Abstract
Membrane lipids modulate the function of membrane proteins. In the case of ion channels, they bias the gating equilibrium, although the underlying mechanism has remained elusive. Here we demonstrate that the N-terminal segment (M0) of the KcsA potassium channel mediates the effect of changes in the lipid milieu on channel gating. The M0 segment is a membrane-anchored amphipathic helix, bearing positively charged residues. In asymmetric membranes, the M0 helix senses the presence of negatively charged phospholipids on the inner leaflet. Upon gating, the M0 helix revolves around the axis of the helix on the membrane surface, inducing the positively charged residues to interact with the negative head groups of the lipids so as to stabilize the open conformation (i.e., the “roll-and-stabilize model”). The M0 helix is thus a charge-sensitive “antenna,” capturing temporary changes in lipid composition in the fluidic membrane. This unique type of sensory device may be shared by various types of membrane proteins.
Keywords: fluorescence measurements, single-channel current, gating kinetics, pH-dependence, activation gate
The cell membrane bears distinctly different kinds of membrane proteins within the matrix of membrane lipids (1), and the lipids are not merely structural building blocks, but substantially modulate the function of membrane proteins (2). The membrane matrix deforms and readily changes its curvature in the manner of an elastic material, and its membrane-embedded proteins are subjected to a variety of physical stresses (3, 4). At the boundary of the matrix, physical effects, such as lateral pressure and tension, modulate the functional properties of the membrane proteins (5–7). In addition to these nonspecific modulating effects, membrane lipids have been suggested to react with specific parts of the membrane proteins. In the fluidic membrane, the lateral diffusion of lipid molecules facilitates the exchange of lipids at the boundary of the membrane proteins, and the membrane matrix is the reaction platform for signal transduction (8).
Data have been reported on the functionally modifying effects of membrane lipids on channel proteins that result in the gating equilibrium being altered (6, 9–13). In voltage-gated channels, the voltage-sensor domain (VSD) primarily senses changes in the membrane electric field, but this sensing is modulated by the lipid composition (9, 11, 13). For the non–voltage-gated (the two-transmembrane-helix inward-rectifier type) channels, such as the KcsA potassium channel from Streptomyces lividans, the underlying mechanism of the effect exerted by lipids is still undergoing extensive investigation, even though the structural information on the channel proteins cocrystallized with lipid molecules has already been reported (14–16).
Here we demonstrate that there is a specialized structural interface in the KcsA potassium channel that senses the membrane milieu and mediates the effect of changes in the lipid composition on channel gating. The N-terminal M0 segment of the KcsA channel is not the membrane spanning region, and functional significance has not been considered. The N-terminal 20 residues around the M0 segment have remained unsolved in the crystal structures of the KcsA channel (14), and this region was even deleted in the course of the successful crystallization of the channel in the open conformation (17). We found that M0 is an amphipathic helix located at the membrane interface and senses changes in the lipid species in the membrane inner leaflet. The M0 helix serves as an unprecedented type of lipid sensor, rendering lipid charge sensitivity. Its unique mode of action upon gating is likely to be shared by various types of channels and membrane proteins in general.
Results
Lipid Sensitivity of the KcsA Channel.
The activation gate of the KcsA potassium channel opens at acidic pH through relaxation of the crossed bundle helices at the cytoplasmic end of the transmembrane domain (18–20). In bacterial-mimetic membranes (e.g., phosphatidylglycerol (PG):phosphatidylethanolamine (PE) = 3:1), the channel is active, whereas in the phosphatidylcholine (PC) membrane, the activity is attenuated, and the presence of negatively charged lipids was demonstrated to be crucial to maintaining channel activity (15, 21, 22). To focus on the effect of the membrane lipid composition on the activation gate of the KcsA channel, the single-channel currents of the noninactivating E71A mutant were examined using planar bilayer membranes (Materials and Methods) (23).
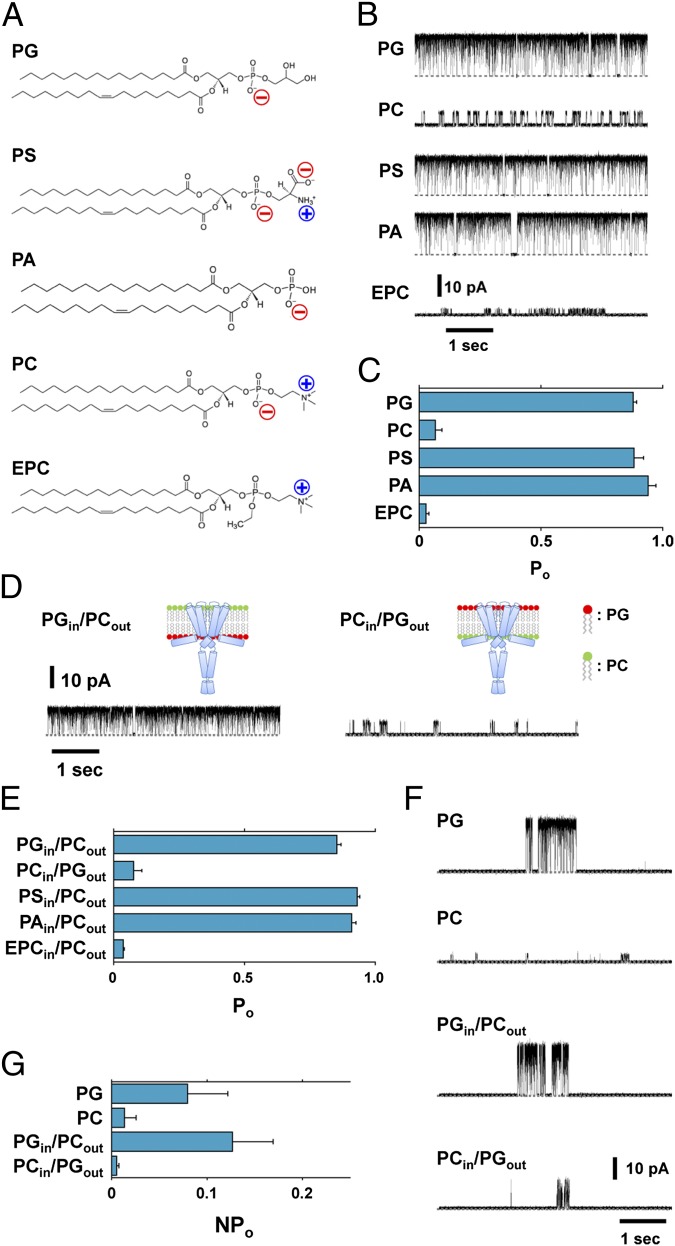
The single-channel currents of the E71A mutant (23) exhibited slow kinetics in the activation gate (Fig. 1B) due to the crossing and relaxing of the bundle helix, as well as frequent brief closures, the mechanism of which is unknown. In the pure PG membrane, the noninactivating KcsA channel stays in the open state most of the time, and the open probability (popen) was shown to be ∼90% (Fig. 1 B and C and Table 1). Other negatively charged lipids that included phosphatidylserine (PS) and phosphatidic acid (PA) (Fig. 1A) rendered the popen value such that it was almost indistinguishable from that in PG. Thus, the net charge on the head group of the phospholipid is important. In contrast, popen was reduced to ∼10% in the PC-containing membrane. Quantitatively similar results were reported for the wild-type (WT) channel (21) (see results for WT in the next section). Furthermore, in the case of the positively charged lipid ethylphosphatidylcholine (EPC) (Fig. 1A), popen was further reduced. These results indicate an electrostatic interaction, rather than a chemical-specific interaction, with the head groups predominating. Kinetic analysis of the single-channel current showed that the mean burst length, representing the lifetime of the open state of the activation gate (24, 25), was significantly shortened in the PC membrane compared with the PG membrane (see Fig. 3D). Changes in the single-channel conductance were evident in membranes having different lipid compositions (Fig. 1B and Table 1). These results are accounted for by the changes in the surface potential introduced by the charged head groups of the phospholipids that exhibit either accumulated or depleted K+ near the membrane surface, either of which changes the conductance (26).
Fig. 1.
Functional effects of membrane lipids on the KcsA channel. (A) Chemical structure of the phospholipids used in this study. (B and C) Single-channel properties of the E71A mutant having different lipid compositions. (B) Single-channel current recordings of different (symmetric) membrane compositions at +100 mV. (C) Open probability in the symmetric membranes. Error bars represent SEM (n = 3–6). (D) Single-channel current recordings in asymmetric membranes at +100 mV. (E) Open probability in asymmetric membranes. Error bars represent SEM (n = 3–8). (F and G) Single-channel properties of the wild-type channel in different membrane compositions. (F) Single-channel current recordings in different (symmetric) membrane compositions at +100 mV. (G) Open probability (Npopen rather than popen) of different compositions of lipids for the WT channels. Error bars represent SEM (n = 3). In these experiments the KcsA channels are oriented such that the cytoplasmic side faces the trans compartment. The solution of each chamber contained 200 mM KCl and the pH was set to 7.5 (cis) or 4.0 (trans). The membrane leaflet that faces the cis compartment is defined as the outer leaflet and that facing trans is the inner leaflet.
Table 1.
Npopen and unitary conductance of WT and mutant channels in the various membrane lipid conditions (n = 3–9)
| Sample | Lipid | Npo | ±SEM | g, pS | ±SD |
| WT | PG | 0.08 | ±0.04 | 160 | ±22 |
| PC | 0.01 | ±0.01 | 27 | ±8 | |
| PGin/PCout | 0.13 | ±0.04 | 212 | ±18 | |
| PCin/PGout | 0.01 | ±0.00 | 119 | ±8 | |
| E71A | PG | 0.88 | ±0.01 | 179 | ±8 |
| PC | 0.07 | ±0.03 | 57 | ±7 | |
| PS | 0.88 | ±0.04 | 169 | ±9 | |
| PA | 0.94 | ±0.03 | 181 | ±7 | |
| EPC | 0.03 | ±0.01 | 22 | ±6 | |
| PGin/PCout | 0.85 | ±0.02 | 106 | ±8 | |
| PCin/PGout | 0.08 | ±0.03 | 57 | ±6 | |
| PSin/PCout | 0.93 | ±0.01 | 159 | ±8 | |
| PAin/PCout | 0.91 | ±0.02 | 170 | ±6 | |
| EPCin/PCout | 0.04 | ±0.00 | 22 | ±7 | |
| PGin/Pcout (pH 4.0in/out) | 0.92 | ±0.03 | 193 | ±7 | |
| PGin/Pcout (pH 4.0in/out) | 0.11 | ±0.06 | 80 | ±7 | |
| PG (pH 3.0) | 0.91 | ±0.00 | 183 | ±7 | |
| PG (pH 5.0) | 0.77 | ±0.02 | 195 | ±9 | |
| PG (pH 6.0) | 0.05 | ±0.01 | 193 | ±8 | |
| PC (pH 3.0) | 0.34 | ±0.03 | 98 | ±6 | |
| E71A C-His Tag | PG | 0.84 | ±0.04 | 202 | ±7 |
| R11Q | PG | 0.59 | ±0.04 | 213 | ±8 |
| K14Q | PG | 0.68 | ±0.05 | 197 | ±9 |
| R11Q/K14Q | PG | 0.26 | ±0.09 | 200 | ±8 |
| R19Q | PG | 0.79 | ±0.04 | 190 | ±9 |
| H20Q | PG | 0.72 | ±0.05 | 191 | ±9 |
| M0-4Q | PG | 0.26 | ±0.02 | 182 | ±11 |
| PG (pH 3.0) | 0.16 | ±0.03 | 148 | ±13 | |
| PG (pH 5.0) | 0.44 | ±0.08 | 181 | ±10 | |
| H25Q/R27Q | PG | 0.89 | ±0.02 | 226 | ±7 |
| Δ10 | PG | 0.80 | ±0.05 | 162 | ±8 |
| Δ22 | PG | 0.33 | ±0.03 | 149 | ±10 |
| R52Q | PG | 0.90 | ±0.01 | 187 | ±8 |
| R64Q | PG | 0.96 | ±0.01 | 164 | ±7 |
| R89Q | PG | 0.91 | ±0.01 | 178 | ±7 |
| PSD-4Q | PG | 0.90 | ±0.00 | 210 | ±8 |
| ΔCPD | PG | 0.75 | ±0.07 | 182 | ±16 |
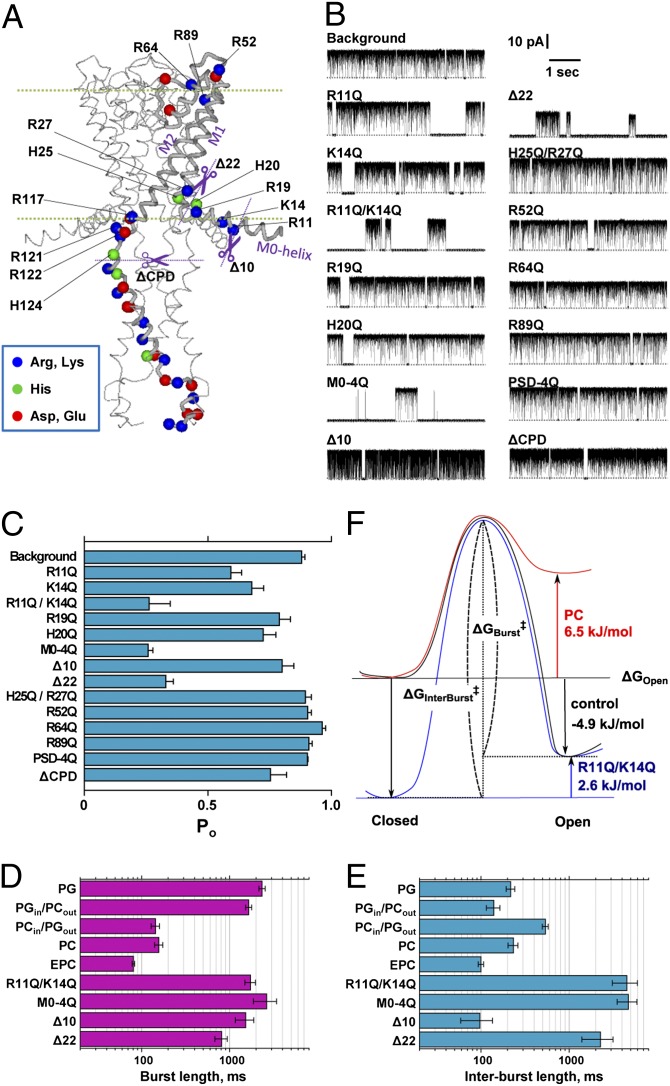
Fig. 3.
Effects of charge neutralization and the deletion mutations on single-channel currents of the E71A mutant channel. (A) Location of the charged amino acid residues in the KcsA channel. For the KcsA mutant (E71A), these charged residues were replaced with glutamine, or the N terminal or the cytoplasmic domain was truncated (purple dotted lines). (B) Representative single-channel current recordings of these mutant channels. Current traces were recorded at 100 mV in the symmetrical PG membrane. (C) Open probability of the mutant KcsA channels. Here, the background represents the KcsA (E71A); M0-4Q indicates the replacement of Arg-11, Lys-14, Arg-19, and His-20 to Gln; PSD-4Q indicates the replacement of Arg-117, -121, -122, and His-124 to Gln; ∆CPD indicates the deletion of the cytoplasmic domain (125–160) by chymotrypsin; ∆10 indicates the deletion of N-terminal 9 amino acids (2–10 aa); ∆22 indicates the deletion of N-terminal 21 amino acids (2–22 aa). Error bars represent SEM (n = 3–9). (D and E) Kinetic analyses of the burst (D) and interburst length (E). Error bars represent ±SEM (n = 3–6). (F) Hypothetical energy profiles for the closed and open transition states. Free energy level of the open state relative to the closed state (ΔGOpen) was calculated from the popen values, and the barrier height or the activation energy (ΔG‡: ΔGBurst‡ and ΔGInterBurst‡) was calculated using the rate constants of the transitions (the reciprocals of the mean burst length and mean interburst length). ΔGBurst‡ was 77.5 kJ/mol, 68.9 kJ/mol, and 74.8 kJ/mol for PG, PC, and R11Q/K14Q. ΔGInterBurst‡ was 69.7 kJ/mol, 69.8 kJ/mol, and 77.4 kJ/mol for PG, PC, and R11Q/K14Q.
Lipids on the Inner Leaflet Govern the Activation Gate.
To address the effect of negatively charged lipids on either the outer or inner leaflet of the membrane, asymmetric membranes obtained through the folding method were used (Materials and Methods) (27). The single-channel current revealed that popen was decreased drastically when the inner leaflet contained PC (Fig. 1D), and popen in this asymmetric membrane (PCin/PGout) was indistinguishable from that in the symmetrical PC membrane (Fig. 1E and Table 1). In contrast, the popen value was ∼90% in the membrane having PG in the inner leaflet, irrespective of the lipid composition on the outer leaflet. Moreover, the single-channel kinetics showed that the burst length in the PGin/PCout (or PCin/PGout) membrane was similar to that in the pure PG (or PC) membrane, respectively (see Fig. 3D). For the WT KcsA channel, Fig. 1 F and G shows that a substantial reduction in Npopen (N: the apparent number of active channels on the membrane; Npopen rather than popen was used because of the low popen value in the WT channel.) was observed in the membrane with PC on the inner leaflet, whether the outer leaflet contained either PC or PG. These results demonstrate unequivocally that negatively charged lipids on the inner rather than the outer leaflet govern the activation of gating by stabilizing the open state. This is in contrast to previous reports (15, 22, 28, 29), which have related the gating feature to the crystal structural data that PGs are bound at the outer half of the membrane-embedded surface of the KcsA channel.
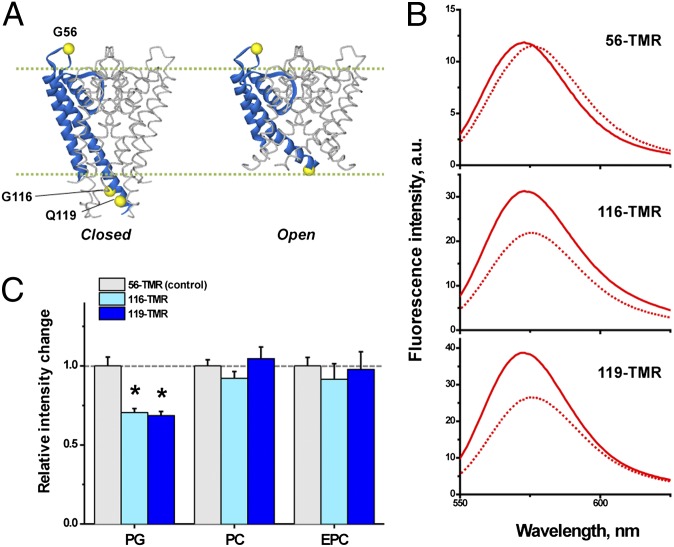
The structural changes of the activation gate in different membrane environments were examined using a fluorescence method. In earlier reports, a fluorescence probe, tetramethylrhodamine (TMR), labeled residues close to the bundle crossing (G116 and Q119, Fig. 2A) (19, 30). Blunck et al. (30) reported that upon the opening of the activation gate, the fluorescence intensity of TMR attached to the 116 and 119 sites decreased substantially because these sites become more hydrophilic in the open conformation. Using the same methods as this group, the TMR-labeled KcsA channel was reconstituted into PG liposomes, and the TMR spectra were compared at pH 7.5 and pH 4 (Fig. 2B and Materials and Methods). Fig. 2C shows that the peak intensity of the fluorescence decreased ∼30% at pH 4.0 relative to pH 7.5 when labeled at the 116 and 119 sites, but not at the 56 site, which is located in the extracellular loop (Fig. 2A). These results indicate that the opening of the activation gate was successfully detected using the fluorescence method.
Fig. 2.
Gating conformational changes monitored with the fluorescence signal. (A) Three TMR-labeled sites of the KcsA channel for the closed (Left) and open (Right) conformations. TMR was reacted with the mutated cysteine residue using a maleimide derivative (TMRM). Note that the experiments were performed for the full-length channel. (B) Fluorescence spectra of the TMR-labeled KcsA channels. KcsA channel was reconstituted in the PG liposomes at pH 7.5 (solid lines) or 4.0 (dotted lines). (C) Relative peak intensity of the fluorescence at pH 4.0 and pH 7.5 for the different lipid compositions. Light gray columns indicate the results of 56-TMR, while those in light blue are 116-TMR, and those in dark blue are 119-TMR. Values were standardized using the control sample (56-TMR) for each lipid condition. Error bars represent SD (n = 4–5, *P < 0.01 by paired t test).
In the PC- and EPC-containing membranes, changes in the relative fluorescence intensity between the two pH values were very small, if any (Fig. 2C and Fig. S1), indicating that the activation gate opens infrequently in these membranes. These fluorescence data demonstrated that the dramatic changes in popen observed in the single-channel currents in the different lipid compositions in the inner leaflet reflect the gating transitions of the activation gate.
M0 Is the Lipid Sensor.
How is the inner lipid composition recognized by the channel, and by what means is the status of the activation gate changed? One may postulate a mechanism in which negatively charged lipids on the inner leaflet interact with the positively charged structural parts of the channel, which in turn impinges on the activation of the gate. First, we introduced single or multiple charge-neutralizing mutations into all of the positive charges in the KcsA (E71A) channel (Fig. 3A) and then measured the single-channel current in the PG membrane for evaluating the popen value (Fig. 3B).
The bulge helix (31) (residues 118–135) in the cytoplasmic domain, bearing the pH-sensing charged residues and located close to the inner membrane, is the most plausible interaction site. Contrary to our expectations, the four positively charged residues (R117, R121, R122, and H124) on the bulge helix did not affect the popen value [Fig. 3 B and C, PSD (pH-sensor domain)-4Q]. In contrast, neutralizing the positive charges on the M0 segment at the N terminal yielded a drastic effect. Either the R11Q or K14Q mutant in the M0 segment reduced the popen value (Fig. 3C). Moreover, the double mutant, R11Q–K14Q, exhibited further decreases in popen. Additional neutralization of up to four positive charges around the M0 segment (M0-4Q), however, did not further decrease the popen value. Note that, despite the low popen value, the M0-4Q mutant retained the normal pH dependency (Table 1). Neutralizing other positively charged residues, including two at the N-terminal end of the M1 helix (H25Q/R27Q) (32), retained high popen values. Also, truncation of the cytoplasmic domain (∆CPD) did not affect popen. The histidine tag attached to either the N terminal or the C terminal did not make a difference to the popen values.
Next, the effects of deletion of portions of the N terminal on gating were examined. Deletion of the N-terminal 22 residues (∆22) decreased popen to a level similar to that of the double mutant R11Q/K14Q, suggesting that the electrostatic effect is dominant in the interaction between the M0 helix and the membrane lipid. When the deletion was restricted to only the terminal 10 residues (∆10) and the two positively charged residues in the rest of the M0 helix were present, a high popen value was maintained (Fig. 3 B and C). The results of the charge-neutralizing mutations and the truncations indicate that two positively charged residues on the M0 helix interact with negatively charged lipid head groups on the inner leaflet in an additive fashion, and thus stabilize the open conformation.
Mechanism of the Lipid Sensitivity.
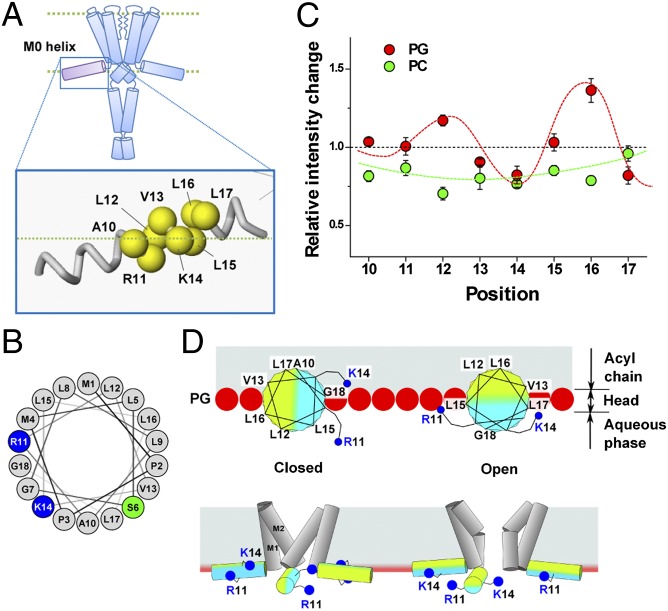
The structure of the N-terminal M0 segment remains unsolved crystallographically, although electron paramagnetic resonance (EPR) measurements have shown that it forms an α-helix, runs radially from the longitudinal axis of the channel, and resides at the membrane interface (Fig. 4A) (33). The M0 helix, defined as the N-terminal 18 residues, has a characteristic amphipathic feature (Fig. 4B) (33): The M0 helix is rich in hydrophobic leucine residues that cover more than a half of one side, whereas on the other side, M0 has two positively charged residues (R11 and K14) and compact glycine and alanine residues. The amphipathic nature of the M0 helix, as well as the positively charged residues of the M0 helix, prompted us to perform an experiment to address how the M0 helix senses the charges of the lipid head group and mediates messages to the activation gate.
Fig. 4.
Conformational changes of the M0 helix upon gating via a monitoring of the fluorescence signal. (A) TMR-labeled sites on the M0 helix. (B) Helical wheel projection of the M0 helix (residues 1–18). (C) Relative intensity changes of the TMR at pH 4.0 and pH 7.5. Red and green circles indicate the results in the PG and PC liposomes. Red curve delineates the periodic changes, with 3.6 aa per turn. This periodicity is typical of an α-helix. Fluorescence values were standardized by means of a control sample (56-TMR) for each lipid condition. Error bars represent ±SD (n = 3–7). (D) Roll-and-stabilize model. (Upper) Helical wheel in the C-terminal half of the M0 helix located at the membrane interface as viewed from the N-terminal side. In the PG membrane, K14 and L17 are exposed to the aqueous environment in the open state, whereas L12 and L16 are turned to the hydrophobic milieu. Red spheres represent the negatively charged head groups of PG, and blue dots represent the positively charged head of the amino acid residues (R11 and K14). (Lower) Amphipathic M0 helix, represented by two-color rods, revolves around the axis by ∼90° upon gating, and the positively charged residues (R11 and K14) interact with the negative head groups of the lipids.
The effect of the M0 helix upon the gating that occurs at different membrane compositions was examined using the aforementioned fluorescence measurement technique. Eight positions on the M0 helix (Fig. 4A) were TMR labeled one by one and the relative changes in the fluorescence intensity were examined at acidic and neutral pH, which primarily represents changes in the hydrophobic milieu of the relevant site. We found surprisingly that the fluorescence intensity in the PG membrane changed periodically along the sequence (Fig. 4C). For instance, three of the C-terminal residues (residues 15–17) are all leucine, and the fluorescence intensity of the L16 site increased and that of the L17 site decreased, whereas that of the L15 site remained unaltered. The reciprocal change in the adjacent sites suggests that M0 undergoes a change in orientation. The overall changes along the sequence can be fitted by a periodic function of 3.6 periodicity. These results support the contention that M0 forms an α-helix. Moreover, the periodic fluorescence change at two different pH levels suggests that the M0 helix is anchored at the membrane interface, where it changes in orientation such that a different side of the helix faces to the hydrophobic core of the membrane under different conditions. This indicates the M0 helix exhibits a revolving motion around the helix axis. The periodic change in the fluorescence intensity gradually became augmented toward the C-terminal end of the M0 helix, suggesting that the relevant segment is buried deep in the hydrophobic core of the membrane.
In contrast to the signal pattern in the PG membrane, the fluorescence intensity in the PC membrane nearly lost its periodicity along the M0 helix (Fig. 4C). It should be noted that the opening of the activation gate is infrequent in the PC membrane as shown by the low popen value of the single-channel recordings (Fig. 1 B and C) and the fluorescence measurements (Fig. 2C). Thus, the measured fluorescence signal of the M0 helix originates predominantly from the closed configuration, and that from the open state is a minor component. Accordingly, the absence of the pattern in the PC membrane indicates that the M0 helix did not revolve substantially upon the pH change, staying mostly closed even at an acidic pH.
To obtain further insights into the revolving changes of the M0 helix, a helical wheel was drawn for the C-terminal half of the M0 helix (Fig. 4D). L12 and L16 face the hydrophilic milieu, and are turned to face the hydrophobic milieu upon the change to acidic pH, whereas V13, K14, and L17 are changed to face in the opposite direction. These two groups are arranged on opposite sides of the helical wheel, so that, upon pH change, the M0 helix revolves clockwise more than 90° around the helix axis. At acidic pH, the M0 helix takes the most stable revolving orientation. The hydrophobic residues are buried deep in the hydrophobic core of the membrane and the positively charged residues in the aqueous phase interact readily with the negatively charged head group at the membrane interface. We call this the roll-and-stabilize model, and to the best of our knowledge this mechanism is unique and has never previously been proposed.
Energetics of the Sensor Mechanism.
Kinetic analyses of the single-channel current (Fig. 3 D and E) as well as the popen values (Fig. 3C and Table 1) have provided mechanistic insight from the energetic point of view. The burst and interburst lengths represent the kinetics of the activation gate and were expressed as the free energy profiles of the closed–open transition (Fig. 3F). Compared with the control condition (the black profile), replacing the charged lipids with neutral lipids decreased both the popen value and the burst length, but did not alter the interburst length (Fig. 3, legend), which is a condition that is characteristic of the destabillization of the open state (the red profile). Thus, in the PG membrane the positively charged residues (R11 and K14) on the M0 helix, when facing the aqueous phase, interact with the negative head groups of the inner membrane so as to stabilize the open state. In the case of charge-neutralizing mutants (R11Q/K14Q), similar decreases in the popen values were found, but with an increased interburst length. This indicates the closed state was substantially stabilized (the blue profile), suggesting that the K14 residue remains unfavorably located in the hydrophobic core of the membrane in the closed conformation. “Snorkeling” is a mechanism by which positively charged residues, with their long side chain, reach the aqueous environment, even if they are buried in the hydrophobic core of the membrane (34, 35), but this seems unlikely in the case of K14 in the closed conformation.
Discussion
In this study we report the discovery of sensory machinery for membrane inner lipids. The M0 helix is a unique amphipathic helix in the KcsA channel, but has not been the focus of attention thus far, presumably due to the persistent lack of crystallographic data for this region (15, 22, 26). The M0 helix bears only positive and not negative charges, and the revolving action is not likely to be initiated in its own right by acidic changes. Rather, the revolving motion of the M0 helix is driven by the conformational change of the membrane-spanning (M1 and M2) helices undergoing the activation gate opening process. Once it has revolved clockwise over 90° around the helix axis, the charged residues, R11 and K14, readily interact with the negatively charged head groups of the inner lipids and stabilize the helical rolling motion, and the anchored helices, in turn, stabilize the open conformation (Fig. 4D).
Only a short stretch (the C-terminal half) of the M0 helix is required for the roll-and-stabilize action, a situation which was revealed by the deletion results (Fig. 3). This short helix is analogous to the interfacial (IF) helix of the two-transmembrane Kir channels in the transmembrane domain. The sequence alignment that is shown in Fig. 5 reveals a certain similarity in the distribution of the positively charged residues that corresponds to R11 and K14 in the M0 helix. These charges are crucial sites for PIP2-binding in the Kir channels (16, 36, 37). In the crystal structure of Kir2.2, the IF helix (residues 61–69) forms an α-helix that corresponds to the C-terminal half of M0 (numbers 11–18), and R65 in the IF helix (corresponding to K14 in M0) interacts with the tether helix in the cytoplasmic domain. On the other hand, the KcsA channel has the least bulky CPD among the K channels, and the open-stabilizing effect of M0 is directed toward the charged lipids on the inner leaflet. Most cell membranes bear negatively charged lipids on the inner leaflet (38, 39), and the M0 helix favors charge sensitivity rather than specific interactions of a chemical nature with the lipid head groups. Thus, the KcsA channel exploits inner membrane lipids as an integral structure in the stabilization of the channel conformation.
Fig. 5.
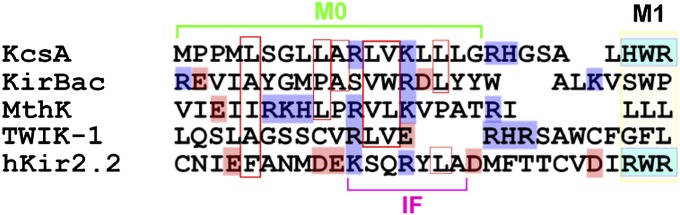
Alignment of the amino acid sequence of the N-terminal region of the KcsA and two-transmembrane channels. Amino acids having a net charge are depicted in color (Right): blue, positive; pink, negative. Numbers indicate the amino acid 28 sequence of KcsA. IF represents the interfacial helix (numbers 61–69) assigned from the crystal structure of Kir2.2 [Protein Data Bank, (PDB) ID code 3SPI]. IF helix corresponds to the C-terminal half of M0 (numbers 11–18), and R65 in the IF helix (corresponding to K14 in KcsA) interacts with the tether helix in the cytoplasmic domain (number 16).
The unprecedented roll-and-stabilize mechanism is operated with a prevalent motif of the amphipathic helix, and one finds similar amphipathic helices in membrane proteins as a linker between transmembrane helices. A candidate is the amphipathic helix of voltage-gated channels (40, 41), and possible involvements of the roll-and-stabilize mechanism should be examined in future. We suggest that this machinery is also implemented in various types of membrane proteins. Our findings help to unveil the underlying mechanism of the lipid–protein interaction.
Materials and Methods
Sample Preparation.
The expression, purification, and reconstitution into liposomes of the wild-type and mutant KcsA channels were described previously (42). The proteoliposomes were made with a protein/lipid weight ratio of 1:2,000 and were passed through a Bio-Spin30 column (Bio-Rad) equilibrated with buffer solution (200 mM KCl, 10 mM Hepes, pH 7.5). The following phospholipids were used in this study (Fig. 1A): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (POPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (sodium salt) (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (sodium salt) (POPA), and 1-palmitoyl-2-oleoyl-sn-glycero-3-ethylphosphocholine (chloride salt) (POEPC).
All lipids were purchased from Avanti Polar Lipids.
Single-Channel Current Recordings of the KcsA Channels.
A planar lipid bilayer method was used for the single-channel current measurements, as reported previously (42). A Teflon sheet of 0.5-mm thickness, with an aperture of <100-μm diameter was used as a partition for the two compartments (cis and trans) of a chamber. A planar lipid bilayer formed on the aperture as a result of an application of 20 mg/mL n-hexadecane (Nacalai) solution of the desired phospholipids. The asymmetric lipid bilayer was formed by the folding method (27). The cis and trans chambers were filled with the electrolyte solution below the aperture. A lipid monolayer was formed on the surface of each solution by dropping an aliquot of the desired phospholipid solution dissolved in hexane (5 mg/mL). After a few minutes, the level of the surface on one side was raised above the aperture by adding the electrolyte solution, followed by raising the surface level on the other side. The electrolyte solution in the cis and trans chambers contained 200 mM KCl buffered by 10 mM Hepes (pH 7.5) and 10 mM succinic acid (pH 4.0), respectively. The reference electrode was placed in the cis chamber, to which KcsA channels embedded in the proteoliposomes were added. The phospholipid composition of the proteoliposomes was set so as to be identical to that of the cis leaflet. The current data were recorded and stored in a personal computer by using pCLAMP software (Molecular Devices) through an Axopatch 200B amplifier and Digidata 1322A digitizer (Molecular Devices). The low pass filter was set to 2 kHz for the cutoff frequency, and the data were sampled at 5 kHz. For the burst length analysis, an algorithm, “Poisson surprise,” was used (43). The observed burst length and the interburst length were converted to the activation energy (ΔGBurst‡ and ΔGInterBurst‡; the double dagger symbol represents the activation energy when used with ΔG) using the following equation:
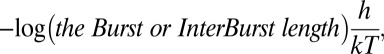 |
where h is the Planck constant, k the Boltzmann constant, and T the absolute temperature (44).
TMR Labeling of the KcsA Channel.
Cysteine-introduced mutants of the KcsA channel, which possesses only one cysteine at the mutated site, were used for labeling of the fluorescent probe. Labeling of these channels with TMR was achieved as follows. Purified cysteine mutants in the buffer solution (0.06% n-dodecyl-β-D-maltoside (DDM), 200 mM KCl, 10 mM Hepes, pH 7.5) were incubated with a 10× molar ratio of tetramethylrhodamine-5-maleimide (TMRM) (AnaSpec) at room temperature. After 2 h, 20 mM of 2-mercaptoethanol were added to complete the reaction. Excess fluorescent dyes were removed by purifying the labeled channels with a Co2+-based affinity gel column. The absorption ratio of 550 nm/280 nm demonstrated that approximately one molecule of TMR dye binds to one KcsA monomer.
Fluorescence Measurement of the TMR-Labeled KcsA Channels.
TMR-labeled channels were reconstituted into liposomes (at a protein/lipid weight ratio of 1:2,000) in an unbuffered 200 mM KCl solution. A half aliquot of the proteoliposome suspension (90 μL of approximately 2 mg lipid/mL) was mixed with 10 μL of a concentrated buffer (0.5 M) of either Hepes or succinic acid to achieve a defined pH (7.5 or 4.0). The suspensions were sonicated thoroughly in a bath sonicator (Sonorex RK31; Bandelin Electronic) to equilibrate internal and external solution of the proteoliposomes. TMR was excited at 532 nm and emission spectra were measured with a fluorescence spectrometer (FP-8200; Jasco). Pairs of data (pH 7.5 and pH 4.0) were obtained from the same preparation of proteoliposome suspension, and fluorescence intensity change was estimated in each dataset. More than five independent datasets were analyzed for each experimental condition.
Supplementary Material
Acknowledgments
We thank Drs. Andy James (University of Bristol), Kunihiko Ishii (RIKEN), Yasushi Sako (RIKEN), Masao Miki (University of Fukui), and Takashi Sumikama (University of Fukui) for discussion. Pacific Edit reviewed the manuscript prior to submission. This work was partially funded by Grants 23370067, 21107508, 21657038, and 20247016 (to S.O.) and 11020017 (to M.I.) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217323110/-/DCSupplemental.
References
- 1.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438(7068):578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 2.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666(1-2):62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Andersen OS, Koeppe RE., 2nd Bilayer thickness and membrane protein function: An energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 4.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459(7245):379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantor RS. Lipid composition and the lateral pressure profile in bilayers. Biophys J. 1999;76(5):2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tillman TS, Cascio M. Effects of membrane lipids on ion channel structure and function. Cell Biochem Biophys. 2003;38(2):161–190. doi: 10.1385/CBB:38:2:161. [DOI] [PubMed] [Google Scholar]
- 7.Imai S, et al. Functional equilibrium of the KcsA structure revealed by NMR. J Biol Chem. 2012;287(47):39634–39641. doi: 10.1074/jbc.M112.401265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, Maxfield FR. Membrane domains. Annu Rev Cell Dev Biol. 2004;20:839–866. doi: 10.1146/annurev.cellbio.20.010403.095451. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt D, Jiang Q-X, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444(7120):775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 10.Gamper N, Shapiro MS. Regulation of ion transport proteins by membrane phosphoinositides. Nat Rev Neurosci. 2007;8(12):921–934. doi: 10.1038/nrn2257. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Ramu Y, Lu Z. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature. 2008;451(7180):826–829. doi: 10.1038/nature06618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475(7356):353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng H, Liu W, Anderson LY, Jiang Q-X (2011) Lipid-dependent gating of a voltage-gated potassium channel. Nat Commun 2:250. [DOI] [PMC free article] [PubMed]
- 14.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414(6859):43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 15.Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41(35):10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 16.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477(7365):495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466(7303):203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu YS, Sompornpisut P, Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol. 2001;8(10):883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 19.Blunck R, Cordero-Morales JF, Cuello LG, Perozo E, Bezanilla F. Detection of the opening of the bundle crossing in KcsA with fluorescence lifetime spectroscopy reveals the existence of two gates for ion conduction. J Gen Physiol. 2006;128(5):569–581. doi: 10.1085/jgp.200609638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu H, et al. Global twisting motion of single molecular KcsA potassium channel upon gating. Cell. 2008;132(1):67–78. doi: 10.1016/j.cell.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 21.Heginbotham L, Kolmakova-Partensky L, Miller C. Functional reconstitution of a prokaryotic K+ channel. J Gen Physiol. 1998;111(6):741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marius P, et al. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys J. 2008;94(5):1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13(4):311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 24.Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating II: Single-channel currents. J Gen Physiol. 2007;130(5):479–496. doi: 10.1085/jgp.200709844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colquhoun D, Hawkes AG. On the stochastic properties of bursts of single ion channel openings and of clusters of bursts. Philos Trans R Soc Lond B Biol Sci. 1982;300(1098):1–59. doi: 10.1098/rstb.1982.0156. [DOI] [PubMed] [Google Scholar]
- 26.Park JB, Kim HJ, Ryu PD, Moczydlowski E. Effect of phosphatidylserine on unitary conductance and Ba2+ block of the BK Ca2+-activated K+ channel: Re-examination of the surface charge hypothesis. J Gen Physiol. 2003;121(5):375–397. doi: 10.1085/jgp.200208746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montal M, Mueller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc Natl Acad Sci USA. 1972;69(12):3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marius P, de Planque MRR, Williamson PTF. Probing the interaction of lipids with the non-annular binding sites of the potassium channel KcsA by magic-angle spinning NMR. Biochim Biophys Acta. 2012;1818(1):90–96. doi: 10.1016/j.bbamem.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vales E, Raja M. The “flipped” state in E71A-K+-channel KcsA exclusively alters the channel gating properties by tetraethylammonium and phosphatidylglycerol. J Membr Biol. 2010;234(1):1–11. doi: 10.1007/s00232-010-9234-9. [DOI] [PubMed] [Google Scholar]
- 30.Blunck R, McGuire H, Hyde HC, Bezanilla F. Fluorescence detection of the movement of single KcsA subunits reveals cooperativity. Proc Natl Acad Sci USA. 2008;105(51):20263–20268. doi: 10.1073/pnas.0807056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uysal S, et al. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci USA. 2009;106(16):6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007;282(20):15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 33.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: Role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117(2):165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strandberg E, Killian JA. Snorkeling of lysine side chains in transmembrane helices: How easy can it get? FEBS Lett. 2003;544(1–3):69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 35.Strandberg E, et al. Lipid dependence of membrane anchoring properties and snorkeling behavior of aromatic and charged residues in transmembrane peptides. Biochemistry. 2002;41(23):7190–7198. doi: 10.1021/bi012047i. [DOI] [PubMed] [Google Scholar]
- 36.Lopes CMB, et al. Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron. 2002;34(6):933–944. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 37.Xie L-H, John SA, Ribalet B, Weiss JN. Activation of inwardly rectifying potassium (Kir) channels by phosphatidylinosital-4,5-bisphosphate (PIP2): Interaction with other regulatory ligands. Prog Biophys Mol Biol. 2007;94(3):320–335. doi: 10.1016/j.pbiomolbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Bretscher MS. Asymmetrical lipid bilayer structure for biological membranes. Nat New Biol. 1972;236(61):11–12. doi: 10.1038/newbio236011a0. [DOI] [PubMed] [Google Scholar]
- 39.Verkleij AJ, et al. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim Biophys Acta. 1973;323(2):178–193. doi: 10.1016/0005-2736(73)90143-0. [DOI] [PubMed] [Google Scholar]
- 40.Lee S-Y, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci USA. 2005;102(43):15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450(7168):376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 42.Iwamoto M, et al. Surface structure and its dynamic rearrangements of the KcsA potassium channel upon gating and tetrabutylammonium blocking. J Biol Chem. 2006;281(38):28379–28386. doi: 10.1074/jbc.M602018200. [DOI] [PubMed] [Google Scholar]
- 43.Legéndy CR, Salcman M. Bursts and recurrences of bursts in the spike trains of spontaneously active striate cortex neurons. J Neurophysiol. 1985;53(4):926–939. doi: 10.1152/jn.1985.53.4.926. [DOI] [PubMed] [Google Scholar]
- 44.Glasstone S, Laidler KJ, Eyring H. The Theory of Rate Processes: The Kinetics of Chemical Reactions, Viscosity, Diffusion and Electrochemical Phenomena. New York: McGraw-Hill; 1941. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.