Abstract
Neutrophils from CCAAT enhancer binding protein epsilon (C/EBPɛ) knockout mice have morphological and biochemical features similar to those observed in patients with an extremely rare congenital disorder called neutrophil-specific secondary granule deficiency (SGD). SGD is characterized by frequent bacterial infections attributed, in part, to the lack of neutrophil secondary granule proteins (SGP). A mutation that results in loss of functional C/EBPɛ activity has recently been described in an SGD patient, and has been postulated to be the cause of the disease in this patient. We have previously demonstrated that overexpression of CCAAT displacement protein (CDP/cut), a highly conserved transcriptional repressor of developmentally regulated genes, suppresses expression of SGP genes in 32Dcl3 cells. This phenotype resembles that observed in both C/EBPɛ−/− mice and in SGD patients. Based on these observations we investigated potential interactions between C/EBPɛ and CDP/cut during neutrophil maturation. In this study, we demonstrate that inducible expression of C/EBPɛ in 32Dcl3/tet cells results in granulocytic differentiation. Furthermore, Northern blot analysis of G-CSF-induced CDP/cut overexpressing 32Dcl3 cells revealed absence of C/EBPɛ mRNA. We therefore hypothesize that C/EBPɛ positively regulates SGP gene expression, and that C/EBPɛ is itself negatively regulated by CDP/cut during neutrophil maturation. We further demonstrate that the C/EBPɛ promoter is regulated by CDP/cut during myeloid differentiation.
CCAAT enhancer binding protein epsilon (C/EBPɛ) is one of a family of basic region/leucine zipper (bZIP) transcription factors that recognizes the consensus DNA-binding sequence 5′-TKNNGYAAK-3′ (Y = C or T, K = T or G) within the regulatory regions of target genes (1, 2). C/EBP family proteins bind DNA as either homo- or heterodimers. This family of transcription factors includes C/EBPα,β,γ,δ,ɛ and CHOP-GADD 153, all of which contain highly homologous C-terminal dimerization (leucine-zipper) domains and DNA-binding (basic-region) motifs, but differ in their N-terminal transactivation domains, with the exception of CHOP-GADD 153, which lacks this domain altogether (reviewed in ref. 2). With the exception of C/EBPɛ, which is expressed at high levels mainly in the late stages of granulopoiesis and in T-lymphocytes, the other C/EBP members are expressed in a wide variety of cells (reviewed in ref. 2). The C/EBP family members are known to exert pleiotropic effects in the tissues in which they are expressed. This effect may be due to their tissue and stage-specific expression, their ability to dimerize with members of their own family and of the Fos/Jun and ATF/CREB families of transcription factors, and their ability to interact with other transcription factors such as NF-κB and Sp-1 (ref. 3 and references therein).
Profound hematopoietic abnormalities have been reported for mice nullizygous for C/EBPα, β, and ɛ. C/EBPɛ−/− mice produce hyposegmented granulocytes that are functionally defective. Mutant mice usually survive 2–5 months and succumb to low pathogenicity bacterial infection (4). The defects manifested in these mice are confined to late-stage gene expression associated with the function of the mature neutrophil. Previous studies (5, 6) have demonstrated that C/EBPɛ−/− mice express absent or low levels of mRNA for several genes including the secondary granule protein (SGP) genes (lactoferrin, neutrophil gelatinase, and neutrophil collagenase). Studies from our laboratory have demonstrated that expression of two SGP genes, lactoferrin and collagenase, in the developing neutrophil are dependent on intact C/EBP binding sites within their gene promoters (ref. 7; A.K.-G. and N.B., unpublished results).
Neutrophils from C/EBPɛ−/− mice have morphological and biochemical features very similar to those observed in patients with neutrophil-specific secondary granule deficiency (SGD). SGD is a rare congenital disorder that is characterized by frequent and severe bacterial infections (8, 9). This condition is marked by defects in neutrophil function including atypical nuclear morphology, impaired bactericidal activity, abnormalities in neutrophil migration, and absence of both neutrophil and eosinophil secondary granule proteins (9, 10). Sequence analysis of archival genomic DNA from one SGD patient revealed a 5-bp deletion within C/EBPɛ, resulting in a truncated mutant protein lacking the dimerization domain and the DNA-binding domain, both necessary for transcriptional activation (11). Lack of functional C/EBPɛ activity has been postulated to be the cause of the observed pathology in this patient (11). We performed similar sequence analysis of the C/EBPɛ locus of another SGD patient. No mutation in the C/EBPɛ cDNA was detected (A.K.-G. and N.B., unpublished results), despite the fact that defects in mRNA expression of secondary granule protein genes and defensins have been previously reported in this patient (12). Sequence analysis of several other SGD patients has also not revealed an abnormality in C/EBPɛ (J.L.-H., unpublished results). These observations suggest that structural abnormalities of the C/EBPɛ gene account for some, but not all, cases of SGD. Whether the defect in other patients lies elsewhere within the C/EBPɛ pathway remains to be determined.
CCAAT displacement protein/cut (CDP/cut) is a highly conserved homeodomain protein homologous to the Drosophila cut protein. CDP/cut has been shown to act as a repressor of developmentally regulated genes including the phagocyte-specific cytochrome heavy chain gene (gp91-phox), which is expressed exclusively in differentiating granulocytes (13, 14). Studies from our laboratory have previously demonstrated that overexpression of CDP/cut in 32Dcl3 myeloid cells blocks G-CSF-induced expression of SGP genes without blocking phenotypic maturation. CDP/cut therefore acts as a coordinate negative regulator of stage-specific expression of the SGP genes (15, 16). Because this phenotype is very similar to that observed in the C/EBPɛ knockout mice (4), and in SGD (11), we chose to examine the role of CDP/cut in the expression of C/EBPɛ during myeloid differentiation.
Materials and Methods
Tissue Culture, Transient Transfections, and Luciferase Assay.
Human erythroleukemic K562 cells, T-lymphocytic Jurkat cells, and NIH 3T3 mouse fibroblast cells were obtained from the American Type Culture Collection. K562 and Jurkat cells were grown in RPMI medium 1640 (GIBCO) and NIH 3T3 cells were grown in DMEM. Both media were supplemented with 10% heat inactivated FCS (Gemini Biological Products, Calabasas, CA), 0.2 mM glutamate, 50 units/ml penicillin, and 50 μg/ml streptomycin. CDP/cut overexpressing 32Dcl3 cells described previously (16), 32Dwt18 cells (a gift from Daniel Link, Washington University, St. Louis), and 32Dcl3/tet cells (a gift from Albert Deisserroth, Yale University, New Haven, CT) were grown in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FCS and 10% WEHI-conditioned medium, as a source of IL-3. The 32Dcl3/tet cells were supplemented with 1 μg/ml of tetracycline (Sigma), and both the 32Dcl3/tet and 32Dcl3/CDP/cut cells were maintained in medium supplemented with 600 μg/ml of G418 (Life Technologies, Gaithersburg, MD). All cells were maintained at 37°C in a humidified 5% CO2 incubator. CDP/cut overexpressing 32Dcl3 cells (16) were induced to undergo neutrophil maturation by addition of 100 ng/ml of granulocyte colony-stimulating factor (G-CSF) (Neupogen, Amgen) in the absence of IL-3. Maturation was monitored by Wright–Giemsa staining.
Approximately 1 × 107 32Dwt18 cells were transiently transfected as previously described by using 10–20 μg of each reporter plasmid and 2 μg pCMVβgal (CLONTECH), an internal control plasmid used to monitor transfection efficiency (7). Transiently transfected cells were divided equally into medium with 1 unit/ml Erythropoietin (Epo) (−IL-3) (Amgen), and IL-3 containing medium as described above. Transfected cells were incubated at 37°C in 5% CO2 for 16–20 h. Jurkat cells were transiently transfected by lipofection using the DMRIE-C (Life Technologies, Gaithersburg, MD) reagent as recommended by the manufacturer, using 4 μg of the reporter plasmid and 0.5 μg of the pCMVβgal plasmid. Cells were harvested 48 h posttransfection. Luciferase activity was then determined by using an assay kit from Promega according to the manufacturer's instructions. NIH 3T3 cells were transfected by using Lipofectamine per the manufacturers' instructions (GIBCO/BRL). Cotransfection experiments included 2 μg of C/EBPɛ reporter plasmids and 0.5 μg of pMT2-CDP expression plasmid (a gift from Ellis Neufeld, Children's Hospital, Boston). Luciferase expression levels were normalized to the levels of β-galactosidase expression (16).
Plasmid Construction, Generation, and Expression of Stable 32Dcl3/tet/C/EBPɛ Wild-Type and Mutant Clones.
An 859-bp full-length C/EBPɛ cDNA as well as a mutant C/EBPɛ cDNA with the previously described 5-bp deletion derived from a patient with SGD (11) were excised with HindIII/BamHI from the pCEpsilon32 and pCEpsilon32(Δ5) plasmids (11), respectively. These fragments were subcloned into the pGEM-72f vector (Promega) also digested with HindIII/BamHI. The cDNAs were further subcloned into EcoRI/BamHI-digested pUHD10-3 (a gift from H. Bujard, University of Heidelberg, Heidelberg, Germany) such that the C/EBPɛ-derived inserts were positioned downstream of three tetracycline-responsive elements. Twenty micrograms of the resulting pTet/C/EBPɛWT and pTet/C/EBPɛmut plasmids were stably cotransfected with 2 μg of pBabepuro plasmid into 32Dcl3/tet cells by electroporation (Bio-Rad gene pulser, 400 mV, 250 μF). Transfected cells were selected in 2 μg/ml of puromycin (Sigma) and the transfected pools cloned by limiting dilution. All clones were maintained in Iscove's modified Dulbecco's medium (IMDM), 10% FCS, 10% WEHI-conditioned medium as a source of IL-3 (17), 1 μg/μl of Tetracycline (Sigma), and 2 μg/ml of puromycin (Sigma) and maintained at 37°C in a humidified 5% CO2 incubator.
The 32Dcl3/tet cells had been previously stably transfected with a plasmid expressing high levels of the tetracycline repressor fused to the activation domain of the VP16 of Herpes Simplex virus. Growth in the presence of tetracycline prevents the expression of the C/EBPɛ cDNAs, because the tetracycline repressor-VP16 fusion protein is bound to tetracycline. Removal of tetracycline from the growth medium frees the tetracycline repressor −VP16 fusion protein to bind to the tetracycline responsive elements, and activates expression of C/EBPɛ (18).
Wild-type and mutant C/EBPɛ expression in the 32Dcl3/tet cells was induced by removal of tetracycline. The cloned cells were washed twice with ice-cold PBS and transferred to Iscove's modified Dulbecco's medium (IMDM), 10% FCS, and 10% WEHI-conditioned medium lacking tetracycline. Induction of wild-type and mutant C/EBPɛ was confirmed by Northern blot analysis.
Northern Blot Analysis.
Northern blot analysis was performed as described (19). Blots were probed with a previously described 600-bp mouse LF probe (20), with a cDNA probe for C/EBPα isolated from the pMSV-C/EBPα plasmid provided by Alan Friedman (Johns Hopkins University, Baltimore, MD), and with an XhoI/HindIII 859-bp C/EBPɛ probe isolated from the described pCMV-C/EBPɛ32 plasmid (21).
Isolation and Subcloning of the C/EBPɛ Promoter and 5′ Regulatory Sequences.
The human C/EBPɛ gene is located on chromosome 14q11.2 (22, 23). The genomic sequence of this portion of the gene has been deposited in the GenBank database (accession no. U48865). The major transcription start site for the 32-kDa isoform has been determined (21). We obtained a BAC clone from the IMAGE consortium, which contains ≈160 kb of human chromosome 14, including the C/EBPɛ locus (R-244E17 in RPCI-11 library). Using C/EBPɛ β− promoter-specific oligomers—Sense: 5′-GAGCTCAGCTTGAAATGGAG-3′; Antisense: 5′-TGGGACATGGCCGGCCCG-3′—and BAC DNA as a template, a 1.8-kb fragment of the C/EBPɛ promoter was obtained by PCR using Platinum Taq DNA polymerase (Life Technologies, Gaithersburg, MD). This promoter fragment was subcloned into the pCR11 vector (Invitrogen) and sequenced to confirm its identity.
The 1.8-kb C/EBPɛ fragment was further subcloned into KpnI/XhoI-digested pGL3Basic promoterless reporter gene vector (Promega). Additionally, a 726-bp fragment of the C/EBPɛ promoter was isolated by digesting pCRII-C/EBPɛ with AvrII/XhoI, and subcloned upstream of the luciferase reporter gene in NheI/XhoI-digested pGL3Basic vector.
Preparation of Nuclear Extracts, Oligonucleotides, and Electrophoretic Mobility-Shift Assays (EMSAs).
Nuclear extracts were prepared essentially as described (16). Complementary oligonucleotides were annealed and labeled at their 5′ ends by using [γ-32P]ATP [6,000 Ci/mmol (1 Ci = 37 GBq); Amersham Pharmacia] and T4 polynucleotide kinase (New England Biolabs). Radiolabeled double-stranded oligonucleotides were separated from unincorporated nucleotide by passage through a Sephadex G-25 spin column (Boehringer Mannheim). Probes were stored at −20°C.
EMSAs were performed by incubating 15 μg of nuclear extracts with 20,000 cpm of double-stranded oligonucleotide in a 20-μl reaction mixture containing 10 mM Hepes-KOH buffer (pH 7.9), 50 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 10% glycerol, 1 μg of acetylated BSA (New England Biolabs), and 0.5 μg of poly(dI-dC) at 25°C for 20 min. For competition analysis, a 100-fold molar excess of unlabeled oligonucleotides was added to the nuclear extracts before the addition of the labeled probe. For the supershift assay anti-CDP serum (a gift from Ellis Neufeld, Children's Hospital, Boston) was preincubated with nuclear extracts for 15 min following the addition of radiolabeled probe. Binding reactions were resolved on a 4% nondenaturing polyacrylamide gel containing 1× TBE (0.089 M Tris-borate, 0.089 M boric acid, and 0.002 M EDTA) and electrophoresed at 150 V for 3 h at 4°C. Gels were exposed to x-ray film with an intensifying screen overnight at −80°C.
The sense oligonucleotides used in EMSA analysis had the following sequence: C/EBPɛ CDP/cut site, 5′-GCACCCATAGCACTTGCTGGTCA-3′; NCAM CDP/cut site, 5′-AGAATTTTAGATTCGGTTCGATTTTCA-3′ (24); E36 CDP/cut site, 5′-CGGATCCGAATTCATCGATAATCGATTAT-3′ (25).
Results
C/EBPɛ Drives Myeloid Differentiation in 32Dcl3 Cells.
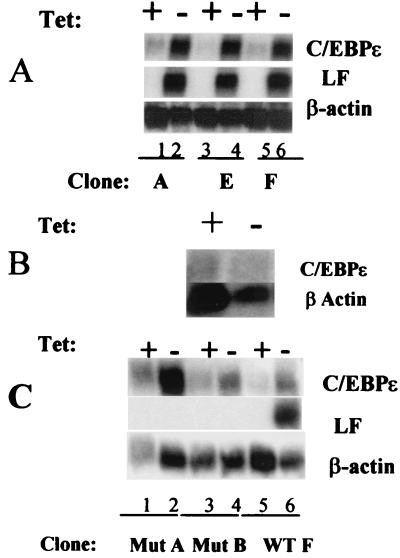
To examine the role of C/EBPɛ in granulopoiesis, 32Dcl3/tet cells were stably transfected with wild-type C/EBPɛ and the mutant C/EBPɛ derived from the described SGD patient (11). Northern blot analysis of three 32Dcl3/tet clones transfected with wild-type C/EBPɛ (A, E, and F) following removal of tetracycline for 4 days revealed that all three clones expressed high levels of C/EBPɛ mRNA (Fig. 1A, lanes 2, 4, and 6). This increase in C/EBPɛ expression was confirmed at the protein level and was accompanied by morphological changes evaluated by Wright–Giemsa staining (data not shown). Induction of lactoferrin mRNA levels was also observed in all three clones (Fig. 1A, lanes 2, 4, and 6). No increase in lactoferrin gene expression was observed when control 32Dcl3/tet cells transfected with the pUHD10-3 vector alone were removed from tetracycline (Fig. 1B). These data indicate that inducible C/EBPɛ expression in 32Dcl3/tet cells is capable of driving granulocytic maturation.
Figure 1.
Northern blot analysis of wild type and mutant C/EBPɛ induction in 32Dcl3/tet cells. (A) Three clones of 32D/tetC/EBPɛ wild-type A, E, and F were grown both in the presence (+, lanes 1, 3, and 5) and absence (−, lanes 2, 4, and 6) of tetracycline for 4 days. Total RNA was extracted from each set of cells and 10 μg was subjected to Northern blot analysis. The blot was sequentially probed with 32P-labeled cDNA for C/EBPɛ, mouse lactoferrin (LF), and β-actin. (B) A similar Northern blot was carried out for the 32Dcl3/tet cells transfected with the pUHD10-3 vector alone. (C) Two clones, MutA and MutB, of 32D/tetC/EBPɛ harboring a mutant C/EBPɛ lacking the basic region/leucine zipper (bZip) domain (SGD mutation) were grown both in the presence (+, lanes 1, 3, and 5) and absence (−, lanes 2, 4, and 6) of tetracycline for 4 days. Total RNA was extracted from each set of cells and subjected to Northern blot analysis. The blot was sequentially probed with 32P-labeled cDNA for C/EBPɛ, mouse lactoferrin (LF), and β-actin.
Induction of mutant C/EBPɛ in 32Dcl3/tet cells resulted in the induced up-regulation of the mutant C/EBPɛ gene in both clones examined (Fig. 1C, MutA and MutB, lanes 2 and 4). Up-regulation of lactoferrin, however, was observed only in the control cells expressing wild-type C/EBPɛ (Fig. 1C, WTF, lane 6). Because the mutant C/EBPɛ cDNA was derived from an SGD patient, this further supports the hypothesis that failure of SGD expression in that patient is related to the mutation in C/EBPɛ. The 32D/tet/C/EBPɛ and the 32D/tet mutC/EBPɛ cells thus provide a useful in vitro model to examine C/EBPɛ function in normal myeloid cells, as well as in SGD cells.
CCAAT Displacement Protein/Cut Down-Regulates C/EBPɛ Expression.
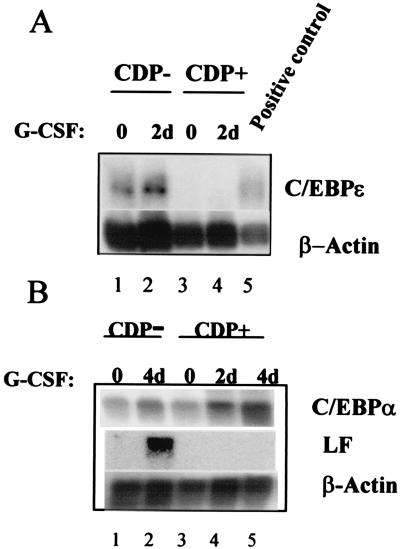
We have previously demonstrated (16) that overexpression of CDP/cut abrogates the expression of G-CSF-induced SGP genes in 32Dcl3 cells without blocking phenotypic maturation. Because this phenotype is very similar to that observed in the C/EBPɛ knockout mice (4) and in the SGD patient, we hypothesized that the expression of C/EBPɛ may be regulated by CDP/cut in the maturing neutrophil. We therefore performed further Northern blot analysis of 32Dcl3 cells constitutively overexpressing CDP/cut (32Dcl3/CDP/cut) before and after 2 days of G-CSF induction. Control 32Dcl3 cells harboring the vector alone (CDP−) expressed inducible levels of the C/EBPɛ transcript (Fig. 2A, lanes 1 and 2). The CDP-overexpressing cells (CDP+), however, did not express basal levels (lane 3) or G-CSF-induced levels (lane 4) of C/EBPɛ, demonstrating that C/EBPɛ expression is repressed in 32Dcl3/CDP/cut cells.
Figure 2.
Expression of C/EBPα but not C/EBPɛ in 32Dcl3 cells overexpressing CCAAT displacement protein (CDP) induced with G-CSF. (A) 10 μg of RNA isolated from uninduced (lanes 1 and 3) and G-CSF-induced (lanes 2, 4, and 5) CDP/cut overexpressing 32Dcl3 (CDP+) and Vector alone 32Dcl3 (CDP−) cells were subjected to Northern blot analysis. The Northern blot was probed with a 32P-labeled cDNA probe for C/EBPɛ (Top). (B) Northern blot analysis of the same RNAs probed by using a 32P-labeled C/EBPα probe revealed G-CSF-induced expression of this transcription factor in both CDP overexpressing 32Dcl3 (CDP+) (Top, lanes 3, 4 and 5) and Vector alone 32Dcl3 (CDP−) cells (Top, lanes 1 and 2). The level of lactoferrin (LF) mRNA was up-regulated only in the G-CSF-induced CDP− cells (Middle, lanes 1 and 2), but not in similarly treated CDP+ cells (Middle, lanes 3, 4, and 5). Equal loading of RNA in each lane of each of the two blots was determined by probing the blots with β-Actin (Bottom).
Interestingly, the level of C/EBPα in 32Dcl3/CDP/cut (CDP+) cells was found to be elevated upon G-CSF-induction (Fig. 2B, lanes 3, 4, and 5), as were the levels in 32Dcl3 vector alone cells (Fig. 2B, lanes 1 and 2). As previously shown (15, 16), lactoferrin expression was up-regulated by G-CSF only in 32Dcl3 vector alone cells (lane 2) and not in the CDP-overexpressing cells (lanes 3, 4, and 5). Hence, increased levels of C/EBPα do not influence lactoferrin gene up-regulation in 32Dcl3/CDP/cut cells. C/EBPɛ has been postulated to be the C/EBP family member responsible for SGP gene expression in the developing neutrophil. Our results confirm this role of C/EBPɛ in modulating SGP expression in an experimental cell line. The increased levels of C/EBPα in the CDP/cut-overexpressing cells (Fig. 2B, lanes 3, 4, and 5) remain unexplained. It is possible that CDP/cut acts as a positive modulator of C/EBPα as previous studies have suggested that CDP/cut can participate in gene activation (26, 27).
A Negative Regulator of the C/EBPɛ Gene Promoter Is Located Between −1.8-kb and −0.726-kb Coordinates.
Given that CDP/cut overexpression in 32Dcl3 cells blocked the expression of C/EBPɛ, we hypothesized that CDP/cut may regulate C/EBPɛ by binding to a CDP/cut recognition site. We analyzed the sequence of a 1.8-Kb fragment of the C/EBPɛ promoter and identified a putative CDP/cut binding site (Fig. 3A). To confirm the function of this site, the 1.8-kb (C/EBPɛ1.8) promoter fragment and a smaller 726-bp fragment (C/EBPɛ0.7) were subcloned into the promoterless pGL3Basic plasmid. Both plasmids were transiently transfected into either Jurkat cells, which have been shown to express C/EBPɛ (23), or into 32Dwt18 cells. The 32Dwt18 cells are a subline of 32Dcl3 cells that have been stably transfected with a chimeric receptor containing the extracellular domain of the erythropoietin receptor and the intracellular domain of the G-CSF receptor. These cells respond to Epo and undergo differentiation along the granulocytic lineage (16). Uninduced 32Dwt18 cells express either no or very low levels of C/EBPɛ (data not shown).
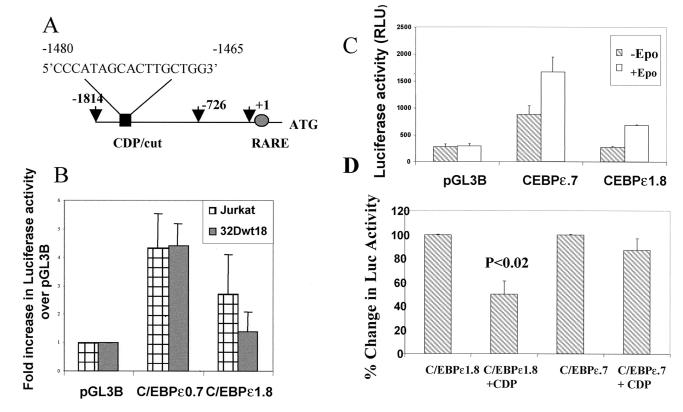
Figure 3.
Presence of a CDP/cut binding site in the C/EBPɛ promoter. (A) A 1.8-kb promoter fragment of the p32 isoform of the C/EBPɛ gene was isolated by using sequence-specific oligos and a BAC clone containing 160 kb of human chromosome 14. A CDP/cut binding site is located between −1480 and −1465 bp from the transcription start site (+1). The −1.8 (−1814) and −0.7 (−726) kb coordinates are indicated by arrows. The presence of a retinoic acid responsive element (RARE) is also indicated. (B) Jurkat and 32Dwt18 cells were transiently transfected with C/EBPɛ gene promoter fragments spanning −0.7 kb (C/EBPɛ0.7) and −1.8 kb (C/EBPɛ1.8) cloned into the promoterless luciferase reporter containing pGL3Basic plasmid. Normalized luciferase values have been represented as a fold increase of luciferase activity over empty pGL3Basic vector. Means ± SE for three experiments performed in duplicate have been illustrated. (C) 32Dwt18 cells were transiently transfected with C/EBPɛ0.7 and C/EBPɛ1.8 promoter plasmids. Half the cells were induced with Epo for 48 h (+Epo), whereas the other half were incubated in medium without Epo (−Epo). The experiment was repeated twice in duplicate. Normalized average luciferase values from one representative experiment have been illustrated. (D) NIH 3T3 cells were transiently cotransfected wtih C/EBPɛ0.7 and C/EBPɛ1.8 promoter plasmids without and with (+CDP) an expression plasmid for CDP/cut. Cells were harvested 24 hr post-transfection. An arbitrary value of 100% has been ascribed to the normalized luciferase values of C/EBPɛ1.8 and C/EBPɛ0.7 plasmids, respectively. Mean ± SE for three experiments performed in duplicate have been illustrated.
Transient transfection analysis of the C/EBPɛ promoter plasmids in Jurkat and 32Dwt18 cells (Fig. 3B) indicate that C/EBPɛ0.7 plasmid, lacking the putative CDP/cut site, exerted an ≈5-fold increase in luciferase activity in both cell lines, compared with the pGL3Basic plasmid alone. The luciferase activity of the C/EBPɛ1.8 plasmid, harboring the CDP/cut site, however, was about 3-fold lower in 32Dwt18 cells than in Jurkat cells. This observation suggests the presence of a repressor element, likely CDP/cut, between the −1.8-kb and 0.726-kb coordinates that binds a repressor in cells not expressing C/EBPɛ (32Dwt18), thereby exerting a repressive effect on reporter gene activity. The higher levels of luciferase activity for the C/EBPɛ1.8 plasmid in the C/EBPɛ-expressing Jurkat cells probably reflect loss of repressor binding to the repressor element in Jurkat cells. We have made a similar observation for CDP/cut binding to the CDP/cut site in the lactoferrin promoter (16).
Induction of 32Dwt18 cells with Epo results in maturation along the neutrophil pathway (16). We have previously demonstrated that C/EBPɛ is up-regulated during this induction (data not shown). Following transient transfection of C/EBPɛ promoter plasmids C/EBPɛ0.7 and C/EBPɛ1.8 into 32Dwt18 cells, Epo induction increased luciferase activity of the C/EBPɛ1.8 plasmid 3-fold over uninduced cells (Fig. 3C) to a level comparable to that observed for Jurkat cells (Fig. 3B). We hypothesize that induction of reporter gene activity is a consequence of derepression at the level of the CDP/cut repressor site. Epo induction increased the levels of reporter gene activity for the C/EBPɛ0.7 plasmid by approximately 2-fold.
In order to demonstrate a more direct role for CDP/cut in down-regulating the C/EBPɛ promoter, C/EBPɛ1.8 and C/EBPɛ0.7 plasmids were cotransfected into NIH 3T3 cells with an expression plasmid for CDP/cut (Fig. 3D). CDP/cut caused a statistically significant (P < 0.02) transrepression of the C/EBPɛ1.8 plasmid. No significant repression was observed for the C/EBPɛ0.7 plasmid, which lacks the CDP/cut site.
CDP/Cut Recognizes and Binds to the C/EBPɛ Promoter.
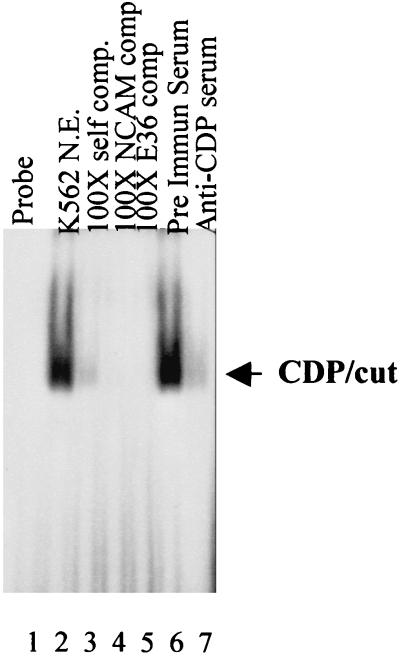
The transient transfection analysis confirms a repressor element, between the −1.8-kb and −726-bp coordinates of the C/EBPɛ promoter. Sequence analysis of this region revealed the potential CDP/cut site, but no other recognized repressor binding sequences. To demonstrate the authenticity of this CDP/cut site, we performed EMSA. A double-stranded oligomer representing the CDP/cut binding site in the C/EBPɛ promoter was 5′ end-labeled with 32P, and incubated with nuclear extracts prepared from K562 cells, which we and others have previously shown to be a good source of CDP/cut (16, 28). Addition of K562 extracts to the CDP/cut probe resulted in the retardation of one protein–DNA complex (Fig. 4, lane 2). This complex could be specifically competed away following the addition of not only a 100-fold molar excess of unlabeled CDP/cut probe (lane 3, self comp), but also by the addition of a 100-fold molar excess of NCAM probe (lane 4) and the same concentration of E36 probe. The NCAM and E36 probes have been shown to bind CDP/cut specifically and with high affinity (24, 25). The competition assay suggested the bound protein to be CDP/cut. This was confirmed by preincubating the K562 nuclear extract with a CDP/cut-specific antiserum before the addition of the 32P-labeled CDP/cut probe, which led to the loss of the protein–DNA complex (lane 7). A similar preincubation with preimmune serum did not alter formation of the protein–DNA complex (lane 6). CDP/cut therefore recognizes and binds to the identified CDP/cut site within the 5′ regulatory sequence of the C/EBPɛ promoter.
Figure 4.
EMSA analysis of the CDP/cut site in the C/EBPɛ promoter. Electrophoretic mobility shift analysis was carried out by using 32P-labeled double-stranded oligos encoding the CDP/cut site in the C/EBPɛ promoter. Addition of nuclear extracts prepared from K562 cells resulted in the formation of a protein–DNA complex (lane 2, see arrow), which was specifically competed away by the addition of a 100-fold molar excess of unlabeled self (lane 3, Self comp.), or known CDP binding oligos—i.e., NCAM (lane 4) and E36 (lane 5). Preincubation of the nuclear extracts with anti-CDP serum (lane 7), but not with preimmune serum (lane 6), resulted in the loss of the protein–DNA complex.
Discussion
We present an in vitro model for the study of the regulation of secondary granule protein gene expression by C/EBPɛ. We demonstrate that inducible expression of wild-type but not SGD-mutant C/EBPɛ in 32Dcl3/tet myeloid cells results in granulocytic differentiation, characterized by both morphologic changes and expression of lactoferrin. Recent studies have demonstrated that inducible overexpression of either C/EBPα or C/EBPɛ in leukemic myelomonocytic U937 cells results in biochemical and morphological maturation along the granulocytic pathway (29, 30). Similar results have been reported in the 32Dcl3 cell line inducibly overexpressing C/EBPα.§ Gene knockout studies in mice place C/EBPα earlier than C/EBPɛ in the transcriptional cascade leading to mature neutrophils. Nevertheless, ambiguity still persists as to which family member is responsible for modulating downstream targets during neutrophil maturation (6), because both family members are expressed in the maturing neutrophil. This observation is further compounded by the fact that C/EBPα and C/EBPɛ have similar affinities for C/EBP consensus binding sites in several myeloid genes including the G-CSF receptor promoter (6), as well as the lactoferrin promoter (A.K.-G. and N.B., unpublished results). Our results confirm that C/EBPɛ, not C/EBPα, is responsible for the up-regulation of the lactoferrin gene during neutrophil development.
We have previously demonstrated that 32Dcl3 myeloid cells constitutively overexpressing CDP/cut fail to undergo G-CSF-induced up-regulation of lactoferrin and other secondary granule protein genes, neutrophil gelatinase, and neutrophil collagenase (15, 16). Here we show that the same cells (32Dcl3/CDP/cut) express high levels of C/EBPα on G-CSF-induction, but that C/EBPɛ is completely absent. We propose that the absence of C/EBPɛ expression is responsible for the lack of lactoferrin expression in these cells. Furthermore, the presence of a functional CDP/cut binding site within the 5′ regulatory sequence of the C/EBPɛ gene promoter is responsible, at least in part, for the observed lack of C/EBPɛ expression in these cells. The process leading to C/EBPɛ expression during neutrophil maturation is a tightly regulated one. Our studies have implicated CDP/cut as being part of the transcriptional machinery involved in this process. It is likely that repression by CDP/cut is relieved by other factors that up-regulate expression of C/EBPɛ. Previous studies have demonstrated that C/EBPɛ is a retinoic acid (RA)-responsive gene in myeloid cells, and that the RA response is mediated through a retinoic acid responsive element (RARE) located in the 5′ untranslated region of the C/EBPɛ gene (30). More recent studies implicate AML1 and the leukemic fusion protein AML1-ETO resulting from the t(8;21) translocation as modulators of C/EBPɛ expression in myeloid cells (31). The relationship between the negative (CDP/cut and AML-1) and positive (RA and AML1-ETO) transcription factors that ultimately results in the expression of C/EBPɛ during neutrophil maturation remains to be defined.
Our results suggest that the role of CDP/cut in SGP gene expression is 2-fold: (i) it directly binds to and represses the promoters of the SGP genes (as we have demonstrated previously for the lactoferrin promoter; refs. 15 and 16) and (ii) it acts indirectly by repressing the expression of C/EBPɛ, a positive regulator of SGP gene expression during neutrophil development. An intriguing question raised by our findings is: How does a ubiquitously expressed repressor such as CDP/cut exert its repressive effects on C/EBPɛ as well as the downstream targets of C/EBPɛ, namely lactoferrin and other SGP genes? We propose that the differential repressive activity of CDP/cut may be inherent in the molecule itself. CDP/cut, a 180–200-kDa evolutionarily conserved polypeptide, has been described as being crucial to the determination of cell fate in Drosophila (32, 33), and more recently in chicken limb development (34). CDP/cut is a homeobox protein containing three highly conserved DNA-binding repeats referred to as cut repeats, each of which is capable of recognizing and binding specific DNA motifs in target genes (14, 25, 35, 36). This observation may explain why the CDP/cut molecule as a whole does not have a well defined consensus DNA-binding sequence (37). A recent study (37) has shown that the cut repeats are incapable of binding DNA as monomers, but that in combination exhibit high DNA-binding affinity.
It has been suggested that CDP/cut activity is restricted to proliferating cells, and that CDP/cut target genes are repressed in proliferating cells and are up-regulated as cells undergo cell cycle arrest and terminal differentiation (13–16, 24, 26, 38). Target genes of CDP/cut include c-myc, c-mos, thymidine kinase (TK), cyclin-dependent kinase (cdk) inhibitor p21WAF1/CIP1, CFTR, TGFβ-type II receptor, gp91phox, MHC class 1 locus (38–44), and, as we have shown, the neutrophil SGP genes (15, 16). Recently, CDP/cut has been shown to function as a repressor of transcription involving chromatin modification through recruitment of histone deacetylases (HDAC; ref. 42), consistent with the notion that transcriptional silencing is associated with hypoacetylated histones (45). Both acetylation of CDP/cut via p300/CBP and phosphorylation of CDP/cut are posttranscriptional modifications that have been postulated to regulate CDP/cut function (46, 47). We speculate that CDP/cut uses a different combination of its four binding elements to bind to the CDP/cut motifs in the promoters of C/EBPɛ and the SGP genes, respectively, during neutrophil maturation. Differential modification, involving either phosphorylation and/or acetylation, of CDP/cut-DNA complexes in the promoters of C/EBPɛ and SGP genes could result in the differential repression exerted by CDP/cut during neutrophil development.
In conclusion, we have demonstrated that C/EBPɛ works to positively regulate the expression of lactoferrin during neutrophil development. Additionally, C/EBPɛ expression is modulated by CDP/cut in the developing neutrophil, at least in part through a bona fide CDP/cut site located in the 5′ regulatory region of the C/EBPɛ promoter. Furthermore, CDP/cut regulates the expression of C/EBPɛ, and not C/EBPα, another C/EBP family member critical to myeloid differentiation (reviewed in ref. 3). We provide definitive evidence that SGP gene expression is directly modulated by C/EBPɛ and not by C/EBPα.
Acknowledgments
We thank members of the Myeloid Stem Cell group at Yale University School of Medicine and Drs. Dan Tenen and Hanna Radomska (Harvard Institutes of Medicine, Boston) for helpful insights and discussions. This work was supported by National Institutes of Health Grants RO1-DK53471 and PO1-HL63357 (to N.B.) and by an Anna G. and Argall L. Hull Cancer Research Award (to A.K.-G.).
Abbreviations
- SGD
secondary granule deficiency
- SGP
secondary granule protein
- C/EBP
CCAAT enhancer binding protein
- CDP
CCAAT displacement protein
- EMSA
electrophoretic mobility shift analysis
- Epo
Erythropoietin
Footnotes
Wang, X., Scott, E., Sawyers, C. E. & Friedman, A. D. (1998) Blood 92, Suppl. 1, 307a (abstr.).
References
- 1.Diehl A M. J Biol Chem. 1998;273:30843–46. doi: 10.1074/jbc.273.47.30843. [DOI] [PubMed] [Google Scholar]
- 2.Lekstrom-Himes J, Xanthopoulos K G. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 3.Tenen D G, Hromas R, Licht J D, Zhang D-E. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 4.Yamanaka R, Barlow C, Lekstrom-Himes J, Castilla L H, Liu P P, Eckhaus M, Decker T, Wynshaw-Boris A, Xanthopoulos K G. Proc Natl Acad Sci USA. 1997;94:13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lekstrom-Himes J, Xanthopoulos K G. Blood. 1999;93:3096–3105. [PubMed] [Google Scholar]
- 6.Verbeek W, Lekstrom-Himes J A, Park D J, My-Chan P, Vuong P T, Kawano S, Babior B M, Xanthopoulos K, Koeffler H P. Blood. 1999;94:3141–3150. [PubMed] [Google Scholar]
- 7.Khanna-Gupta A, Zibello T A, Simkevich C, Rosmarin A G, Berliner N. Blood. 2000;95:3734–3741. [PubMed] [Google Scholar]
- 8.Ambruso D R, Sasada M, Nishiyama A, Kubo A, Komiyama A, Allen R H. J Clin Immunol. 1984;4:23–30. doi: 10.1007/BF00915283. [DOI] [PubMed] [Google Scholar]
- 9.Tamura A, Agematsu K, Mori T, Kawai H, Kuratsuji T, Shimane M, Tani K, Asano S, Komiyama A. Int J Hematol. 1994;59:137–142. [PubMed] [Google Scholar]
- 10.Rosenberg H F, Gallin J I. Blood. 1993;82:268–273. [PubMed] [Google Scholar]
- 11.Lekstrom-Himes J A, Dorman S E, Kopar P, Holland S M, Gallin J I. J Exp Med. 1999;189:1847–1852. doi: 10.1084/jem.189.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston J J, Boxer L A, Berliner N. Blood. 1992;80:2088–2091. [PubMed] [Google Scholar]
- 13.Lievens P M, Donady J J, Tufarelli C, Neufeld E J. J Biol Chem. 1995;270:12745–12750. doi: 10.1074/jbc.270.21.12745. [DOI] [PubMed] [Google Scholar]
- 14.Neufeld E J, Skalnik D G, Lievens P M, Orkin S H. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 15.Lawson N D, Khanna-Gupta A, Berliner N. Blood. 1998;91:2517–2524. [PubMed] [Google Scholar]
- 16.Khanna-Gupta A, Zibello T, Kolla S, Neufeld E J, Berliner N. Blood. 1997;90:2784–2795. [PubMed] [Google Scholar]
- 17.Ymer S, Tucker W Q J, Sanderson C J, Hapel A J, Campbell H D, Young I G. Nature (London) 1985;317:255–258. doi: 10.1038/317255a0. [DOI] [PubMed] [Google Scholar]
- 18.Bujard H. J Gene Med. 1999;1:372–374. doi: 10.1002/(SICI)1521-2254(199909/10)1:5<372::AID-JGM61>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 19.Khanna-Gupta A, Kolibaba K, Zibello T A, Berliner N. Blood. 1994;84:294–302. [PubMed] [Google Scholar]
- 20.Johnston J J, Rintels P, Chung J, Sather J, Benz E J, Jr, Berliner N. Blood. 1992;79:2998–3006. [PubMed] [Google Scholar]
- 21.Yamanaka R, Kim G D, Radomska H S, Lekstrom-Himes J, Smith L T, Antonson P, Tenen D G, Xanthopoulos K G. Proc Natl Acad Sci USA. 1997;94:6462–6467. doi: 10.1073/pnas.94.12.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koike M, Chumakov A M, Takeuchi S, Tasaka T, Yang R, Nakamaki T, Tsuruoka N, Koeffler H P. Leuk Res. 1997;21:833–839. doi: 10.1016/s0145-2126(97)00072-6. [DOI] [PubMed] [Google Scholar]
- 23.Antonson P, Stellan B, Yamanaka R, Xanthopopoulos K G. Genomics. 1996;35:30–38. doi: 10.1006/geno.1996.0319. [DOI] [PubMed] [Google Scholar]
- 24.Valarche I, Tissier-Seta J P, Hirsch M R, Martinez S, Goridis C, Brunet J F. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 25.Andres V, Chiara M D, Mahdavi V. Genes Dev. 1994;8:245–257. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 26.Barberis A, Superti-Furga G, Busslinger M. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- 27.El-Hodiri H M, Perry M. Mol Cell Biol. 1995;15:3587–3596. doi: 10.1128/mcb.15.7.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Soudant N, Drachman J G, Kaushansky K, Nepveu A. Leukemia. 2000;14:863–873. doi: 10.1038/sj.leu.2401764. [DOI] [PubMed] [Google Scholar]
- 29.Radomska H S, Huettner C S, Zhang P, Cheng T, Scadden D T, Tenen D G. Mol Cell Biol. 1998;7:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park D J, Chumakov A M, Vuong P T, Chih D Y, Gombart A F, Miller W H, Koeffler H P. J Clin Invest. 1999;103:1399–1408. doi: 10.1172/JCI2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu K, Kitabyasi I, Kamada N, Tatsuo A, Suzukawa K, Ohki M. Blood. 2000;96:288–296. [PubMed] [Google Scholar]
- 32.Blochlinger K, Bodmer R, Jack J, Jan L Y, Jan Y N. Nature (London) 1988;333:629–635. doi: 10.1038/333629a0. [DOI] [PubMed] [Google Scholar]
- 33.Blochlinger K, Jan L Y, Jan Y N. Genes Dev. 1991;5:1124–1135. doi: 10.1101/gad.5.7.1124. [DOI] [PubMed] [Google Scholar]
- 34.Tavares A T, Tsukui T, Izpisua Belmonte J C. Development. 2000;127:5133–5144. doi: 10.1242/dev.127.23.5133. [DOI] [PubMed] [Google Scholar]
- 35.Aufiero B, Neufeld E J, Orkin S H. Proc Natl Acad Sci USA. 1994;91:7757–7761. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada R, Dufort D, Denis-Larose C, Nepveu A. J Biol Chem. 1994;269:2062–2067. [PubMed] [Google Scholar]
- 37.Moon N S, Berube G, Nepveu A. J Biol Chem. 2000;275:31325–31334. doi: 10.1074/jbc.M002912200. [DOI] [PubMed] [Google Scholar]
- 38.Skalnik D G, Strauss E C, Orkin S H. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 39.Dufort D, Nepveu A. Mol Cell Biol. 1994;14:4251–4257. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim E C, Lau J S, Rawlings S, Lee A S. Cell Growth Differ. 1997;8:1329–1338. [PubMed] [Google Scholar]
- 41.Croqueret O, Berube G, Nepveu A. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld E J, LeLeiko N S, Walsh M J. J Biol Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 43.Jackson R J, Antonia S J, Wright K L, Moon N S, Nepveu A. Arch Biochem Biophys. 1999;371:290–300. doi: 10.1006/abbi.1999.1459. [DOI] [PubMed] [Google Scholar]
- 44.Snyder S R, Wang J, Waring J F, Ginder G D. J Biol Chem. 2000;275:35112–35120. [Google Scholar]
- 45.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Aufiero B, Schiltz R L, Walsh M J. Proc Natl Acad Sci USA. 2000;97:7166–7171. doi: 10.1073/pnas.130028697. . (First Published June 13, 2000; 10.1073/pnas.130028697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coqueret O, Martin N, Berube G, Rabbat M, Litchfield D W, Nepveu A. J Biol Chem. 1998;271:31452–31457. doi: 10.1074/jbc.273.5.2561. [DOI] [PubMed] [Google Scholar]