Summary
This past decade has seen the identification of numerous conserved genes that extend lifespan in diverse species, yet the number of compounds that extend lifespan is relatively small. A class of compounds called STACs, which were identified as activators of Sir2/SIRT1 NAD+-dependent deacetylases, extend the lifespans of multiple species in a Sir2-dependent manner and can delay the onset of age-related diseases such as cancer, diabetes and neurodegeneration in model organisms. Plant-derived STACs such as fisetin and resveratrol have several liabilities, including poor stability and relatively low potency as SIRT1 activators. To develop improved STACs, stilbene derivatives with modifications at the 4′ position of the B ring were synthesized using a Horner-Emmons-based synthetic route or by hydrolyzing deoxyrhapontin. Here, we describe synthetic STACs with lower toxicity toward human cells, and higher potency with respect to SIRT1 activation and lifespan extension in Saccharomyces cerevisiae. These studies show that it is possible to improve upon naturally occurring STACs based on a number of criteria including lifespan extension.
Keywords: lifespan, longevity, resveratrol, SIRT1, sirtuins, STACs
Introduction
Dietary restriction (DR) delays aging and extends the maximum lifespan of a wide range of organisms (Koubova & Guarente, 2003). In mammals, DR delays numerous age-associated diseases including cancer, neurodegeneration, atherosclerosis and type II diabetes (Weindruch, 1996; Lane et al., 2001; Roth et al., 2001). The search for small molecule mimetics that can deliver the health benefits of DR is an active area of research, but no safe and effective DR mimetic has been found (Ingram et al., 2004). An approach we have taken to finding DR mimetics has been to screen libraries for compounds that modulate the activity of longevity regulatory proteins, and to test these for their ability to extend lifespan in relatively simple organisms such as baker's yeast and the nematode Caenorhabditis elegans before moving on to mammals (Wood et al., 2004).
There are a number of reasons for taking this approach. First, DR works on almost all organisms that have been tested. Therefore, we expect bona fide DR mimetics to extend the lifespans of species as different as yeast, worms, flies and mice. Second, simple organisms are far more amenable to genetic manipulation and their lifespans are relatively short – less than 10 days in the case of actively dividing yeast – therefore, the pathways through which lifespan-extending compounds are working can be identified more readily. Third, the pathways that control the DR response are far better defined in these organisms than in mammals. Finally, small molecules can have effects that are more complex than simple activation or inhibition, such as altering substrate specificity, and understanding how they extend lifespan may reveal additional longevity pathways (Viswanathan et al., 2005).
In Saccharomyces cerevisiae, C. elegans, and Drosophila melanogaster, increased copy number or expression of the SIR2 gene extends lifespan 30–50% (Kaeberlein et al., 1999; Tissenbaum & Guarente, 2001; Rogina & Helfand, 2004). With regards to S. cerevisiae, this refers to replicative lifespan, which is the number of daughter cells produced by a single mother before senescence, rather than the more recently defined chronological lifespan, which measures survival time in the absence of nutrients (Bitterman et al., 2003). In wild-type S. cerevisiae and Drosophila, lifespan extension by DR requires SIR2, implying that the gene is intimately involved in mediating its effects. In the absence of the FOB1 gene, which suppresses the rDNA hyper-recombination defect of a sir2 mutant, glucose restriction can extend the lifespan of some sir2 strains (Kaeberlein et al., 2004; Lin & Guarente, 2006) revealing that there is redundancy in the DR pathway, which we have shown is mediated, at least in part, by the Sir2 homolog Hst2 (Lamming et al., 2005). Although it is not known whether the closest mammalian Sir2 homolog, SIRT1, extends lifespan in mammals, this gene is known to modulate several distinct physiologies known to be important for lifespan including cell survival, energy metabolism, and resistance to stresses (Luo et al., 2001; Vaziri et al., 2001; Araki et al., 2004; Brunet et al., 2004; Cohen et al., 2004; Yeung et al., 2004; Moynihan et al., 2005; Bordone et al., 2006).
We recently reported the design, synthesis and characterization of SIRT1 inhibitors that are derivatives of splitomycin (Bedalov et al., 2001; Mai et al., 2005). Although SIRT1 inhibitors may be useful reagents in laboratory settings and possibly in the treatment of diseases such as cancer (Lim, 2006), we have placed most of our effort on identifying and optimizing molecules that activate Sir2/SIRT1, based on the hypothesis that they should extend lifespan and possibly mimic the health benefits of DR. To this end we performed in vitro screens for small molecules that could modulate human SIRT1 and identified 18 SIRT1-activating compounds or STACs, which were of three structural classes: chalcones, flavones and stilbenes (Howitz et al., 2003). The most potent of these was 3,5,4′-hydroxystilbene (resveratrol). Resveratrol stimulated SIRT1 activity ∼tenfold by lowering the Km for both NAD+ and the peptide substrate. Resveratrol also provided the largest yeast lifespan extension, ∼60%, and this was entirely dependent on the SIR2 gene (Howitz et al., 2003), as was the lifespan extension in C. elegans and D. melanogaster (Wood et al., 2004), although we do not discount the possibility that the beneficial effects of resveratrol in mammals are mediated through a potentially large number of cellular factors in addition to SIRT1 (Baur & Sinclair, 2006).
Keijer and colleagues recently identified three additional STACs from plants, but none of them was more potent than resveratrol (de Boer et al., 2006). Interestingly, resveratrol has been shown to provide protection from numerous diseases in rodent models including cancer and diabetes, but the pathways through which it exerts these effects are not known (Bhat & Pezzuto, 2002; Aggarwal et al., 2004; Su et al., 2006).
Activation of SIRT1 by STACs is detectable in a variety of in vitro assays including a fluorescent peptide-based assay (Howitz et al., 2003), a high performance liquid chromatography (HPLC)-based assay (de Boer et al., 2006) and a radiolabelled peptide assay (A. Suave & U. Cornell, personal communication). Activation of SIRT1 is not observed in some in vitro assays that use 4–20 amino acid peptide substrates, leading one group to conclude that activation is an in vitro artifact (Kaeberlein et al., 2005). Although the reason for this discrepancy has not yet been fully elucidated, a likely explanation is that not all peptide substrates fully mimic the endogenous situation, where the targets of deacetylation are full-length proteins (Borra et al., 2005; de Boer et al., 2006).
With regards the in vivo evidence for activation, STACs such as resveratrol and fisetin extend lifespan in yeast, C. elegans and Drosophila, and this is proportional to their level of activation in vitro (Howitz et al., 2003; Bauer et al., 2004; Jarolim et al., 2004; Wood et al., 2004; Viswanathan et al., 2005). Importantly, STACs do not extend lifespan in the absence of SIR2. In C. elegans, resveratrol and fisetin can also mimic the protective effects of additional copies of Sir2, but they only work if the Sir2 gene is present (Parker et al., 2005). Activation of mammalian SIRT1 by STACs has also been demonstrated in a variety of ways using a number of experimental systems, including showing increased deacetylation of a target peptide in HEK293 and HT29 cells (Howitz et al., 2003; de Boer et al., 2006), increased deacetylation of the SIRT1 target RelA/p65 (Yeung et al., 2004), increased deacetylation and nuclear localization of the SIRT1 targets FOXO1 (Daitoku et al., 2004; Yang et al., 2005) and FOXO4 (Brunet et al., 2004), the SIRT1-dependent mobilization of fat stores from cultured adipocytes (Picard et al., 2004), suppression of axonal degeneration (Araki et al., 2004), deacetylation of RelA/p65 and the protection of human neurons against Aβ peptide (Chen et al., 2005), sirtuin-dependent protection of striatal mouse neurons overexpressing mutant huntingtin (Wood et al., 2004; Yeung et al., 2004; Parker et al., 2005), reduced neurotoxicity from mutant superoxide dismutase (an amyotrophic lateral sclerosis model) and mutant p35 (an Alzheimer's mouse model; L. H. Tsai & D. A. Sinclair, unpublished), SIRT1-dependent protection of cardiomyocytes from poly(ADP-ribose)polymerase (PARP) -induced apoptosis (Pillai et al., 2005), and SIRT1-dependent neuroprotection following ischemia (Raval et al., 2006).
Unfortunately, resveratrol has limitations as a laboratory tool and a proof-of-concept molecule, which include low bioavailability in mammals, low solubility, and sensitivity to light and to oxidation (Bhat et al., 2001; Halliwell, 2003; Aggarwal et al., 2004; Walle et al., 2004; de Boer et al., 2006). These limitations led us to design molecules with greater potency, stability, and a greater ability to extend the lifespan of model organisms. Here we describe the synthesis and characterization of a series of stilbene derivatives and show that it is possible to find STACs that are superior to resveratrol in terms of their stability in solution, toxicity, ability to activate SIRT1, and the extent to which they extend the lifespan of yeast cells.
Results
Previous studies examining structure-activity relationship of SIRT1 activation by stilbenes from plant sources showed that hydroxyls on the 3 and 5 position of the A ring and the trans conformation of the rings are crucial structural elements, and that the 4′ position is amenable to modification (Howitz et al., 2003). We therefore focused our efforts on synthesizing resveratrol analogs with replacements at the 4′ position of the stilbene backbone using the synthesis schemes outlined in Fig. 1. Modifications included thiomethyl, methyl, ethyl, methoxy, and acetoxy groups (Fig. 2). Between 5 and 10 mg of each compound was obtained. Because of the limiting amounts of each compound that were available, SIRT1 was chosen as a representative sirtuin to test activation, based on its large dynamic range and the previously demonstrated correlation between activation of SIRT1 and its orthologs of other species (Howitz et al., 2003).
Fig. 1.
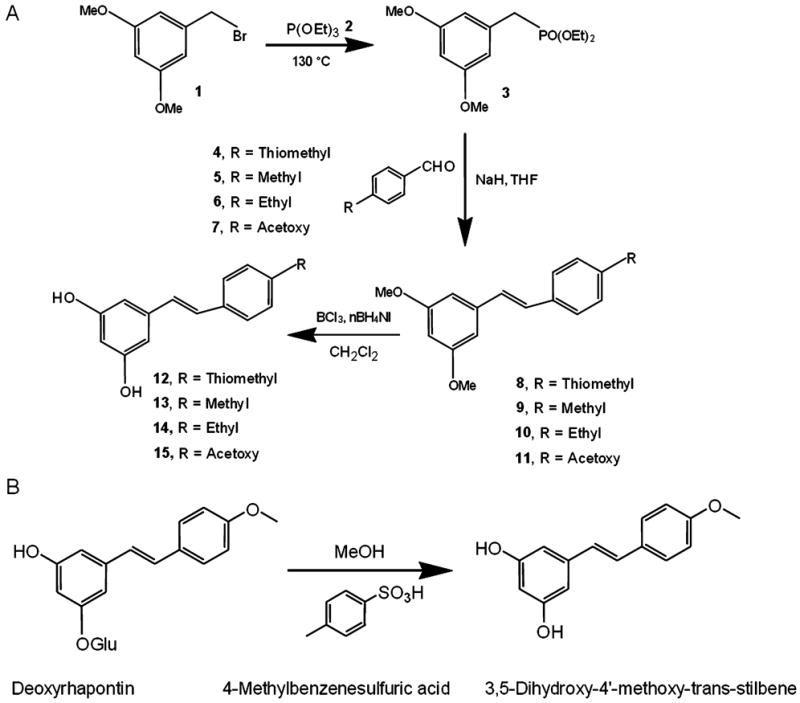
Synthesis steps for resveratrol derivatives. (A) Resveratrol Derivatives 1, 2, 3, and 5 were synthesized by a modified Horner-Emmons-based route using a diethyl phosphonate to generate stilbene (Andrus et al., 2003). 1-(Bromomethyl)-3,5-dimethoxybenzene 1 was treated with neat triethyl phosphate 2 using an Arbuzov reaction to produce diethyl (3,5-dimethoxyphenyl)methylphosphonate 3 in high yield. Coupling with 4-(methylthio)benzaldehyde 4; 4-methylbenzaldehyde 5; 4-ethylbenzaldehyde 6; 4-formylphenyl acetate 7 using sodium hydride as base in THF gave the protected stilbene 8, 9, 10, 11 in 65% yield. Boron trichloride was then used to give resveratrol derivatives 12, 13, 14, 15. (B) Derivative 4 (3,5-dihydroxy-4′-methoxy-trans-stilbene) was synthesized by hydrolysis of deoxyrhapontin (3-hydroxy-5[(E)-2-(4-methoxyphenyl)ethenyl]phenyl hexopyranoside) treated with 4-methyl benzensulfuric acid in methanol.
Fig. 2.
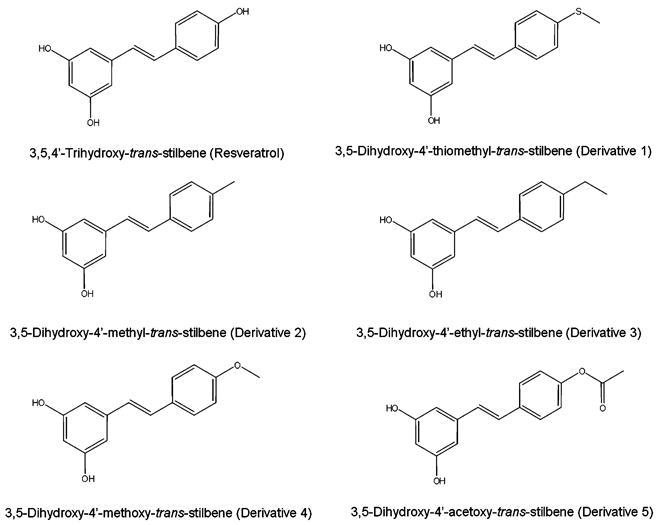
Chemical structures of resveratrol and synthetic resveratrol derivatives used in this study.
In agreement with previous reports (Howitz et al., 2003; de Boer et al., 2006), resveratrol (50 μm) stimulated the activity of SIRT1 ∼12-fold (Fig. 3). Derivative 1 (3,5-dihydroxy-4′-thiomethyl-trans-stilbene), in which the 4′-hydroxyl was replaced with a thiomethyl group, was the most effective, stimulating SIRT1 activity 18-fold, while Derivative 4 (3,5-dihydroxy-4′-methoxy-trans-stilbene), bearing a chemically similar methoxy group activated SIRT1 11-fold (Fig. 3). Derivative 2 (3,5-dihydroxy-4′-methyl-trans-stilbene) and Derivative 3 (3,5-dihydroxy-4′-ethyl-trans-stilbene), substituted with hydrophobic methyl and ethyl groups at the 4′ positions, activated SIRT1 16- and 14-fold, respectively. Derivative 5 (3,5-dihydroxy-4′-acetoxy-trans-stilbene), bearing an ester group with a relatively large volume at the 4′ position was the least potent, stimulating SIRT1 only threefold. Based on this structure–activity relationship, it appears that small, hydrophobic substituents at the 4′ position can enhance the potency of stilbenes.
Fig. 3.
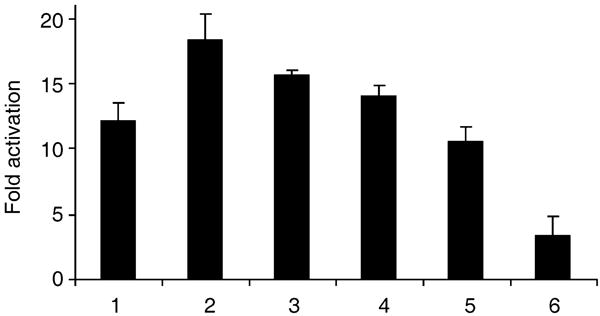
Fold activation of SIRT1 by resveratrol and synthesized resveratrol derivatives. (1) Resveratrol; (2) 4′-thiomethyl (Derivative 1); (3) 4′-methyl (Derivative 2); (4) 4′-ethyl (Derivative 3); (5) 4′-methoxy (Derivative 4); (6) 4′-acetoxy (Derivative 5). Fold activations are calculated as the ratio of the deacetylation rate in samples containing the tested compounds to that of the samples with no activating compound added. Data from three independent experiments are shown ± standard deviation.
Solubility of resveratrol derivatives
The low solubility of resveratrol in aqueous buffers (approximately 400 μm in distilled water or PBS) is a significant limitation to delivering high doses in animal studies. Using the limited quantities of derivatives that were available we were able to determine their relative solubility in aqueous solution. The solubility of Derivative 2 was threefold greater than resveratrol (> 1.2 mm). Thus, modification of the 4′ position is a viable strategy for improving the solubility of resveratrol. In contrast, the solubility of Derivative 3 was not appreciably different, and those of Derivatives 1, 4, and 5 were lower than resveratrol (100–200 μm) (data not shown).
Stability of resveratrol derivatives
Although resveratrol is stable when lyophilized and stored under nitrogen, it has a half-life of only ∼4–5 days when dissolved in dimethyl sulfoxide (DMSO) or ethanol and stored at room temperature and exposed to ambient light. This presumably limits how effective it is in extending lifespan in yeast experiments that are conducted over a period of 2 weeks. As shown in Fig. 4, the half-life of Derivative 5 is increased, suggesting that its effects may last longer despite its low initial potency in vitro. This experiment demonstrates that modification of the 4′ position of resveratrol is a feasible synthetic strategy to improve stability.
Fig. 4.
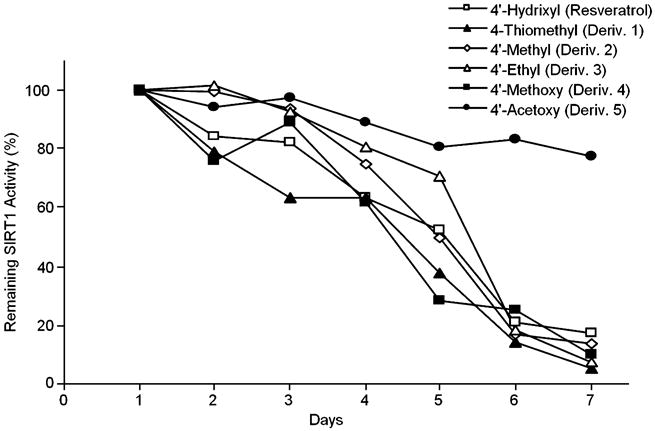
Stability of resveratrol and synthesized derivatives. Resveratrol or derivatives were dissolved in ethanol at a concentration of 2.5 mm and stored on at room temperature with ambient lighting. Aliquots were taken daily and assessed for the ability to activate SIRT1. The parent compound decayed with a half-life of ∼4.5–5 days and all derivatives were similar with the exception of Derivative 5 (3,5-hydroxy-4′-acetoxy-stilbene) which displayed significantly improved stability (half-life ∼10 days).
Concentration-dependent toxicity of resveratrol derivatives to HEK 293 cells
Resveratrol has been reported to have a biphasic effect on cell survival. Lower concentrations are beneficial, whereas higher concentrations become toxic (Aggarwal et al., 2004). A significant concern of ours was that improved SIRT1 activation might be counterbalanced by increased cytotoxicity, rendering them unsuitable for treatment of living cells or multicellular organisms. In order to test the cytotoxicity of our derivatives, we measured the survival of HEK 293 cells over a period of 96 h. At 25 μm, resveratrol and all five of the synthesized derivatives had little to no effect on the growth of HEK 293 cells. At higher concentrations, Derivatives 3, 4 and 5 showed similar toxicity to resveratrol, but the other derivatives, 1 and 2, were less toxic (Fig. 5 and data not shown). After 48 h, at a concentration of 50 μm, less than 10% of the cells were alive in the dishes containing resveratrol or Derivative 3, whereas 38% of the cells remained alive when treated with Derivative 2. Interestingly, 61% of the cells remained alive when treated with Derivative 1, the most potent activator, demonstrating that the replacement of the 4′-hydroxyl with a thiomethyl group reduces toxicity to cells while enhancing the ability of this compound to activate SIRT1. Thus, cytotoxicity can be uncoupled from the ability to stimulate SIRT1. Large-scale synthesis of this compound is in progress with a view to testing its pharmacokinetic properties and against cancer and age-related diseases in mouse models.
Fig. 5.
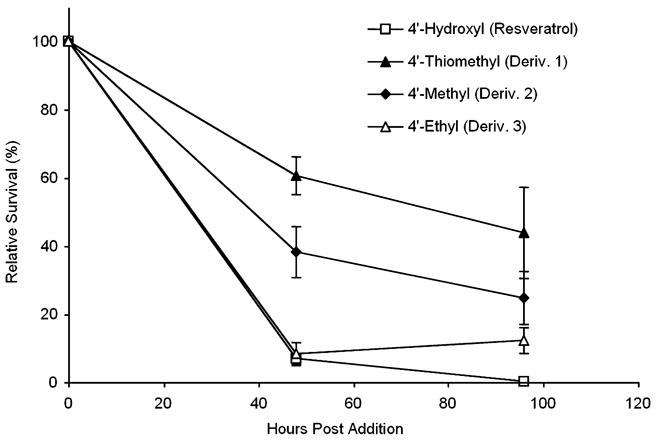
Toxicity of resveratrol derivatives to mammalian cells. HEK293 cells were seeded on six-well plates and allowed to attach overnight. The following day, resveratrol, Derivative 1 (3,5-hydroxy-4′-thiomethyl-stilbene), Derivative 2 (3,5-hydroxy-4′-methyl-stilbene), or Derivative 3 (3,5-hydroxy-4′-ethyl-stilbene) was added at a concentration of 50 μm. Cells in replicate wells were periodically washed to remove any debris or detached cells, trypsinized, and counted using a Beckman-Coulter Z2 particle counter. The proportion surviving is plotted as a percentage of cells counted in the corresponding ethanol (vehicle)-treated wells.
Resveratrol derivatives extend yeast replicative lifespan
The ability of STACs to increase SIRT1 activity in vitro has been shown to correlate with yeast lifespan extension: resveratrol being the most potent activator and generating the longest lifespan extension, and correspondingly lower activation potential and lifespan extension for compounds such as butein (3,4,2′,4′-tetrahydroxychalcone) and fisetin (3,7,3′,4′-tetrahydroxyflavone) (Howitz et al., 2003). We therefore sought to test whether our derivatives could extend yeast lifespan. For this experiment, we chose Derivatives 1, 3, and 5, which activated SIRT1 to varying extents. As shown in Fig. 6 (A,B), all of the three resveratrol derivatives tested extended yeast mean and maximum lifespan significantly (P value for Derivative 1 vs. no treatment < 0.05; P values for Derivative 3 and 5 vs. no treatment < 0.001). To our surprise, however, Derivative 5 extended lifespan the furthest (68%), despite its low in vitro potency. This may be attributable to the improved stability of Derivative 5 in solution (Fig. 4) or in vivo once it has been taken up by the yeast cells.
Fig. 6.
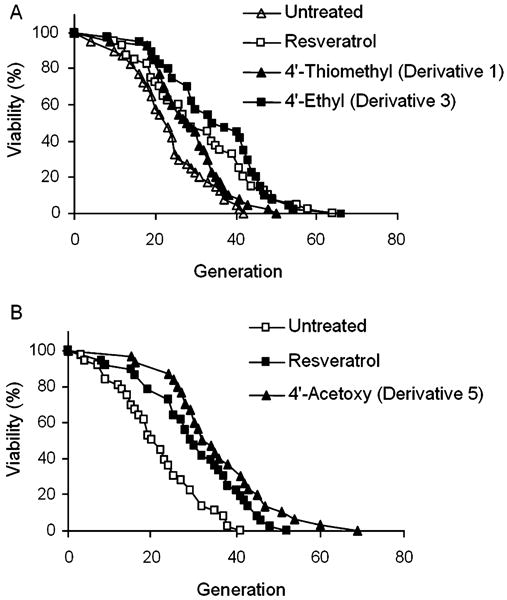
Lifespan analyses of Saccharomyces cerevisiae (PSY316ATα) when grown in the presence of resveratrol derivatives. Replicative lifespan was performed on at least 30 cells by micromanipulating mother cells and counting how many daughters were produced. Resveratrol and the derivatives were dissolved in yeast glucose medium at a concentration of 10 μm. Mean lifespans for treated and untreated cells were (A), 31.2 (Resveratrol); 28.6 (Derivative 1); 35.3 (Derivative 3); 23.3 (Untreated) and (B), 30.3 (Resveratrol); 36.0 (Derivative 5); 21.4 (Untreated).
Discussion
Our previous work with STACs demonstrated that the 3- and 5-hydroxl groups of the A ring as well as the trans conformation are crucial structural elements for SIRT1 activation (Howitz et al., 2003). In contrast, modification of the 4′ position on the B ring, which is the residue that accounts for the majority of free radical scavenging ability (Stojanovic et al., 2001), did not interfere with SIRT1 activation in five synthetic derivatives of resveratrol, although this activity was significantly diminished in Derivative 5, which bore a relatively bulkier acetoxy functional group at the 4′ position.
Although there is no co-crystal structure for human SIRT1 in complex with a STAC, crystal structures of yeast Hst2, human SIRT2 and two bacterial Sir2 homologs have been solved. These Sir2 homologs possess a similar catalytic core composed of ∼270 amino acids (Finnin et al., 2001; Min et al., 2001; Zhao et al., 2003a,b, 2004a,b). In Hst2, the highly conserved β1-α2 loop in the N-terminus undergoes significant structural rearrangements to facilitate the ordered NAD+ reactions of nicotinamide cleavage and ADP-ribose transfer to acetate (Marmorstein, 2004) and has been suggested as a candidate site to which sirtuin activators such as resveratrol might bind (Zhao et al., 2004b). We speculate that resveratrol and its analogs might bind to the N-terminus of SIRT1 to induce a conformational change that lowers the Km for the substrate. This model is supported by our studies showing that the mutations SIRT1-E230K and Sir2-D223K within the N-terminal sequences close to the catalytic cores of SIRT1 and Sir2, prevent activation by resveratrol without affecting basal activity (our unpublished data). The electronegativity of the substituted functionality at the 4′ position in the derivatives may play a role in determining binding affinity since substitution of sulfur in Derivative 1 with a more electronegative oxygen atom in Derivative 4 decreased the in vitro sirtuin activating activity from 18-fold to 11-fold. In order to elucidate the exact binding site of STACs to sirtuins, co-crystallization of resveratrol with sirtuins is in progress.
The stability of a molecule is an important consideration when conducting lifespan studies in which the molecule is exposed to water, light and oxygen. According to Stojanovic and co-workers, the 4′-hydroxyl group in resveratrol plays a central role in its antioxidant activity against biologically generated free radicals (Stojanovic et al., 2001). This implies that resveratrol could lose its activity due to redox reactions at this position. Substitution with different functionalities at the 4′ position in resveratrol might lower the redox potentials of these derivatives, thereby increasing the half-life in solution. This hypothesis was supported by the enhanced stability of Derivative 5, substituted with a 4′-acetoxy group.
The activity of a STAC in vivo will depend on many factors such as its rate of uptake and metabolism. For this reason, in vitro activity may not always correlate with the ability to activate sirtuins in cells. For example, in vivo esterases are known to readily cleave off acetyl groups from acetyl-containing molecules to release acetate (White & Hope, 1984). For this reason drugs are often acetylated in a prodrug form as a means of enhancing its properties, such as stability, without diminishing its biological activity (Takahashi et al., 1992). Derivative 5, which is acetylated at the 4′ position, was the least efficacious of the STACs in activating SIRT1 in vitro, although the lifespan extension it produced was the largest achieved by treatment with a STAC to date, possibly because it remains more stable in the yeast medium than other STACs and is cleaved by esterases once it enters the cell, releasing free resveratrol and acetate and providing a steady stream of the active compound, but clearly more experiments will be required to confirm this.
Although resveratrol has some liabilities as a pharmacological agent, one of its best attributes is its lack of toxicity. Cell toxicity is induced in mammalian cells upon prolonged exposure to doses typically in excess of 30 μm, whereas serum resveratrol concentrations peak well below 10 μm in rodents following a high dose of up to 50 mg kg−1 and decay with a half-life of ∼8–14 min (Asensi et al., 2002; Marier et al., 2002). No adverse effects have been detected in rats using doses as high as 300 mg kg−1. However, even minor changes to a non-toxic molecule can alter its toxicity so if any of these molecules are to be employed in higher animals, toxicity is an important consideration. At high concentrations, Derivative 1 was considerably less toxic than the other derivatives and even the parent compound. The observation that substitution of a less electronegative sulfur atom for the oxygen in the 4′-hydroxyl of resveratrol decreased toxicity while simultaneously increasing in vitro catalytic efficiency may be an important consideration in the design of STACs that achieve high in vivo concentrations.
In summary, we have explored the effects of altering the structure of resveratrol on stability, toxicity, and potency of SIRT1 activation. We show that it is possible to improve upon numerous characteristics of this molecule and extend yeast lifespan further than can be achieved using natural molecules. We consider Derivative 1 (with its high potency and lack of toxicity) and Derivative 5 with its improved stability and potential as a prodrug form of resveratrol as the two most promising candidates to explore in studies of lifespan extension in higher organisms.
Experimental procedures
Compound synthesis
Resveratrol Derivatives 1–3 and 5 were synthesized by a modified Horner-Emmons-based route using a diethyl phosphonate to generate stilbene (Andrus et al., 2003). Briefly, 1 -(bromomethyl)-3,5-dimethoxybenzene 1 purchased from Sigma-Aldrich (St. Louis, MO, USA) was treated with neat triethyl phosphate 2 by Arbuzov reaction to produce diethyl (3,5-dimethoxyphenyl) methylphosphonate 3 in high yield. Coupling with 4-(methylthio) benzaldehyde 4; 4-methylbenzaldehyde 5; 4-ethylbenzaldehyde 6; 4-formylphenyl acetate 7 (from Sigma-Aldrich) using sodium hydride as base in tetrahydrofuran (THF) gave the protected stilbene 8, 9, 10, 11 in 65% yield. Boron trichloride was then used to give resveratrol derivatives 12, 13, 14, 15. Derivative 4 (3,5-dihydroxy-4′-methoxy-trans-stilbene) was synthesized by hydrolysis of deoxyrhapontin (3-hydroxy-5[(E)-2-(4-methoxyphenyl)ethenyl]phenyl hexopyranoside) treated with 4-methyl benzensulfuric acid in methanol. The numbers given in bold above are the identification numbers of structures in Figure 1.
SIRT1 assays
SIRT1 assays were performed as previously described using a fluorescently labeled peptide substrate (Howitz et al., 2003). To assess the stability of derivatives each compound was dissolved in ethanol at 2.5 mm and stored at room temperature exposed to ambient light. An aliquot (1 μL) was added to a SIRT1 assay to give a final concentration of 50 μm and the reaction was incubated for 60 min at 37 °C before addition of the developer reagent. Assays were incubated for an additional 20 min at room temperature to allow the development of fluorescence, and then read for 0.1 s using a Victor 3 fluorometer (PerkinElmer, Downers Grove, IL, USA) with excitation and emission wavelengths of 360 and 450 nm, repsectively.
Solubility assays
Resveratrol and derivatives were dissolved at 200 mm in DMSO, then serially diluted twofold in distilled water. Precipitation was judged by eye, then all samples were heated to 50 °C for 5 min and allowed to cool and precipitate a second time in order to confirm that the crystallization was not due to the dilution process. The final concentration of DMSO was less than 1% in all solutions. Identical results were obtained using PBS instead of distilled water.
Toxicity assays
HEK293 cells were routinely cultured in Dulbecco's modified eagle medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum. Stock solutions of resveratrol and derivatives were made freshly in ethanol (at 1000×) at the start of each experiment. Cells grown in six-well dishes were treated with resveratrol or the indicated derivative in regular growth medium for the indicated periods of time. Images were captured using an RT Monochrome Spot camera attached to a Nikon Eclipse TE2000-U microscope with Spot (version 3.5.9) software. Cell counts were obtained by trypsinization and quantification using a Beckman-Coulter Z2 particle counter.
Yeast lifespan analyses
Lifespan measurements were performed using PSY316ATα as previously described (Bitterman et al., 2002). All compounds for lifespan analyses were dissolved in 95% ethanol. Plates were used within 24 h of preparation. Cells were pre-incubated on their respective medium for at least 15 h and equilibrated on the plates for a minimum of 4 h before micromanipulation. At least 30 cells were examined in each experiment. The statistical significance of lifespan differences were determined using JUMP 5.1 software by the Wilcoxon rank sum test. Differences are stated to be significant when the confidence was higher than 95%.
Acknowledgments
We wish to thank BIOMOL for advice and assistance with compound synthesis. The Sinclair laboratory is supported by RO1 grants from National Institutes of Health and National Institute on Aging, and the Glenn Laboratories for the Molecular Biology of Aging. H. Yang was supported by a Harvard/Hartford Advanced Research Award. J. A. Baur was supported by American Heart Association, Award number 0425834T. David A. Sinclair is a cofounder and advisor to Sirtris Pharmaceuticals, Cambridge, MA.
References
- Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:2783–2840. [PubMed] [Google Scholar]
- Andrus M, Liu J, Meredith E, Nartey E. Synthesis of resveratrol using a direct decarbonylative Heck approach from resorcylic acid. Tetrahedron Lett. 2003;44:4819–4822. [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Asensi M, Medina I, Ortega A, Carretero J, Band MC, Obrador E, Estrela JM. Inhibition of cancer growth by resveratrol is related to its low bioavailability. Free Radic Biol Med. 2002;33:387–398. doi: 10.1016/s0891-5849(02)00911-5. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Goupil S, Garber GB, Helfand SL. An accelerated assay for the identification of lifespan-extending interventions in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:12980–12985. doi: 10.1073/pnas.0403493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci USA. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KPL, Kosmeder JW, 2nd, Pezzuto JM. Biological effects of resveratrol. Antioxid Redox Signal. 2001;3:1041–1064. doi: 10.1089/152308601317203567. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Pezzuto JM. Cancer chemopreventive activity of resveratrol. Ann N Y Acad Sci. 2002;957:210–229. doi: 10.1111/j.1749-6632.2002.tb02918.x. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer VC, de Goffau MC, Arts IC, Hollman PC, Keijer J. SIRT1 stimulation by polyphenols is affected by their stability and metabolism. Mech Ageing Dev. 2006;127:618–627. doi: 10.1016/j.mad.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L. SIRT1 protects against microglia-dependent beta amyloid toxicity through inhibiting NF-kappa B signaling. J Biol Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Oxidative stress in cell culture: an under-appreciated problem? FEBS Lett. 2003;540:3–6. doi: 10.1016/s0014-5793(03)00235-7. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Ingram DK, Anson RM, de Cabo R, Mamczarz J, Min Z, Mattison J, Lane MA, Roth GS. Development of calorie restriction mimetics as a prolongevity strategy. Ann N Y Acad Sci. 2004;1019:412–423. doi: 10.1196/annals.1297.074. [DOI] [PubMed] [Google Scholar]
- Jarolim S, Millen J, Heeren G, Laun P, Goldfarb DS, Breitenbach M. A novel assay for replicative lifespan in Saccharomyces cerevisiae. FEMS Yeast Res. 2004;5:169–177. doi: 10.1016/j.femsyr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17:313–321. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lane MA, Black A, Handy A, Tilmont EM, Ingram DK, Roth GS. Caloric restriction in primates. Ann N Y Acad Sci. 2001;928:287–295. doi: 10.1111/j.1749-6632.2001.tb05658.x. [DOI] [PubMed] [Google Scholar]
- Lim CS. SIRT1: Tumor promoter or tumor suppressor? Med Hypotheses. 2006;67:341–344. doi: 10.1016/j.mehy.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Guarente L. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2006;2:e33. doi: 10.1371/journal.pgen.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Mai A, Massa S, Lavu S, Pezzi R, Simeoni S, Ragno R, Mariotti FR, Chiani F, Camillom G, Sinclair DA. Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. J Med Chem. 2005;48:7789–7795. doi: 10.1021/jm050100l. [DOI] [PubMed] [Google Scholar]
- Marier JF, Vachon P, Gritsas A, Zhang J, Moreau JP, Ducharme MP. Metabolism and disposition of resveratrol in rats: extent of absorption, glucuronidation, and enterohepatic recirculation evidenced by a linked-rat model. J Pharmacol Exp Ther. 2002;302:369–373. doi: 10.1124/jpet.102.033340. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem Soc Trans. 2004;32:904–909. doi: 10.1042/BST0320904. [DOI] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Neri C. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Raval AP, Dave KR, Perez-Pinzon MA. Resveratrol mimics ischemic preconditioning in the brain. J Cereb Blood Flow Metab. 2006;26:1141–1147. doi: 10.1038/sj.jcbfm.9600262. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth GS, Ingram DK, Lane MA. Caloric restriction in primates and relevance to humans. Ann N Y Acad Sci. 2001;928:305–315. doi: 10.1111/j.1749-6632.2001.tb05660.x. [DOI] [PubMed] [Google Scholar]
- Stojanovic S, Sprinz H, Brede O. Efficiency and mechanism of the antioxidant action of trans-resveratrol and its analogues in the radical liposome oxidation. Arch Biochem Biophys. 2001;391:79–89. doi: 10.1006/abbi.2001.2388. [DOI] [PubMed] [Google Scholar]
- Su HC, Hung LM, Chen JK. Resveratrol, a red wine antioxidant, possesses an insulin-like effect in streptozotocin-induced diabetic rats. Am J Physiol Endocrinol Metab. 2006 doi: 10.1152/ajpendo.00487.2005. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tamagawa S, Haginaka J, Yasuda H, Katagi T, Mizuno N. Stereoselective hydrolysis of O-acetyl propranolol as prodrug in rat tissue homogenates. J Pharm Sci. 1992;81:226–227. doi: 10.1002/jps.2600810307. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Walle T, Hsieh F, DeLegge MH, Oatis JE, Jr, Walle UK. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab Dispos. 2004;32:1377–1382. doi: 10.1124/dmd.104.000885. [DOI] [PubMed] [Google Scholar]
- Weindruch R. The retardation of aging by caloric restriction: studies in rodents and primates. Toxicol Pathol. 1996;24:742–745. doi: 10.1177/019262339602400618. [DOI] [PubMed] [Google Scholar]
- White KN, Hope DB. Characterization of aspirin hydrolase of guinea-pig liver cytoplasm. Biochim Biophys Acta. 1984;785:132–137. doi: 10.1016/0167-4838(84)90137-7. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol. 2003a;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure. 2003b;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J Mol Biol. 2004a;337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]
- Zhao K, Harshaw R, Chai X, Marmorstein R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc Natl Acad Sci USA. 2004b;101:8563–8568. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]


