Abstract
Increased transforming growth factor beta (TGF-β) signaling has been implicated in the pathogenesis of syndromic presentations of aortic aneurysm, including Marfan syndrome (MFS) and Loeys-Dietz syndrome (LDS)1-4. However, the location and character of many of the causal mutations in LDS would intuitively infer diminished TGF-β signaling5. Taken together, these data have engendered controversy regarding the specific role of TGF-β in disease pathogenesis. Shprintzen-Goldberg syndrome (SGS) has considerable phenotypic overlap with MFS and LDS, including aortic aneurysm6-8. We identified causative variation in 10 patients with SGS in the proto-oncogene SKI, a known repressor of TGF-β activity9,10. Cultured patient dermal fibroblasts showed enhanced activation of TGF-β signaling cascades and increased expression of TGF-β responsive genes. Morpholino-induced silencing of SKI paralogs in zebrafish recapitulated abnormalities seen in SGS patients. These data support the conclusion that increased TGF-β signaling is the mechanism underlying SGS and contributes to multiple syndromic presentations of aortic aneurysm.
Keywords: Aortic aneurysm, Shprintzen-Goldberg syndrome, Marfan syndrome, Loeys-Dietz syndrome, TGF-β signaling, SKI
The TGF-β family of cytokines influences a diverse repertoire of cellular processes, including cell proliferation, differentiation, survival and synthetic activity. One of three TGF-β ligand isoforms (TGF-β1, -β2 or -β3) initiates signaling by binding to the TGF-β receptor complex, composed of type 1 and type 2 receptor subunits (TβRI and TβRII, respectively) 11. The complex then transmits the signal through the canonical (SMAD dependent) pathway or noncanonical mitogen activated protein kinase (MAPK) cascades, prominently including extracellular signal-regulated kinase (ERK).
The ability of the nuclear SMAD proteins to precisely regulate gene transcription is further modulated by the recruitment of transcriptional co-activators such as p300 and CREB binding protein (CBP), and co-repressors such as the Sloan Kettering Institute proto-oncoprotein, SKI12. The SKI family of proteins, which also includes the SKI-like protein SKIL, negatively regulate SMAD-dependent TGF-β signaling by impeding SMAD2/3 activation, preventing nuclear translocation of activated R-SMAD/Co-SMAD complexes and inhibiting TGF-β target gene output by competing with p300/CBP for SMAD binding and recruiting transcriptional repressor proteins, such as mSin3A and HDAC1 (Supplementary Figure 1)13-15. A role for SKI in the regulation of MAPK cacades has not been described16,17.
Dysregulated TGF-β signaling has been implicated in the pathogenesis of syndromic presentations of aneurysm. Excessive canonical and noncanonical TGF-β signaling is seen in the aortic wall and other diseased tissues in mouse models of MFS, a systemic connective tissue disorder caused by mutations in the gene (FBN1) encoding the extracellular matrix protein fibrillin-11,3,18-20. Furthermore, pharmacologic antagonism of TGF-β and/or ERK signaling in mouse models of MFS has been shown to ameliorate multiple disease manifestations, including aortic aneurysm, suggesting that high TGF-β signaling drives disease progression3,4.
An excess of canonical TGF-β signaling has also been demonstrated in the aortic wall of patients with LDS2. However, this has been difficult to reconcile with the character of the underlying mutations. LDS is predominantly caused by heterozygous missense substitutions in the kinase domains of either TβRI or TβRII (encoded by the TGFBR1 and TGFBR2 genes, respectively)2,5. When mutant proteins are expressed in cells naïve for the corresponding receptor subunit, there is failure to propagate SMAD-dependent TGF-β signaling21. More rarely, LDS-like phenotypes can be caused by haploinsufficiency for SMAD3 or the TGFβ2 ligand, positive effectors of TGF-β signaling22,23. Notably, heterozygous loss-of-function mutations in SMAD3 or TGFB2 also associate with high TGF-β signaling in the aortic wall of patients and mouse models22,23. Taken together, these apparently contradictory data have engendered considerable controversy regarding the precise role of TGF-β in the pathogenesis of aortic aneurysm.
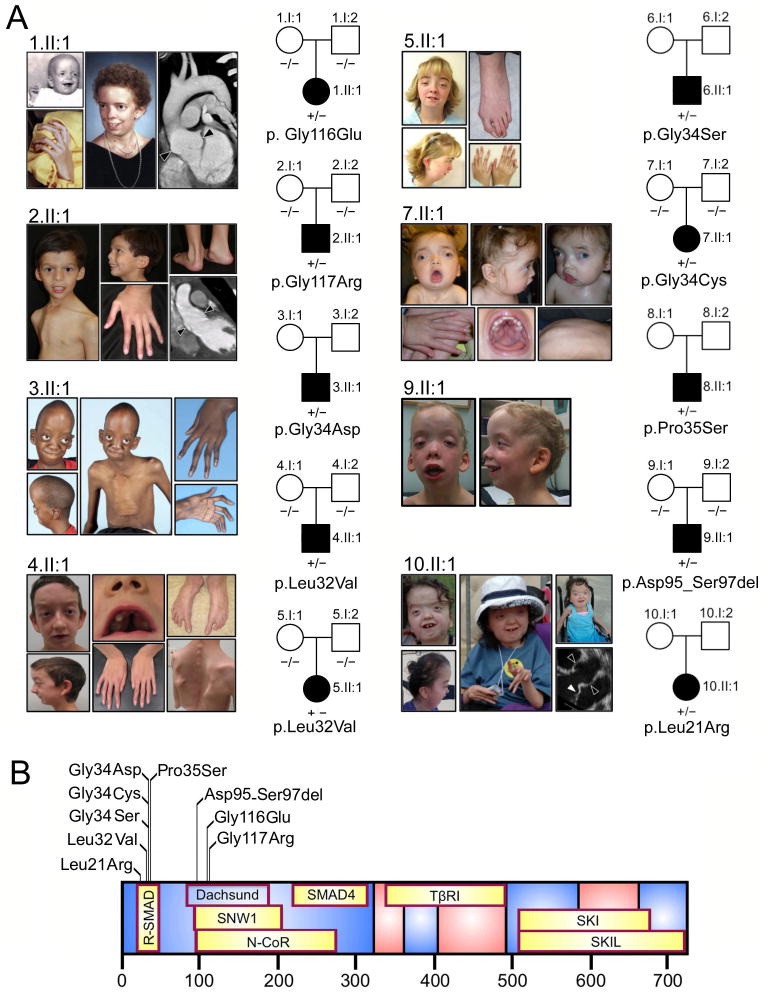
Shprintzen-Goldberg syndrome is a systemic connective tissue disorder of unknown etiology that includes virtually all the craniofacial, skeletal, skin and cardiovascular manifestations of MFS and LDS, with the additional findings of mental retardation and severe skeletal muscle hypotonia6,8. We hypothesized that aberrant TGF-β activity also underlies SGS and that identification of the genetic basis would inform our understanding of other syndromic presentations of aneurysm2. We performed whole exome sequencing for a single affected SGS child-unaffected parent trio. An average of 6.9 gigabases of sequence was generated per individual as paired-end 75 base pair reads, out of which >98.9% mapped to the human reference genome (UCSC hg19). This revealed only one variant, a heterozygous missense change in exon 1 of the SKI gene (NM_003036; c.347G>A, p.Gly116Glu) that was not present in SNP databases, was predicted to be damaging (PolyPhen-2 score: 0.999, SIFT score: 0.05), was not present in either parent, and was a strong functional candidate based upon a described relationship to TGF-β signaling (Supplementary Figure 2)24,25. We subsequently sequenced SKI in 11 other sporadic SGS patients (Table 1 and Figure 1A) and identified heterozygous variants in 9 including 8 missense mutations and one 9 base pair deletion (Supplementary Figure 3). These mutations were absent from dbSNP 134, the 1000 genomes project and over 10,000 exomes reported on the NHLBI Exome Variant Server, and confirmed to be de novo when testing of the unaffected parents was possible (5/9). Collectively, 10 mutations in 10 patients with SGS were identified in SKI by a combination of whole exome and Sanger sequencing, including a recurrent mutation in 2 unrelated probands. Each mutation substituted or deleted residues that show complete evolutionary conservation among members of the SKI gene family (Supplementary Figure 4)26. No mutations were identified upon sequencing of SKIL (NM_005414) in the 2 remaining patients. The mutations in SKI clustered in two distinct regions at the N-terminus of the protein (Figure 1B). The first region is located in the SMAD2/3 binding domain of SKI (residues 17-45), while the second region localizes to a portion of the Dachshund-homology domain (DHD) of the SKI protein that mediates binding to SNW1 and N-CoR, proteins that are essential to the transforming activity of SKI and to the recruitment of transcriptional co-repressors such as histone deacetylases, respectively15,27,28. One of the mutations (p.Leu21Arg) substitutes an amino acid that was previously shown to be essential for SKI-SMAD3 interaction29. Two of the mutations (p.Gly116Glu and p.Gly117Arg) substitute immediately adjacent glycine residues at positions that contribute to an exposed β hairpin loop in the DHD30. In silico analysis suggests that both glycine substitutions maintain the β-turn; while there is variation in where the algorithm defines the margins of the adjacent β sheets, the overall structure of the DHD is fully overlapping for the reference and mutant sequences (Supplementary Figure 5). It is therefore possible that the principle consequence of these mutations is disruption of molecular interactions at the protein surface31. In this light, it is interesting to note that residues 125-131, which lie immediately distal to the β-turn, have been shown to influence binding of SMAD2 and SMAD3 to their more proximal N-terminal binding site30,32.
Table 1. Clinical Manifestations in Shprintzen-Goldberg Syndrome (SGS) and Related Disorders.
| SGS Ref.7 | SGS Ref.8 | Pt.1 43y F | Pt.2 6y M | Pt.3 16y M | Pt.4 12y M | Pt.5 22y F | Pt.6 21y M | Pt.7 2y F | Pt.8 6y M | Pt.9 5y M | Pt.10 4y F | SGS (SKI neg.) | Classic MFS | Classic LDS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Features typical of SGS | |||||||||||||||
| Craniofacial | |||||||||||||||
| Craniosynostosis | 7/7 | 4/7 | + | + | + | + | + | + | + | + | + | + | 1/2 | − | + |
| Dolichocephaly | 16/17 | 11/14 | + | + | + | + | + | + | − | + | + | + | + | + | |
| Hypertelorism | 15/16 | 8/13 | + | + | + | + | + | + | + | + | + | + | 0/2 | − | + |
| Down-slanting eyes | 14/16 | 11/14 | + | + | + | + | + | + | + | + | + | + | 2/2 | + | + |
| Proptosis | 13/17 | 11/12 | + | − | + | + | + | − | + | + | + | + | 1/2 | − | − |
| Malar hypoplasia | 15/15 | 7/8 | + | + | + | + | + | + | + | + | + | + | 2/2 | + | + |
| High/narrow palate | 17/17 | 12/12 | + | + | + | + | + | + | + | + | + | + | 2/2 | + | + |
| Micrognathia | 16/17 | 13/14 | + | + | + | + | + | + | + | + | + | + | 2/2 | + | + |
| Low set ears | 13/13 | 12/14 | + | + | + | + | + | + | + | + | + | + | 2/2 | − | − |
| Skeletal | |||||||||||||||
| Arachnodactyly | 15/17 | 14/14 | + | + | + | + | + | + | + | + | + | + | 2/2 | + | + |
| Camptodactyly | 10/17 | 6/14 | + | + | + | + | + | + | − | − | + | + | + | + | |
| Scoliosis | 7/14 | 11/13 | + | + | + | + | + | + | + | − | − | + | 2/2 | + | + |
| Pectus deformity | 15/17 | 13/13 | + | + | + | + | + | + | + | − | + | + | 1/2 | + | + |
| Joint hypermobility | 9/14 | 10/10 | + | + | + | + | + | + | + | + | + | + | 2/2 | + | + |
| Joint contracture | 8/17 | 2/10 | + | + | + | + | + | + | − | + | + | + | 1/2 | − | − |
| C1/2 spine malform. | 4/16 | − | − | + | + | + | + | − | + | ||||||
| Neuromuscular | |||||||||||||||
| Hypotonia | 14/16 | 11/12 | + | + | + | + | + | + | + | + | + | + | 1/2 | − | − |
| Developmental delay | 15/17 | 12/14 | + | + | + | + | + | + | + | + | + | + | 2/2 | − | − |
| Mitral valve prolapse | 4/11 | 6/17 | + | + | + | − | − | − | + | + | 0/2 | + | + | ||
| Aortic root dilatation1 | 5/17 | 3/11 | + | + | + | + | + | + | + | − | − | + | 0/2 | + | + |
| Other features of MFS or LDS | |||||||||||||||
| Dural ectasia | + | + | + | + | + | ||||||||||
| Ectopia lentis | 1/152 | − | − | − | − | − | − | − | − | − | − | 0/2 | + | − | |
| Cleft palate | 2/11 | − | − | − | + | − | − | + | + | − | − | 0/2 | − | + | |
| Broad/Bifid uvula | − | + | − | − | + | 1/2 | − | + | |||||||
| Club foot deformity | − | − | + | − | − | − | + | − | − | 0/2 | − | + | |||
| Arterial tortuosity | − | − | − | + | − | + | |||||||||
| Other aneurysm | +3 | − | − | +4 | − | − | + | ||||||||
Aortic root z-score ≥ 2.0 (range 2.1-5.7).
Atypical patient with FBN1 mutation.
Splenic artery aneurysm.
Splenic artery aneurysm with spontaneous rupture. Blank cells, not determined.
Figure 1.
SKI mutations in patients with Shprintzen-Goldberg syndrome (SGS). A) Clinical features and mutations seen in SGS patients. Illustrated features involve the craniofacial (abnormal head shape due to craniosynostosis, widely-spaced eyes, small and receding chin and high-arched palate), skeletal (long fingers, joint contractures, chest wall deformity, spine curvature, foot deformity) and cardiovascular (aortic root aneurysm indicated by black arrowheads and mitral valve prolapse indicated by white arrowhead) systems. Pedigrees indicate that all cases were sporadic (affected, black symbols; unaffected, open symbols). Mutation status is indicated below each individual (−/−, mutation negative; +/−, heterozygous; blank, not available for testing). Permission to publish photographs was obtained from the affected individuals or their parents. B) Location of mutations in relation to known binding sites (highlighted in yellow) of SKI binding partners. The position of the Dachshund homology domain is indicated.
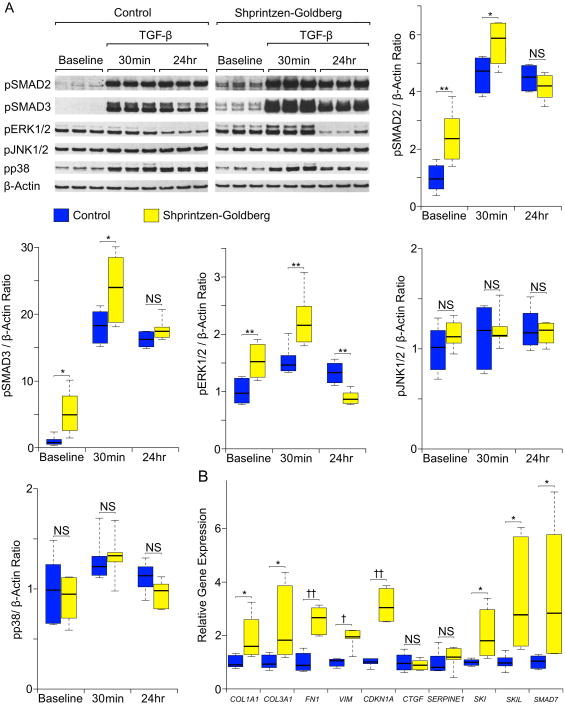
In order to assess the functional consequence of SKI mutations identified in SGS, we monitored TGF-β signaling in primary dermal fibroblasts derived from 2 SGS patients and 2 controls. Western blot analysis showed excessive SMAD2/3 and ERK1/2 phosphorylation (pSMAD2/3 and pERK1/2) in patient cells compared to controls, both at baseline and after acute (30 minute) stimulation with exogenous TGF-β2 (Figure 2A). This implies a loss of suppression of TGF-β dependent signaling cascades in SGS cells. In contrast, there was no difference in the activation of JNK or p38 between patient and control cells at baseline or in response to TGF-β2. These signaling alterations directly parallel previous findings in the aortas of both patients and mouse models of MFS and LDS4,23. We also observed a significant increase in mRNA expression for TGF-β dependent genes in SGS fibroblasts, including COL1A1, COL3A1, FN1, VIM and CDKN1A (encoding types I and III collagen, fibronectin, vimentin and p21, respectively) (Figure 2B). This is in keeping with prior work showing that transcription of these genes is normally suppressed by SKI33. TGF-β also induces transcription of genes encoding negative regulators of itself, including SKI, SKIL, and SMAD7, as part of an autoregulatory loop. We observed increased mRNA expression for each of these genes in SGS cells compared to controls, suggesting that SKI normally suppresses the expression of itself as well as other negative regulators of TGF-β signaling. Interestingly, the TGF-β-regulated target genes CTGF and SERPINE1 showed equal expression in SGS and control cells, suggesting either gene-specific differences in their sensitivity to attenuation of SKI function or to the regulatory influence of other transcriptional repressors such as SKIL or SMAD7.
Figure 2.
Assessment of TGF-β signaling in dermal fibroblasts. A) Western blot analysis of pSMAD2, pSMAD3, pERK1/2, pJNK1/2, pp38 and β-Actin (loading control) at steady state (baseline) and in response to TGF-β2. Representative Shprintzen-Goldberg syndrome (SGS) and control blots are shown (for a single SGS patient and single control with 3 biological replicates for each). Graphs display Western blot quantification of 3 biological replicates in each of 2 SGS patients and 2 controls. All data are normalized to β-Actin. The control baseline value for each graph was set to 1.0; all other values represent fold change in comparison to this sample. B) Relative mRNA expression for TGF-β target genes (normalized to β-Actin), as assessed by quantitative polymerase chain reaction analysis. Quantification was performed on 3 biological replicates in each of 2 SGS patients and 2 controls. The upper and lower margins of the box define the 75th and 25th percentiles respectively; the internal line defines the median and the whiskers define the range. NS, not significant; * p<0.05; ** p<0.01; † p<0.001; †† p<0.0001.
Despite the near-complete phenotypic overlap between LDS and SGS, it is notable that the aneurysm phenotype in SGS is less penetrant, less diffuse (generally restricted to the aortic root) and less severe than that seen in LDS. We reasoned that this might relate to the temporal and regional expression pattern of SKI. To test this hypothesis, we performed a developmental survey of SKI expression in wild-type mice (Figure 3A). At embryonic day 13.5, SKI is robustly expressed throughout the vessel wall in the proximal ascending aorta, with less expression in the descending segment, and localizes to both the cytoplasm and nucleus. At birth, aortic expression is somewhat reduced, with predominant expression in the cytoplasm. In adult mice, SKI expression is further reduced in the medial layer of the aortic root, with exclusive cytoplasmic localization; in the more distal ascending aorta SKI expression is largely excluded from the central zone of the aortic media despite some residual expression in the intimal and adventitial layers. Taken together, these data support the hypothesis that the requirement for SKI for proper regulation of TGF-β signaling within the arterial media may predominate in the very proximal aorta at early stages of development.
Figure 3.

Assessment of SKI in model systems. A) Murine SKI expression at embryonic day 13.5 (E13.5) is high in the mitral valve (MV), tricupsid valve (TV), pulmonary trunk (Pu), pulmonary valve (PuV), bronchus (Br) and proximal aorta (Ao; both nucleus and cytoplasm). Lower expression is seen in the descending aorta (dAo), esophagus (E), right and left ventricles (RV and LV) and left atrium (LA). Red, SKI; green, myosin heavy chain; blue, nuclei. At birth (P0), SKI is expressed in the aortic root (AoR), distal ascending aorta (ascAo) and endothelial surface of the mitral and aortic (AoV) valves, with less nuclear expression than at E13.5. In adulthood, SKI is expressed exclusively in the cytoplasm of the AoV and the media of the AoR; in the ascAo SKI is expressed in the intima (I) and adventitia (A), but excluded from the central media (M). B) Disruption of skia or skib expression in zebrafish. Top panels, assessment of cardiovascular anatomy at 3 days post-fertilization (dpf). Uninjected embryos display proper cardiac looping with normal relation between the atrium (A, dotted outline) and ventricle (V), and a distinct bulbous arteriosus (arrow) defining the outflow tract (OFT). skia morphants show incomplete looping with an irregular OFT, while skib morphants show failure of looping and an ill-defined OFT. Asterisk, atrioventricular cushion. Lower panels, alcian blue staining of cartilage at 6 dpf. skia and skib morphants display craniofacial cartilage deficits and prominent mandibular malformation. Meckel's cartilage, m; palatoquadrates, pq; ceratohyales, ch; ceratobranchial arches, cb. Scale bars, μm.
We show that the multisystem manifestations of SGS are caused by primary mutations in a prototypical repressor of TGF-β signaling, and that this associates with a cell-autonomous increase in SMAD2/3 activation at steady-state and in the acute-phase response to TGF-β ligand as well as with increased transcriptional output of TGF-β responsive genes. It remains to be determined whether the increased ERK1/2 activation seen in SGS cells manifests loss of a previously unrecognized primary function of SKI or secondary cellular events. The conclusion that release of the suppressive effects of SKI on TGF-β signaling underlies SGS is consistent with prior observation of CNS patterning alterations, skeletal muscle hypoplasia and craniofacial defects (all cardinal manifestations of SGS) in homozygous Ski-targeted mice34. It is further strengthened by our ability to replicate SGS-like defects after morpholino based knockdown of the 2 paralogs of mammalian SKI (skia and skib) in zebrafish. Morphant skia and skib zebrafish embryos showed significant craniofacial cartilage deficits including shortened and flat Meckel's cartilage, irregular lengths of palatoquadrates, shortened ceratohyales and depletion of ceratobranchial arches (Figure 3B, Supplementary Table 1). These deficits manifest in larval fish as maxillary hypoplasia, malformed ethmoid plate, micrognathia and microcephaly, and are frequently accompanied by ocular hypertelorism and spinal malformations. Furthermore, ski morphant embryos displayed severe cardiac anomalies, characterized by partial to complete failure in cardiac looping and malformations of the outflow tract (Figure 3B, Supplementary Table 1). Thus, in comparison to Ski-null mice, the zebrafish morphants more closely recapitulated the human SGS craniofacial phenotype and uniquely implicated a requirement for SKI in early cardiovascular development. Prior demonstration of a prominent role for SKI in both neurogenesis and myogenesis reconciles the highly penetrant developmental delay and hypotonia seen in SGS, respectively35,36. It is also notable that a recurrent mutation in SMAD4 that strongly impairs TGF-β transcriptional responses by the R-SMAD/Co-SMAD complex has recently been shown to cause Myhre syndrome, a condition characterized by short bones and aortic stenosis, a striking contrast to the bone overgrowth and aortic dilatation seen in SGS37. Patients with the recurrent 1p36 deletion syndrome are haploinsufficient for SKI and show some phenotypic overlap with SGS including hypotonia, developmental delay, craniofacial dysmorphism, vertebral abnormalities and structural heart disease38,39. Possible reasons for the features that distinguish these two conditions include the involvement of contiguous genes in the 1p36 deletion syndrome and/or the putative dominant-negative potential of mutant forms of SKI in SGS, that might retain their ability to form homodimeric complexes due to structural preservation of the SKI-interacting domain at their C-terminus, with functional deficits imposed by N-terminal mutations that selectively perturb R-SMAD and/or N-CoR interactions. Substitution of neighboring cysteine residues (p.Cys1223Tyr and p.Cys1221Tyr) in an epidermal growth factor-like domain of fibrillin-1 have been reported in association with an SGS-like phenotype including all features of MFS and SGS-specific features such as craniosynostosis and developmental delay40,41. While it remains to be determined if this tentative phenotype-genotype correlation manifests a predominant consequence of FBN1 genotype, modifier loci or chance, it seems notable that both fibrillin-1 and SKI are recognized regulators of TGF-β signaling.
In conclusion, this manuscript provides evidence that mutations predicted and observed to enhance TGF-β signaling are sufficient to cause human phenotypes that have variably been associated with low TGF-β signaling states, including craniosynostosis, altered palatogenesis and aortic aneurysm. The development of individualized treatment strategies will require further work to determine whether low and high signaling states achieve the same phenotypic consequence or, as we propose, if complex compensatory events during tissue morphogenesis and homeostasis culminate in a common pathogenic mechanism for apparently disparate disease etiologies42.
Online Methods
Participants
Patients were recruited from the Connective Tissue Clinic at Johns Hopkins Hospital (H.C.D.), Radboud University Hospital/Antwerp University Hospital (B.L.L. and G.M.), The Nemours Children's Clinic (P.H.A.) and Genetic Medicine Central California (C.J.C.). All skin biopsies and research protocols were collected in compliance with the Institutional Review Board at each respective institution after informed consent was obtained. The diagnosis of SGS was made after exclusion of other known congenital syndromes on the basis of distinguishing phenotypic features. Echocardiograms were performed and interpreted as previously described43. Aortic root aneurysm was defined by a maximal aortic root z-score ≥ 2.0.
Whole Exome sequencing
Genomic DNA was extracted from peripheral blood lymphocytes using standard protocols. DNA fragmentation was performed using a Covaris S2 system (Covaris, Woburn, MA, USA), exon capture was performed using the Agilent SureSelect 38 Mb Human All Exon Target Enrichment system (Agilent Technologies, Inc., Santa Clara, CA, USA) and DNA sequencing was performed on an Illumina GAIIx instrument (Illumina, Inc, San Diego, CA, USA), all using standard protocols for a 75 bp paired-end run.
Bioinformatic analysis
Reads were mapped to the human reference genome (UCSC hg19) using the Burrows-Wheeler Aligner and a variant list created using SAMtools and annotated using ANNOVAR44-46. Local realignment and recalibration of base call quality scores was performed using the genome analysis toolkit47,48. Duplicates were marked using Picard (http://picard.sourceforge.net). We selected for novel variants (absent from dbSNP 134 or 1000 genome project databases), focusing only on exonic non-synonymous, splice site and indel variants. Variants were viewed directly using the integrated genome viewer and excluded if reads were only present in one direction, if ambiguously mapped reads were present or if an indel occurred within 3 bp of the end of the read49.
Mutation validation and Sanger sequencing of candidate genes
The polymerase chain reactions were performed using a DNA Engine Dyad thermal cycler (Bio-Rad, Hercules, CA, USA). Phusion Flash High Fidelity PCR Master Mix was used in accordance with the manufacturer's instructions for each primer set (Dharmacon, Inc, Lafayette, CO, USA). Primer sequences and reaction conditions are listed in Supplementary Table 2. Cycle sequencing was performed using BigDye Terminator v3.1 kit and an ABI 3730 ×l DNA Analyzer/sequencing machine in accordance with the manufacturer's instructions (Life Technologies, Carlsbad, CA, USA). The samples were purified using QIAquick PCR purification kit (Qiagen, Inc, Valencia, CA, USA).
Cell culture
Primary human dermal fibroblasts were derived from forearm skin biopsies from 2 control individuals and 2 patients with SGS. They were cultured in Dulbecco's modified eagle medium with 10% fetal bovine serum in the presence of antibiotics and passaged at confluence. All cell experiments were conducted in serum-starved media, and stimulation was performed using 5 ng/mL recombinant human TGF-β2 (302-B2, R&D Systems, Minneapolis, MN, USA). Cells were collected at baseline, 30 minutes and 24 hours after stimulation.
Protein and RNA analysis
All protein and RNA was extracted, processed and analyzed using reagents and protocols as described previously4,23. The following pre-validated probes were used for mRNA analysis: Hs01060665 (β-ACTIN), Hs00164004 (COL1A1), Hs00943809 (COL3A1), Hs00287359 (FN1), Hs00185584 (VIM), Hs00355782 (CDKN1A), Hs01026927 (CTGF), Hs01126606 (SERPINE1), Hs00161707 (SKI), Hs01045418 (SKIL) and Hs00998193 (SMAD7) (Life Technologies, Carlsbad, CA, USA). In the Western blot analysis separate membranes were used for the control and SGS patients due to the large number of samples. The membranes were processed together at all stages, and exposed to the same piece of imaging film simultaneously, allowing for valid comparisons.
Developmental survey of SKI expression in mouse
Immunohistochemical analysis was performed using protocols as described previously50. Samples were incubated with an anti-mouse SKI polyclonal antibody diluted to 1:250 (EMD Millipore Corporation, Billerica, MA, USA) and myocyte-specific mouse monoclonal MF20-c diluted to 1:50 (Developmental Studies Hybridoma Bank, Iowa City, IA, USA). Elastin was detected using autofluorescence at 488 nm. Nuclei were stained with Hoechst dye diluted to 1:10,000 (Life Technologies, Carlsbad, CA, USA).
Zebrafish maintenance, imaging and staining
Adult AB and Tg(kdrl:G-RCFP; fluorescently marked endothelium) zebrafish lines were maintained according to standard methods51,52. Embryos were injected with previously published skia (12 ng) or skib (14 ng) morpholino antisense oligonucleotides (MO) at the 1-2 cell stage and analyzed at 2, 3, 5, 6, and 8 days post fertilization (dpf) for G-RCFP expression via fluorescence microscopy53. Morpholino oligonucleotides are listed in Supplementary Table 2. Prior work had established the specificity of these MOs via concomitant use of random MO and mRNA rescue experiments53. Cartilage staining was performed as previously described on zebrafish embryos fixed in 4% paraformaldehyde at 5, 6 and 8 dpf54. All experiments were performed in accordance with ethical permits by the Johns Hopkins Animal Care and Use Committee.
Statistical Analysis
All quantitative data are shown as box and whisker plots produced using the R statistical package. The upper and lower margins of the box define the 75th and 25th percentiles respectively; the internal line defines the median and the whiskers define the range. Statistical analysis was performed using two tailed t tests in Excel (Microsoft, Redmond, WA, USA). A p value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was funded by grants to H.C.D. from the National Institutes of Health (RO1-AR41135 and PO1-AR049698), the Bloomberg Fund of the National Marfan Foundation, the Smilow Center for Marfan Syndrome Research, the Howard Hughes Medical Institute and the Baylor-Hopkins Center for Mendelian Genetics (1U54HG006542). A.J.D. was supported as a Research Associate by the Howard Hughes Medical Institute. J.J.D. was supported by a Victor A. McKusick Fellowship from the National Marfan Foundation; and M.E.L. was supported by a NHLBI K08 Award (HL107738-01) and by a Fellow-to-Faculty Award from the National Marfan Foundation. A.S.M. was supported in part by the NHLBI (1R01HL111267). M.J.C. was supported by the National Institute for Health Research (NIHR) through Barts NIHR Cardiovascular Biomedical Research Unit. This study was also supported in part by funding from the Fund for Scientific Research, Flanders (Belgium) [G.0458.09; G.0221.12]; European Grant Fighting Aneurysmal Disease [EC-FP7]; Special Research Fund of Ghent University [BOF10/GOA/005]; B.L.L. is senior clinical investigator of the Fund for Scientific Research, Flanders (FWO, Belgium); D.S. was supported by a Ph.D. grant from the Agency for Innovation by Science and Technology. The authors would also like to thank Elizabeth Gerber and Hans Bjornsson for their contributions to the cell culture experiments.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Dietz HC, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 2.Loeys BL, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37:275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- 3.Habashi JP, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–121. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holm TM, et al. Noncanonical TGFbeta signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science. 2011;332:358–361. doi: 10.1126/science.1192149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeys BL, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 6.Shprintzen RJ, Goldberg RB. A recurrent pattern syndrome of craniosynostosis associated with arachnodactyly and abdominal hernias. J Craniofac Genet Dev Biol. 1982;2:65–74. [PubMed] [Google Scholar]
- 7.Greally MT, et al. Shprintzen-Goldberg syndrome: a clinical analysis. Am J Med Genet. 1998;76:202–212. [PubMed] [Google Scholar]
- 8.Robinson PN, et al. Shprintzen-Goldberg syndrome: fourteen new patients and a clinical analysis. Am J Med Genet A. 2005;135:251–262. doi: 10.1002/ajmg.a.30431. [DOI] [PubMed] [Google Scholar]
- 9.Akiyoshi S, et al. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- 10.Luo K, et al. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 12.Wotton D, Massague J. Smad transcriptional corepressors in TGF beta family signaling. Curr Top Microbiol Immunol. 2001;254:145–164. [PubMed] [Google Scholar]
- 13.Prunier C, et al. The oncoprotein Ski acts as an antagonist of transforming growth factor-beta signaling by suppressing Smad2 phosphorylation. J Biol Chem. 2003;278:26249–26257. doi: 10.1074/jbc.M304459200. [DOI] [PubMed] [Google Scholar]
- 14.Reed JA, et al. Cytoplasmic localization of the oncogenic protein Ski in human cutaneous melanomas in vivo: functional implications for transforming growth factor beta signaling. Cancer Res. 2001;61:8074–8078. [PubMed] [Google Scholar]
- 15.Nomura T, et al. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 17.Pearson G, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 18.Neptune ER, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 19.Ng CM, et al. TGF-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J Clin Invest. 2004;114:1586–1592. doi: 10.1172/JCI22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohn RD, et al. Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat Med. 2007;13:204–210. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuguchi T, et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet. 2004;36:855–860. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van de Laar IM, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]
- 23.Lindsay ME, et al. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nature Genetics. 2012 doi: 10.1038/ng.2349. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura N, et al. Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res. 1989;17:5489–5500. doi: 10.1093/nar/17.14.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin BY, Lam SS, Correia JJ, Lin K. Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control. Genes Dev. 2002;16:1950–1963. doi: 10.1101/gad.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahl R, Wani B, Hayman MJ. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene. 1998;16:1579–1586. doi: 10.1038/sj.onc.1201687. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, et al. Competition between Ski and CREB-binding protein for binding to Smad proteins in transforming growth factor-beta signaling. J Biol Chem. 2007;282:11365–11376. doi: 10.1074/jbc.M700186200. [DOI] [PubMed] [Google Scholar]
- 30.Wilson JJ, Malakhova M, Zhang R, Joachimiak A, Hegde RS. Crystal structure of the dachshund homology domain of human SKI. Structure. 2004;12:785–792. doi: 10.1016/j.str.2004.02.035. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueki N, Hayman MJ. Direct interaction of Ski with either Smad3 or Smad4 is necessary and sufficient for Ski-mediated repression of transforming growth factor-beta signaling. J Biol Chem. 2003;278:32489–32492. doi: 10.1074/jbc.C300276200. [DOI] [PubMed] [Google Scholar]
- 33.Wang LH, et al. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res. 2007;13:102–110. doi: 10.1158/1078-0432.CCR-06-1467. [DOI] [PubMed] [Google Scholar]
- 34.Berk M, Desai SY, Heyman HC, Colmenares C. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyons GE, et al. Protooncogene c-ski is expressed in both proliferating and postmitotic neuronal populations. Dev Dyn. 1994;201:354–365. doi: 10.1002/aja.1002010407. [DOI] [PubMed] [Google Scholar]
- 36.Colmenares C, Stavnezer E. The ski oncogene induces muscle differentiation in quail embryo cells. Cell. 1989;59:293–303. doi: 10.1016/0092-8674(89)90291-2. [DOI] [PubMed] [Google Scholar]
- 37.Le Goff C, et al. Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat Genet. 2012;44:85–88. doi: 10.1038/ng.1016. [DOI] [PubMed] [Google Scholar]
- 38.Heilstedt HA, et al. Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet. 2003;72:1200–1212. doi: 10.1086/375179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battaglia A, et al. Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics. 2008;121:404–410. doi: 10.1542/peds.2007-0929. [DOI] [PubMed] [Google Scholar]
- 40.Sood S, Eldadah ZA, Krause WL, McIntosh I, Dietz HC. Mutation in fibrillin-1 and the Marfanoid-craniosynostosis (Shprintzen-Goldberg) syndrome. Nat Genet. 1996;12:209–211. doi: 10.1038/ng0296-209. [DOI] [PubMed] [Google Scholar]
- 41.Kosaki K, et al. Molecular pathology of Shprintzen-Goldberg syndrome. Am J Med Genet A. 2006;140:104–108. doi: 10.1002/ajmg.a.31006. author reply 109-110. [DOI] [PubMed] [Google Scholar]
- 42.Lindsay ME, Dietz HC. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature. 2011;473:308–316. doi: 10.1038/nature10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooke BS, et al. Angiotensin II blockade and aortic-root dilation in Marfan's syndrome. N Engl J Med. 2008;358:2787–2795. doi: 10.1056/NEJMoa0706585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Norris RA, et al. Expression of the familial cardiac valvular dystrophy gene, filamin-A, during heart morphogenesis. Dev Dyn. 2010;239:2118–2127. doi: 10.1002/dvdy.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westerfield M. The Zebrafish Book A Guide for the Laboratory Use of Zebrafish (Danio rerio) University of Oregon Press; Eugene, OR: 1995. [Google Scholar]
- 52.Cross LM, Cook MA, Lin S, Chen JN, Rubinstein AL. Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol. 2003;23:911–912. doi: 10.1161/01.ATV.0000068685.72914.7E. [DOI] [PubMed] [Google Scholar]
- 53.Cui Z, Clark KJ, Kaufman CD, Hackett PB. Inhibition of skiA and skiB gene expression ventralizes zebrafish embryos. Genesis. 2001;30:149–153. doi: 10.1002/gene.1052. [DOI] [PubMed] [Google Scholar]
- 54.Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.