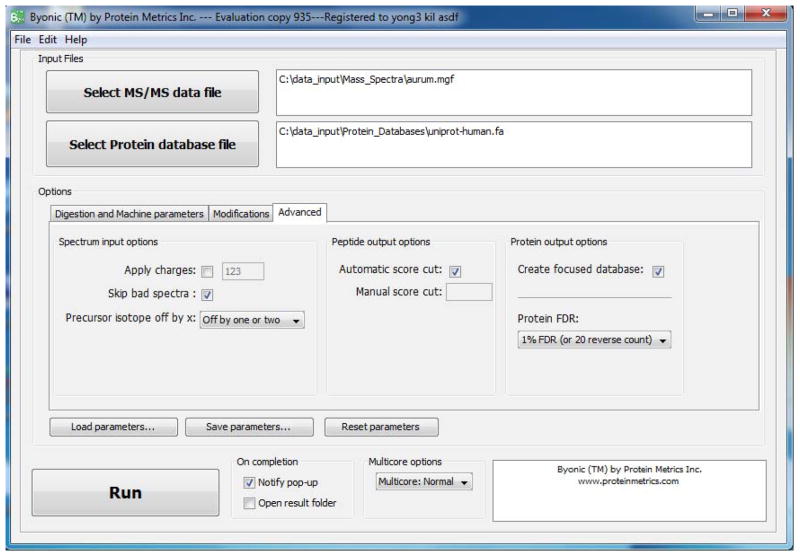
Optionally choose spectrum input options. The Advanced tab, shown in Figure 3, controls a variety of parameters that depend upon the MS instrument set-up and data preprocessing. For example, on many MS instruments, precursor ion charges are uncertain for some or all spectra; to handle this case, the Apply charges box allows the user to override the charge assignments in the spectrum data file and assign charges manually. If this box is not checked, Byonic will use the assigned charge for all spectra with assigned charges and use +1, +2, +3 for all CID spectra and +2, +3, +4 for all ETD spectra without assigned charges.
Similarly, on many instruments the nominal precursor mass or m/z may actually be the mass or m/z of a 13C isotope peak rather than of the base (all 12C monoisotopic) peak. The pulldown menu labeled Precursor isotope off by x provides three options: No error check, which uses only the assigned precursor; Off by one or two, the default, which allows the assigned precursor to be up to 2 Da too high; and Off by one or more, which allows the assigned precursor to be up to 1 Da too high for a precursor with mass in the range 1000 – 2000 Da, up to 2 Da too high for a precursor in the range 2000 – 3000 Da, and so forth.
-
10
Optionally choose filtering options for peptide-spectrum matches (PSMs). Byonic by default automatically filters both PSMs and proteins, but the user can filter PSMs by Byonic score by unchecking the checkbox labeled Automatic score cut and typing the minimum acceptable score into the box labeled Manual score cut. A reasonable score threshold is one that leaves only a few decoy PSMs in the output list; for most searches this threshold will be in the range 200 – 400, lower in the case of high-accuracy precursor masses, and higher in the case of very wide searches such as wildcard searches. For searches without a wildcard, false PSMs with Byonic scores over 400 are rare (less than 1 in 1000 spectra) and false PSMs with Byonic scores over 500 are very rare (less than 1 in 10,000).
-
11
Optionally create a focused database. Checking the box labeled Create focused database on the Advanced tab directs Byonic to output a new FASTA file (labeled focused and appearing in the output objs directory) containing only the proteins found in the search, along with suitable decoys (labeled >Reverse) for unbiased FDR estimation. The focused database can then be used for subsequent wide searches, including more modifications and/or a wildcard. Of course, the user can also create focused databases outside of Byonic by editing existing FASTA files.
-
12
Optionally choose filtering options for proteins using the pulldown menu labeled Protein FDR. By default, Byonic cuts its protein list at 1% protein False Discovery Rate or after 20 decoy proteins, whichever comes last. (Byonic uses whichever comes last, because FDR of 1% is an unreasonable choice for samples containing fewer than 100 proteins.) The user can ask for a slightly more aggressive cut, 2% False Discovery Rate or 50 decoy proteins, or ask for no protein cut—show all proteins (and hence all matched spectra). Even with the least aggressive option, Byonic cuts its protein list not at the last confident protein, but rather some ways into the “noise”. This is a reasonable design for a search engine, because users with detailed knowledge of their samples may recognize correct or possibly correct proteins amidst the noise of incorrect identifications.
Figure 3.
GUI view with the Advanced tab open.
Bottom Pane
-
13
Run the program. In the bottom pane of the input GUI, there is a button to run the program, a pulldown menu that controls the number of computer cores of the CPU that Byonic will use, and checkboxes that determine what happens upon completion. Byonic is parallelized and by default will use all but two cores of a multi-core computer. Simple searches (say a fully tryptic search of 10,000 spectra with only a few modifications enabled) will take only a few minutes, but highly complex searches may run overnight.
-
14
Optionally save all inputs. Due to Byonic’s many options and capabilities, writing modification rules and setting parameters can sometimes be a nontrivial task. For this reason, Byonic allows the user to save all inputs using the button labeled Save parameters and load a previously saved input, which can then be edited, using the button labeled Load parameters. Reset parameters blanks out the modification rules and restores defaults. Steps 13 and 14 can be performed in the opposite order: the user can save parameters before or after executing the search.
BASIC PROTOCOL 2 VIEWING THE SEARCH RESULTS IN EXCEL
Byonic writes its outputs in two different formats: as an Excel spreadsheet (extension .xlsx) for viewing, searching, sorting, and importing into other programs; and as a customized database (extension .byprot), which can then be viewed and explored interactively by Byonic’s output Viewer, a separate program from Byonic the search engine. In this protocol, we describe the Excel spreadsheet.
Necessary Resources
Hardware
Windows PC or workstation. Editions for Mac and Unix will be released later.
Software
Microsoft Excel
Steps
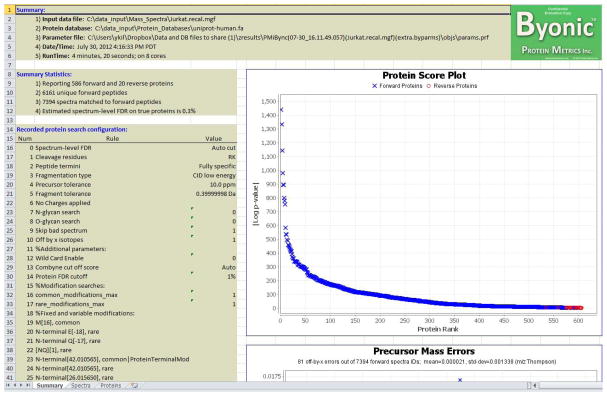
Open the Excel output file. Each time Byonic runs, it creates a folder with a name beginning PMiBync to hold its outputs. (The rest of the output folder name gives the date and time that Byonic ran, the name of the spectrum file, and the name of protein database.) The Excel file is at the topmost level within the output folder, and its first sheet should look like the one shown in Figure 4.
Examine the results in Sheets 1, 2, and 3 of the Excel output file. Sheet 1 is the summary, which gives vital statistics such as the numbers of proteins, peptides, and spectra. Sheet 1 also shows a plot of proteins ranked according to their probability scores with target and decoy proteins distinguished by color, blue and red respectively, and a plot of precursor m/z measurement errors. The two other Excel sheets give the Protein view and the Spectrum view.
Consult the section below entitled Guidelines for Understanding Results for help in interpreting the information shown in the spreadsheet.
Figure 4.
Byonic output (Summary worksheet) as viewed in Excel.
BASIC PROTOCOL 3 VIEWING THE SEARCH RESULTS IN THE INTERACTIVE VIEWER
Byonic’s output Viewer allows interactive exploration of Byonic’s search results. It is especially valuable for visual assessment of interesting identifications.
Necessary Resources
Hardware
Windows PC or workstation. Editions for Mac and Unix will be released later.
Software
Byonic Viewer.
Steps
Launch the interactive viewer by double-clicking on the blue Byonic viewer icon. A splash screen will be shown for a few seconds followed by a four-paned window as shown in Figure 5. The four-paned view will initially be blank.
Click on the File button in the upper left, and then on the Open Project button to select a Byonic output file (extension .byprot) to view. Alternatively one can directly open a Byonic output and launch the viewer by double-clicking on a file with extension .byprot.
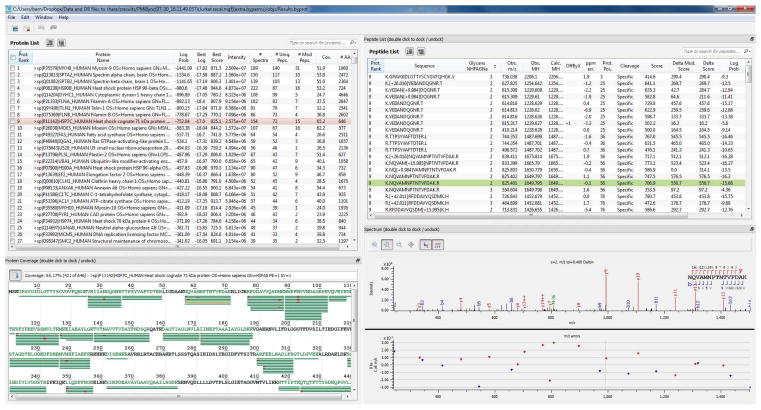
Notice that the four panes provide successively more detailed views of the Byonic search results. The upper left pane shows identified protein groups, listed from most confident on down. (A protein with the same numerical rank as the one above it is a “grouped” protein, generally a close homologue, that explains exactly the same spectra as the first protein in the group.) Clicking on a protein in the upper left pane populates the lower left and upper right panes with the coverage map and peptide-spectrum matches (PSMs) for the selected protein. Clicking on a green bar in the coverage map or on a PSM in the upper right pane then populates the lower right pane with the corresponding annotated mass spectrum. In Figure 5, the selected protein is HSP7C_HUMAN Heat shock cognate 71 kDa and the selected spectrum matches the 1650-Da peptide NQVAMNPTNTVFDAK.
The user can make a personalized layout by dragging the boundaries between the four panes where there are rows of dots. Users can dock (attach panes) and undock (detach panes) by double clicking; this can be especially useful for double-headed displays with two or more computer monitors. The user can also rearrange and sort columns as well as hide/show columns and adjust their widths for optimum viewing. The spectrum pane includes a vertical-line cursor, which allows the user to line up identified peaks with their associated m/z errors. When the cursor is positioned exactly over a spectrum peak, the reported m/z and intensity of that peak are shown inside curly braces. There are buttons that turn on/off the peak and cleavage diagram annotations. Finally, there are buttons for zooming in (magnifying glass with a +), zooming out (magnifying glass with a −), and panning (rosette of compass directions), along with a 1:1 reset button.
Consult the Guidelines for Understanding Results for help in interpreting the information shown in the interactive viewer.
Figure 5.
Byonic output as viewed in Byonic’s interactive Viewer.
SUPPORT PROTOCOL 1 INSTALLING BYONIC
Byonic is currently available from Protein Metrics Inc. (San Carlos, CA) as a standalone software application. Later it will become available as a component of larger software packages.
Necessary Resources
Hardware
Windows PC or workstation, preferably with 64-bit architecture and a minimum of 8 GB RAM. Editions for Mac and Unix will be released later.
Software
Byonic and Java 6 or 7.
Request a 30-day free trial of Byonic by sending e-mail to info@proteinmetrics.com. You will receive a pointer to a password-protected .zip file containing the download package; the e-mail will include the password to unzip the file.
The download includes an installer, user’s manual, FAQ document, and example data files, protein databases, and Byonic parameter files. Double-click on the installer and follow the standard installation instructions.
Byonic must be registered before it can be run. To register Byonic, go to the Help tab on the top toolbar of the Byonic GUI and then to Register. Follow the instructions to receive the activation code by e-mail.
GUIDELINES FOR UNDERSTANDING RESULTS
As seen in Figures 4 and 5, Byonic presents its results in two lists: proteins and peptide-spectrum matches (PSMs). In both Excel and the Byonic Viewer, these lists are searchable and sortable spreadsheets with a substantial number of columns, for example, 20 columns in the Excel spreadsheet for PSMs. Here we explain the less obvious data fields.
Protein List
Byonic outputs a protein list ranked by the base-10 logarithm of the protein p-value. The protein p-value is the likelihood of the PSMs to this protein (or protein group) arising by random chance, according to a simple probabilistic model. A log p-value of −3.0 corresponds to a protein p-value of 0.001, or one chance in a thousand, so that in a search against a database containing 10,000 independent proteins, we expect to see only about 10 log p-values better than −3.0 arising at random. Byonic’s log p-values are only as accurate as the probabilistic model, however, so the user should also check the ranking of the proteins relative to the decoy (>Reverse) proteins. A protein with log p-value at least 2.0 smaller than the log p-value of the top decoy protein is a confident identification. Proteins lower on the list lie in the “gray zone”: these are not confident identifications and the user must decide whether or not to believe them based on p-value, number of distinct peptides, single best score, ranking relative to other decoys, and outside knowledge.
Byonic’s protein list includes a number of columns that reflect the quality of the PSMs for each protein:
Log Prob – Log base 10 of the protein p-value
Best Log Prob – Best (most negative) log p-value of an individual PSM
Best Score – Best (largest) Byonic score of a PSM
Total Intensity – Sum of all peak intensities over all MS/MS spectra
# of spectra – Total number of PSMs, including duplicate PSMs
# of unique peptides – Total number of PSMs, discounting duplicates. (The same modification differently placed counts as a distinct PSM.)
Coverage % – Percent of the protein sequence covered by PSMs
Byonic joins proteins P1, P2, … into a protein group if exactly the same spectra match all the proteins in the group. An “ambiguous” PSM, meaning one matching a peptide that is found in two or more proteins, is always assigned to the higher-ranking of two proteins it matches. Thus, if P1 has separate evidence but P2 does not, P2 will not be shown, and if P1 has a lot of separate evidence but P2 has only a little, then P1 will be ranked according to all its evidence, but P2 will be ranked according to only its separate evidence. For this reason, as well as many other reasons, none of Byonic’s outputs (# of spectra, total intensity, etc.) is an accurate measure of protein abundance.
PSM List
As initially presented, Byonic’s PSM list is organized by protein, and left to right (that is N- to C-terminus) by starting position within proteins. The user can also sort the list by other columns. For example, the user might sort by Byonic score to compare the scores of especially interesting PSMs such as phosphopeptides to the scores of decoy peptides. The PSM list includes the following columns, as well as some others that require no explanation:
Off-by-x Error – [MObserved – MComputed], where MObserved is the observed M+H (singly charged) precursor mass and MComputed is the computed M+H precursor mass, and [ ] means closest integer.
Mass Error (ppm) – 106 × (MObserved – MComputed) / (MComputed). The ppm mass error is computed after correcting for off-by-x errors.
Starting Position – Position within the protein of the N-terminal residue of the peptide.
Cleavage – Digestion specificity, where Specific means fully specific, Nragged means nonspecific at the N-terminus, Cragged (or Semi) means nonspecific at the C-terminus, and Non means nonspecific at both termini.
Score – Byonic score, the primary indicator of PSM correctness. Byonic scores reflect the absolute quality of the peptide-spectrum match, not the relative quality compared to other candidate peptides. Byonic scores range from 0 to about 1000, with 300 a good score, 400 a very good score, and PSMs with scores over 500 almost sure to be correct.
Delta – The drop in Byonic score from the top-scoring peptide to the next distinct peptide. In this computation, the same peptide with different modifications is not considered distinct.
DeltaMod – The drop in Byonic score from the top-scoring peptide to the next peptide different in any way, including placement of modifications. DeltaMod gives an indication of whether modifications are confidently localized; DeltaMod over 10.0 means that there is high likelihood that all modification placements are correct. A low DeltaMod (often zero) indicates a PSM with uncertain modification placement, and manual inspection of the annotated spectrum may in some cases resolve the ambiguity.
Log Probability – The log p-value of the PSM. This is the log of the probability that the PSM with such a score and delta would arise by chance in a search of this size (size of the protein database, as expanded by the modification rules). A log p-value of −3.0 should happen by chance on only one of a thousand spectra. Caveat: it is very hard to compute a p-value that works for all searches and all spectra, so read Byonic p-values with a certain amount of skepticism.
# of unique peptides – Total number of PSMs for the protein that “owns” this PSM, discounting exact duplicates. The same peptide with the same modification differently placed counts as a distinct PSM.
COMMENTARY
Background Information
The central problem of mass spectrometric analysis of protein samples, no matter whether the sample contains a complex proteome or a single therapeutic protein, is the identification of molecular ions. Over the past 20 years, researchers have proposed and implemented a number of data analysis strategies, including de novo sequencing, database search, sequence tagging, spectral library searching, within-sample spectrum-spectrum comparison (“spectral networks analysis”), and hybrids and combinations of these approaches. Each of these approaches has its strengths. For example, de novo sequencing can obtain sequence or partial sequence on high-quality spectra of peptides from organisms with unsequenced genomes. Spectral library searching and within-sample spectrum-spectrum comparison can often identify low-quality spectra of peptides that have been previously observed. Sequence tagging, a hybrid of de novo sequencing and database search, offers a speed-up over pure database search by limiting attention to candidate peptides matching a short subsequence.
Despite this proliferation of approaches, the workhorse of peptide identification has remained conventional database search, as implemented in programs such as SEQUEST, Mascot, X!Tandem, because database search offers the most uniform level of sensitivity, without favoring peptides that fragment completely, peptides that have been previously observed, or peptides that occur in more than one modification state. Database search compares observed spectra to theoretically predicted spectra, rather than to previously observed spectra. The success of this strategy depends upon an empirical phenomenon: for all the major fragmentation methods, peptides tend to break rather predictably at the peptide bonds, thereby yielding sequence information rather than, say, a random collection of side-chain fragments. By contrast, MS identification of small molecules, which generally fragment unpredictably, employs the spectral library approach.
Current database search programs, however, have not kept pace with the rapid innovation in MS instruments. For example, Mascot’s algorithm considers 10 fragment peaks matched out of 30 predicted to be equally likely, regardless of whether the fragment mass tolerance is 1.0 Da or 0.01 Da, an accuracy that is routinely achievable on many modern MS/MS instruments. SEQUEST and Mascot, in fact, all of the major search engines except Byonic, have practical or actual limits on the number of modification types that can be considered at one time. Finally, no search engine except Byonic supports glycopeptide analysis, which has only recently become much more feasible due to a combination of high mass accuracy and new fragmentation methods such as ETD.
Critical Parameters
Byonic compares each mass spectrum in the data set to each candidate peptide in the protein database that fits the search criteria. Byonic, like other database search programs, makes hard-edged decisions on precursor mass tolerances, digestion specificity, and modifications considered—the program will not identify a peptide with precursor error 11 ppm if the tolerance is set to 10 ppm—so it behooves the user to make wise choices of parameter settings.
Set mass tolerances appropriate for the type of instrument, for example, 10 ppm precursor tolerance for a high resolution instrument and 0.4 Dalton fragment tolerance for ion trap fragmentation. Preview’s mass error plots can help the user choose these tolerances. Preview’s m/z recalibration can remove systematic errors so that data can be run with tighter tolerances, for example, 5 ppm instead of 10 ppm tolerance for high resolution precursors. Tight tolerances offer significant advantage for difficult searches, for example, resolving nearly isobaric modifications such as sulfation and phosphorylation, or identifying glycopeptides with poor fragmentation. Tolerances can be set in either Da or ppm, as appropriate for the instrument.
Set digestion specificity based on the prevalence of nonspecific digestion and the complexity of the search. If the modification complexity of the search is high, as in wildcard, glycosylation, or oxidative footprinting searches, it is best to avoid the extra complexity of searching for nonspecific digestion, unless the nonspecific digestion rate is high (say, over 20% of all peptides). For tryptic digests, allowing a non-specific N-terminus increases the search size about 10-fold; allowing non-specific digestion at either, but not both, termini gives a 20-fold increase; and allowing fully non-specific digestion blows up the search 100-fold.
Set modifications based upon prevalence reported by Preview and the goal of the study. If the goal of the study is phosphorylation site identification, enable up to 3 or even 4 phosphorylation sites per peptide, and avoid other modifications unless they are prevalent. If the goal of the study is simply protein identification, perhaps as an initial search to produce a focused database, it is best to enable only the most common modifications (for example, oxidized methionine and deamidated asparagine). Be especially alert to over-alkylation; in some samples, over-alkylation is so common that the majority of peptides carry iodoacetamide artifacts. Some modifications are costly (for example, sodiation on any residue as opposed to just glutamic and aspartic acid), but others (such as pyro-glu on N-terminal glutamine) barely increase the size of the search space.
Users may wish to “bracket” their searches, as a photographer brackets light exposure, by running the same data with narrow, medium, and wide searches, spanning a 10- to 100-fold range of search sizes.
Troubleshooting
Like any complicated software, Byonic makes certain assumptions that must be met in order to obtain optimal performance. Byonic assumes centroided MS/MS spectra; profile-mode spectra will give few if any identifications. Input slip-ups, for example, setting the fragment tolerance to 20 Dalton instead of 20 ppm, will cause Byonic to produce essentially random results. Currently Byonic does very little error checking of user inputs. In the future, Byonic will pop up warning messages: Do you really mean 20 Daltons!?
Complicated searches (many spectra and many modifications) run on computers with modest amounts of random access memory (RAM) may cause Byonic to run out of memory space and crash. This problem can usually be resolved by reducing the maximum number of MS/MS spectra handled by a single thread. To do this, edit the saved parameter file (params.byparms in the obj subfolder of the output folder, or saved under a user-defined name with the Save parameters button) by reducing the value of the parameter max_chunk_size, for example reducing 5000 to 2000. Then use the Load parameters button to resubmit the job to Byonic.
Byonic performance may degrade if the input MS/MS spectra have been de-charged and/or de-isotoped. Byonic handles isotope peak series internally. De-charging and/or de-isotoping the spectra beforehand using, for example, Mascot Distiller, destroys valuable information. De-isotoping is an especially bad idea for ETD spectra, which often have peaks (c-1 and z+1 peaks) that lead de-isotoping algorithms astray.
Acknowledgments
The research that led to Byonic was funded in part by NIH grant R21 GM085718.
LITERATURE CITED
- Bern M, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectreometry. Anal Chem. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- Bern M, Finney G, Hoopmann MR, Merrihew G, Toth MJ, MacCoss MJ. Deconvolution of mixture spectra from ion-trap data-independent-acquisition tandem mass spectrometry. Anal Chem. 2009;82:833–841. doi: 10.1021/ac901801b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M, Saladino J, Sharp JS. Conversion of methionine into homocysteic acid in heavily oxidized proteomics samples. Rapid Commun Mass Spectrom. 2010;24:768–772. doi: 10.1002/rcm.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Kil YJ, Ueberheide B, Chait BT, Tayo L, Cruz L, Lu B, Yates JR, Bern M. Constrained de novo sequencing of cone snail toxins. J Proteome Res. 2012 doi: 10.1021/pr300312h. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvatova O, Foley BL, Bern MW, Sharp JS, Orlando R, Woods RJ. Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1. J Am Soc Mass Spectrom. 2008;19:1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- Eng J, McCormack AL, Yates JR. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Kil YJ, Becker C, Sandoval W, Goldberg D, Bern M. Preview: a program for surveying shotgun proteomics tandem mass spectrometry data. Anal Chem. 2011;83:5259–5267. doi: 10.1021/ac200609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Guo T, Park JE, Li X, Meng W, Datta A, Bern M, Lim SK, Sze SK. Elucidating in vivo structural dynamics in integral membrane protein by hydroxyl radical footprinting. Mol Cell Proteomics. 2009;8:1999–2010. doi: 10.1074/mcp.M900081-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]