Abstract
Lipin 1 is a coregulator of DNA-bound transcription factors and a phosphatidic acid (PA) phosphatase (PAP) enzyme that catalyzes a critical step in the synthesis of glycerophospholipids. Lipin 1 is highly expressed in adipocytes, and constitutive loss of lipin 1 blocks adipocyte differentiation; however, the effects of Lpin1 deficiency in differentiated adipocytes are unknown. Here we report that adipocyte-specific Lpin1 gene recombination unexpectedly resulted in expression of a truncated lipin 1 protein lacking PAP activity but retaining transcriptional regulatory function. Loss of lipin 1-mediated PAP activity in adipocytes led to reduced glyceride synthesis and increased PA content. Characterization of the deficient mice also revealed that lipin 1 normally modulates cAMP-dependent signaling through protein kinase A to control lipolysis by metabolizing PA, which is an allosteric activator of phosphodiesterase 4 and the molecular target of rapamycin. Consistent with these findings, lipin 1 expression was significantly related to adipose tissue lipolytic rates and protein kinase A signaling in adipose tissue of obese human subjects. Taken together, our findings identify lipin 1 as a reciprocal regulator of triglyceride synthesis and hydrolysis in adipocytes, and suggest that regulation of lipolysis by lipin 1 is mediated by PA-dependent modulation of phosphodiesterase 4.
Lipin 1 is a bifunctional intracellular protein that is emerging as a critical regulator of metabolism (1, 2). Lipin 1 dephosphorylates phosphatidic acid (PA) to form diacylglycerol (DAG) (3), which is the penultimate step in triglyceride (TG) synthesis and a pivotal step in the synthesis of glycerophospholipids. In addition to its role as a metabolic intermediate, PA functions as an intracellular messenger to regulate signaling cascades and vesicular transport processes (4, 5). Intracellular DAG also affects insulin signaling by activating protein kinase C isoenzymes, and may serve as a mechanistic link between tissue lipid accumulation and insulin resistance (6).
Interestingly, lipin 1 also regulates metabolism by acting as a coregulatory protein that translocates to the nucleus and regulates the activity of DNA-bound transcription factors (7). Lipin 1 interacts with members of the peroxisome proliferator-activated receptor (PPAR) family (7, 8) and enhances their activity by recruiting other coactivator proteins. Lipin 1 also has been shown to repress the activity of nuclear factor of activated T cells c4 (NFATc4) to inhibit cytokine production in adipocytes (9). Although lipin 1’s effects on PPARs and NFATc4 are not dependent on its PAP activity (7), the repression of sterol regulatory element-binding protein 1 transcriptional activity by lipin 1 requires both lipin 1 phosphatidic acid phosphohydrolase (PAP) enzymatic activity and translocation to the nucleus (10). Thus, lipin 1 acts as a scaffold protein that can confer context-specific transactivating or transrepressing effects on gene transcription.
Naturally occurring mutations in lipin 1 have been identified as the genetic defects underlying the phenotype of two distinct lines of phenotypically similar fatty liver dystrophic (fld) mice (11). Fld mice are characterized by the near absence of adipose tissue (12). Lipin 1 is highly expressed in adipose tissue, and its expression is induced early in the adipocyte differentiation program (13). Lipin 1 is required for the process of adipogenesis. Treatment of embryonic fibroblasts from fld mice with an adipogenic mixture in vitro fails to induce the expression of adipogenic markers and neutral lipid droplet formation (13), and both lipin 1-mediated PAP activity and nuclear localization are required for induction of adipogenesis in fibroblasts (14). Lipin 1 likely serves to coordinate the process of adipogenesis by regulating the transcription of genes encoding adipogenic factors in the nucleus and increasing the capacity for glyceride storage in the cytoplasm by acting as a PAP enzyme.
Because constitutive loss of lipin 1 in fld mice results in a global failure of adipogenic differentiation, the metabolic effects of lipin 1 in differentiated adipocytes in vivo remain unknown. We evaluated the effects of lipin 1 deficiency in differentiated adipocytes using an adiponectin promoter-driven Cre (15). Phenotypic characterization of these mice confirmed the important role of lipin 1-mediated PAP activity in adipocyte intermediary fat metabolism and revealed an unanticipated role in regulating adipocyte lipolysis.
Results
Generation of Mice with Adipocyte-Specific Loss of Lipin 1-Mediated PAP Activity.
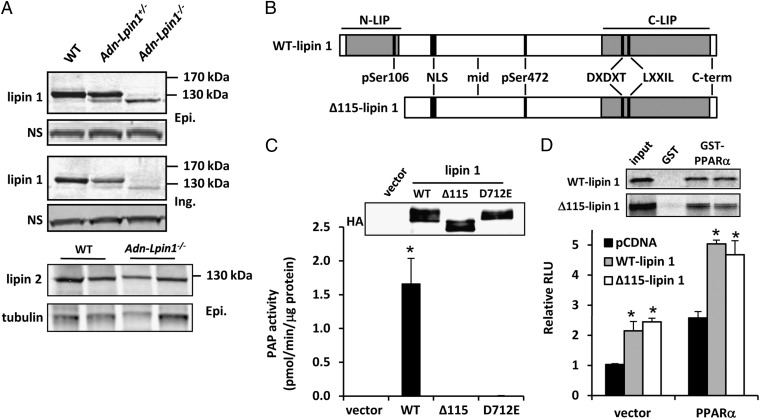
Mice with LoxP sites flanking exons 3 and 4 of the Lpin1 gene (Lpin1flox/flox mice) (5) were mated with mice expressing Cre recombinase driven by the promoter of the gene encoding adiponectin (15). Lipin 1 mRNA, as measured by primers annealing to the deleted exons, was reduced by 95% in adipose tissue, but not in other tissues (Fig. S1A). However, Western blot analyses using an antibody directed against the C terminus of the lipin 1 protein and protein lysates from white adipose tissue (WAT; epididymal or inguinal) depots of AdnCre+-Lpin1−/− or AdnCre+-Lpin1+/− mice detected a band, albeit significantly less abundant, that migrated ∼10% faster than the WT protein (Fig. 1A). This faster-migrating band was not observed in lysates from fld tissues and does not correspond to lipin 2 (Fig. 1A).
Fig. 1.
A truncated lipin 1 protein is expressed in Adn-Lpin1−/− mice. (A) Western blot analysis for lipin 1 and lipin 2 in WAT [epididymal (Epi.) and inguinal (Ing.)] of 6- to 8-wk-old male WT, Adn-Lpin1+/−, and Adn-Lpin1−/− mice. (B) Schematic depicting the domain structure of full-length and Δ115-lipin 1. The approximate binding locations of the antibodies used in Fig. S1 are shown. DXDXT, PAP catalytic site; LXXIL, nuclear receptor interaction domain; NLS, nuclear localization sequence/polybasic domain; N-LIP, N-terminus lipin homology domain; C-LIP, C-terminus lipin homology domain. (C) Graph showing Mg2+-dependent PAP activity of the indicated proteins overexpressed in 293 cells and immunopurified with HA antibody. Western blots to detect overexpressed protein are shown. (D) Graph showing luciferase activity in transfection studies using the PPAR-responsive acyl CoA oxidase-thymidine kinase luciferase reporter construct and the indicated expression constructs. GST-pull down images are shown above graph.
Hepatic lysates from fasted WT mice and Lpin1flox/flox mice expressing Cre under control of the albumin promoter (Alb-Lpin1−/−) were subjected to immunoprecipitation with an antibody against the C terminus of lipin 1 and immunoprecipitates blotted with an antibody against amino acids 377–397 of lipin 1. The protein precipitated by the C-terminal antibody was also detected by a second lipin 1-specific antibody (Fig. S1B). Two alternative translational start sites were found in-frame in exon 5 of the Lpin1 gene that would encode a protein lacking the first 100 or 115 amino acids (Fig. S2). An antibody against phospho-Ser106 lipin 1 detected WT lipin 1, but not the truncated protein precipitated from liver lysates from insulin-injected Alb-Lpin1−/− mice (Fig. S1B). In contrast, anti–phospho-Ser472 lipin 1 readily detected lipin 1 in WT and Alb-Lpin1−/− lysates. An expression vector driving expression of a cDNA beginning at exon 5 without an enforced start codon also produced a protein that was not detected by the phospho-Ser106 lipin 1 antibody, but was detected by anti–phospho-Ser472 (Fig. S1B). These data suggest that the mutant protein is lacking the 115 N-terminal amino acids that compose the entire highly conserved N-terminus lipin (N-LIP) domain (Fig. 1B) and are consistent with the presence of consensus Kozak sequence for the second, but not the first, alternative start site (Fig. S2).
Although the catalytic site for PAP enzymatic activity is in the C terminus of the protein, PAP activity of Δ115-lipin 1 was indistinguishable from that of a previously identified catalytically inactive mutant (lipin 1 D712E) when the overexpressed proteins were isolated by HA-tag immunoprecipitation (Fig. 1C), consistent with other N-terminal deletion mutants (16). However, the Δ115-lipin 1 protein retained the ability to interact with and coactivate PPARα in GST pull-down assays and PPAR-responsive acyl CoA oxidase-thymidine kinase luciferase reporter assays, respectively (Fig. 1D). The expression of several genes known to be directly regulated by lipin 1 (e.g., Pepck, Fabp4, Tnfa) (8, 9) was not affected in adipose tissue of Adn-Lpin1−/− mice (Fig. S1C). Although PPARγ expression was significantly reduced in Adn-Lpin1−/− mice, many known PPARγ target genes (e.g, Cd36, Fsp27, Lpl) were not affected. In addition, other known direct lipin 1 target genes (Acadm and Ppara) were actually induced in Adn-Lpin1−/− mice. Taken together, these findings suggest that Δ115-lipin 1 is enzymatically inactive but retains at least some of its function as a transcriptional coactivator.
Adn-Lpin1−/− Mice Exhibit Diminished Adiposity.
Adn-Lpin1−/− mice were born at the expected frequency. Like fld mice (17, 18), Adn-Lpin1−/− mice exhibited neonatal hepatic steatosis, whereas Alb-Lpin1−/− mice did not (Fig. S3). However, also like fld mice, hepatic steatosis in Adn-Lpin1−/− mice resolved before weaning, and intrahepatic TG content in 6-wk-old mice was not different from that in littermate controls.
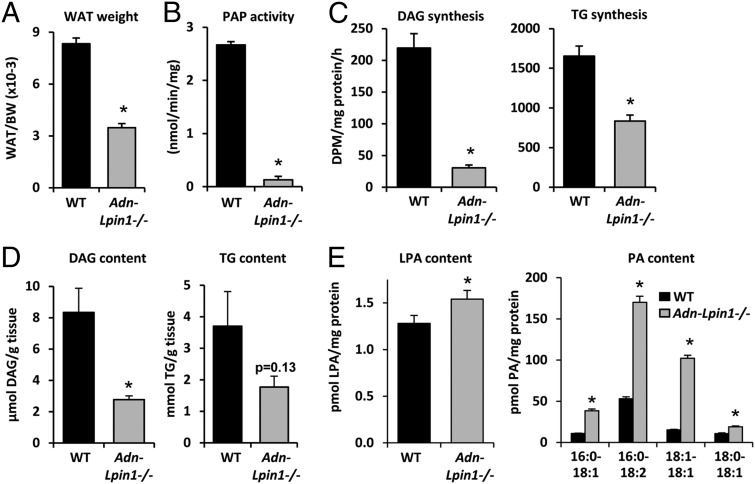
At age 6 wk, there was no difference in body weight between Adn-Lpin1−/− mice and littermate WT control mice (WT, 21.8 ± 1.3 g; Adn-Lpin1−/−, 21.8 ± 1.2 g); however, the WAT mass-to-body weight ratio was markedly diminished in Adn-Lpin1−/− mice compared with WT (Fig. 2A). As expected, adipose tissue PAP activity was diminished by 95% in Adn-Lpin1−/− mice (Fig. 2B), and incorporation of 3H-oleate into DAG, TG, and phosphatidylethanolamine was markedly reduced in fat pad explants from Adn-Lpin1−/− mice compared with WT controls, whereas labeled phosphatidylcholine (PC) synthesis was unchanged (Fig. 2C and Fig. S1D). Unexpectedly, rates of labeled phosphatidylglycerol and cardiolipin synthesis were also significantly reduced in the fat of Adn-Lpin1−/− mice (Fig. S1D). The steady-state DAG content of epididymal WAT was also significantly reduced and TG content tended to be reduced in Adn-Lpin1−/− mice (Fig. 2D). The lysophosphatidic acid (LPA) and PA contents of epididymal WAT were increased in Adn-Lpin1−/− mice compared with WT controls (Fig. 2E). These data suggest that loss of lipin 1 in adipocytes disrupts TG synthesis capacity, leading to diminished fat pad weight, DAG content, and TG content, along with reciprocal accumulation of phospholipid intermediates upstream of lipin 1 in the TG synthesis pathway.
Fig. 2.
Adn-Lpin1−/− mice are lean and have defects in TG synthesis. (A) Graph showing the ratio of epididymal plus inguinal WAT to body weight in 6- to 8-wk-old male WT and Adn-Lpin1−/− mice (n = 8 per group). *P < 0.05 vs. WT. (B) Graph depicting PAP activity in epididymal adipose tissue of 6- to 8-wk-old male WT and Adn-Lpin1−/− mice (n = 3 per group). *P < 0.05 vs. WT. (C) Graphs showing rates of DAG (Left) and TG (Right) synthesis from 3H-oleate in epididymal adipose tissue explants isolated from 6- to 8-wk-old male WT and Adn-Lpin1−/− mice (n = 4 per group). *P < 0.05 vs. WT. (D) Graphs showing DAG (Left) and TG (Right) content of epididymal adipose tissue of 6- to 8-wk-old male WT and Adn-Lpin1−/− mice (n = 5 per group). (E) Graphs showing LPA (Left) and PA (Right) content of epididymal adipose tissue of 6- to 8-wk-old male WT and Adn-Lpin1−/− mice (n = 5 per group). Specific PA species are indicated below the graph for PA content. *P < 0.05 vs. WT.
Loss of Lipin 1 Leads to Diminished Lipolysis and Protein Kinase A Activity.
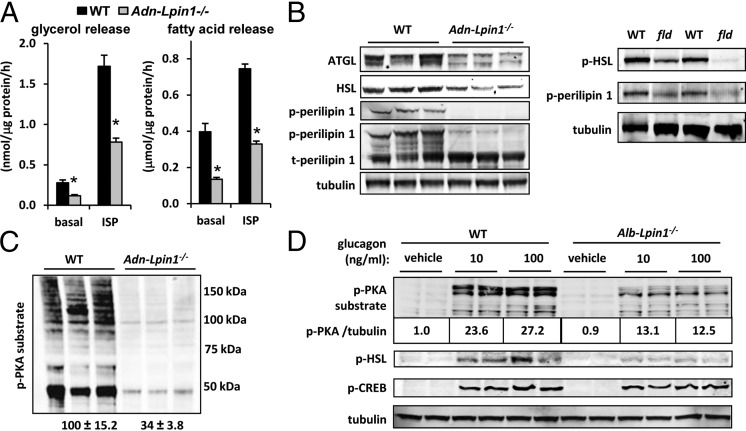
We next examined the effects of lipin 1 deficiency on lipolysis. Basal and isoproterenol-stimulated glycerol and fatty acid release rates were markedly diminished in fat explants from Adn-Lpin1−/− mice compared with WT controls (Fig. 3A). Adn-Lpin1−/− mice also exhibited reduced expression of adipose tissue TG lipase (ATGL) and hormone-sensitive lipase (HSL) (Fig. 3B). Lipolytic rates are tightly regulated by the protein kinase A (PKA)-mediated phosphorylation of HSL and perilipin 1. The phosphorylation of PKA sites of HSL and perilipin 1 was diminished in adipose tissue from Adn-Lpin1−/− mice and fld mice compared with matched WT littermates (Fig. 3B). Western blot analysis with a pan-phosphoserine PKA substrate antibody showed that phosphorylation of PKA substrates was broadly and markedly reduced by more than 70% in Adn-Lpin1−/− mice, suggesting a global attenuation of PKA activity (Fig. 3C). Glucagon-stimulated phosphorylation of PKA substrates, HSL, and cAMP response element-binding protein was also significantly attenuated in hepatocytes isolated from mice with liver-specific lipin 1 deficiency (Alb-Lpin1−/− mice) (Fig. 3D), confirming that this effect of lipin 1 deficiency is relevant in other cell types as well.
Fig. 3.
Impaired lipolysis in Adn-Lpin1−/− mice. (A) Graphs showing rates of basal and isoproterenol (ISP)-stimulated glycerol (Left) and fatty acid (Right) release by adipose tissue explants from 6- to 8-wk-old male WT and Adn-Lpin1−/− mice (n = 8 per group). *P < 0.01 vs. WT explants. (B and C) Representative Western blot images for the indicated proteins and phosphoproteins in epididymal adipose tissue of 6- to 8-wk-old male WT, Adn-Lpin1−/−, and fld mice. (D) Representative Western blot images using the indicated antibodies and lysates from hepatocytes isolated from 6- to 8-wk-old male WT and Alb-Lpin1−/− mice stimulated with the indicated concentrations of glucagon (ng/mL). The values below p-PKA substrate image represent the normalized quantification of phospho-PKA substrate blots corrected to tubulin abundance.
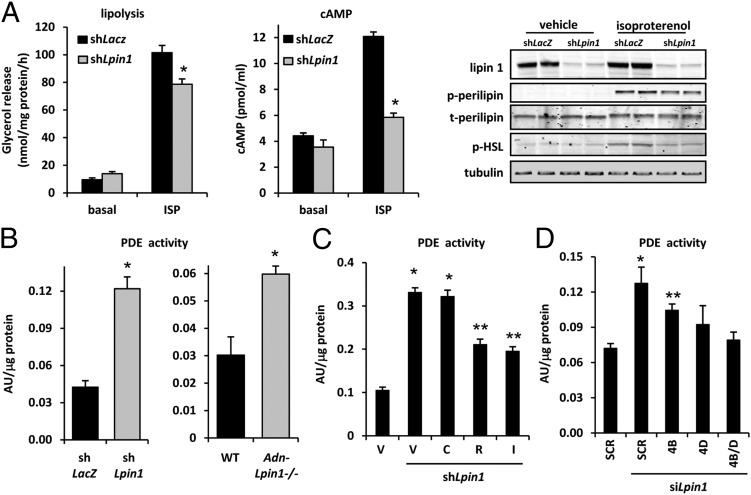
Knockdown of lipin 1 in 3T3-L1 adipocytes, which abundantly express lipin 1, led to diminished rates of β-adrenoreceptor agonist (isoproterenol)-stimulated lipolysis (Fig. 4A). Isoproterenol-induced increases in cAMP levels or phosphorylation of HSL, perilipin 1, or PKA substrates also were attenuated by lipin 1 knockdown (Fig. 4A); however, activation of adenylate cyclase by forskolin overcame the effects of lipin 1 shRNA (Fig. S4A). PKA signaling in adipocytes can be highly regulated by the activity of cAMP-degrading phosphodiesterase (PDE) enzymes. PDE activity was significantly increased in 3T3-L1 adipocytes by lipin 1 shRNA and in adipose tissue lysates of Adn-Lpin1−/− mice compared with WT controls (Fig. 4B). Adipocyte PDE activity is catalyzed primarily by PDE3B and PDE4 family members, and both PDE3 and PDE4 inhibitors reduce basal PDE activity in 3T3-L1 adipocytes (19). However, whereas an inhibitor of PDE3 (cilostamide) failed to suppress the increased PDE activity after lipin 1 knockdown (Fig. 4C), a PDE4 inhibitor (rolipram) and a general PDE inhibitor [3-isobutyl-1-methylxanthine (IBMX)] significantly reduced PDE activity in lipin 1 shRNA-treated cells. Several genes encode PDE4 enzymes (PDE4A, PDE4B, PDE4C, and PDE4D). In differentiated 3T3-L1 adipocytes, PDE4B and PDE4D are the most highly expressed enzymes (Fig. S4B); thus, we designed and used siRNAs to PDE4B and PDE4D. Knockdown of PDE4B significantly attenuated the increase in PDE activity observed with lipin 1 siRNA, and the use of PDE4D or PDE4B and 4D siRNAs together completely abolished this increase (Fig. 4D), suggesting that PDE4B and PDE4D activity is affected by lipin 1.
Fig. 4.
Adipocyte lipin 1 deficiency impairs lipolytic rate and enhances PDE activity. (A) (Left and Center) Graphs showing rates of basal (n = 4 per group) and isoproterenol-stimulated (n = 8 per group) glycerol release (Left) and cAMP content (Center; n = 3 per group) in 3T3-L1 adipocytes infected with adenovirus to knockdown lipin 1 (shLpin1) or control shRNA (shLacZ). *P < 0.01 vs. controls. (Right) Representative Western blot images for the indicated proteins or phosphoproteins. (B) Graphs depicting PDE activity in lysates from 3T3-L1 adipocytes infected with adenovirus to knockdown lipin 1 (shLpin1; Left) or in 6- to 8-wk-old male WT and Adn-Lpin1−/− male mice (Right). *P < 0.05 vs. controls. (C) Graph showing PDE activity in lysates from 3T3-L1 adipocytes infected with adenovirus to knockdown lipin 1 (shLpin1) and treated with vehicle (V) PDE3 (cilostamide; C), PDE4 (rolipram; R), or general PDE (IBMX; I) inhibitors (n = 6). *P < 0.01 vs. shLacZ control group; **P < 0.01 vs. shLacZ and shLpin1 DMSO groups. (D) Graph showing PDE activity in lysates from 3T3-L1 adipocytes transfected with siRNAs to knockdown lipin 1 (siLpin1), PDE4B (4B), and/or PDE4D (4D) with a scramble (SCR) siRNA control (n = 6). *P < 0.01 vs. scramble control group; **P < 0.05 vs. scramble control group and scramble control with Lpin1 siRNA.
Role for PA in Controlling Cyclic Nucleotide Phosphodiesterase Activity.
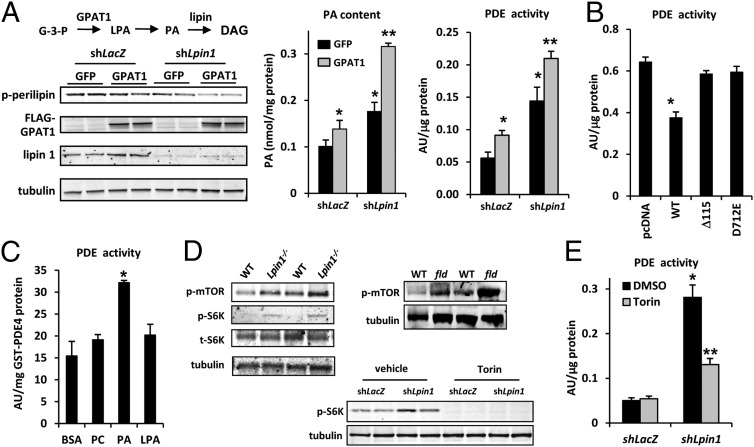
Previous work has linked PA accumulation to increased PDE4 activity in HeLa cells (20), but this “signaling” pool of PA has been thought to be regulated by the actions of phospholipase D (PLD) and other factors. In 3T3-L1 adipocytes, we found that knockdown of lipin 1 elicited a much greater increase in PDE activity than overexpression of PLD (Fig. S4C). In another approach to raising PA levels, we overexpressed glycerol-3-phosphate acyltransferase 1 (GPAT1) to elevate endogenous PA levels in a physiological manner (Fig. 5A). This also raised LPA levels, whereas Lpin1 shRNA treatment did not (Fig. S4D). Consistent with a role for PA in regulating PKA activity, GPAT1 overexpression with concomitant lipin 1 knockdown significantly and additively reduced phosphorylation of perilipin 1 (Fig. 5A) in 3T3-L1 adipocytes. The effect of lipin 1 knockdown on PDE activity was also accentuated by concomitant overexpression of GPAT1, which significantly increased PDE activity itself (Fig. 5A). In HEK293 cells, overexpression of WT lipin 1 led to reduced PDE activity (Fig. 5B); however, overexpression of Δ115-lipin 1 or catalytically inactive mutant (D712E) did not affect PDE activity. Taken together, these data suggest that accumulation of PA in adipocytes activates PDE and suppresses PKA signaling.
Fig. 5.
Altered PA content due to lipin 1 deficiency regulates PDE activity. (A) (Left, Upper) Schematic showing the initial steps in synthesis of glycerolipids from glycerol-3-phosphate (G-3-P). (Left, Lower) Representative Western blot images for the indicated proteins or phosphoproteins in 3T3-L1 adipocytes infected with the indicated adenovirus constructs. (Center and Right) Graphs showing the PA content (Center) and PDE activity (Right) of 3T3-L1 adipocytes infected with adenovirus to knockdown lipin 1 (shLpin1) with or without GPAT1 overexpression. *P < 0.01 vs. control group; **P < 0.01 vs. control group and P < 0.08 vs. shLpin1 alone. (B) Graph depicting PDE activity in HEK293 cells transfected with expression vectors to overexpress WT-lipin 1, Δ115-lipin 1, and lipin 1 D712E. *P < 0.01 vs. control group. (C) Graph showing PDE activity when GST-PDE4 was incubated with BSA, PC, PA, or LPA (n = 3). *P < 0.01 vs. control group. (D) Representative Western blot images for the indicated proteins or phosphoproteins in adipose tissue from 6- to 8-wk-old male WT, Adn-Lpin1−/−, and fld mice, or in 3T3-L1 adipocytes infected with adenovirus to knockdown lipin 1 or control shRNA in the presence or absence of Torin. (E) Graph showing PDE activity of 3T3-L1 adipocytes infected with adenovirus to knockdown lipin 1 (shLpin1) (or shLacZ control) in the presence or absence of Torin. n = 6. *P < 0.01 vs. control group; **P < 0.01 vs. control group and shLpin1 cells treated with DMSO.
Potential Allosteric and Signaling-Mediated Regulation of PDE4 Activity.
PA has been shown to enhance PDE4 activity via allosteric regulation (21). Consistent with this finding, incubation of GST-PDE4 with PA, but not with PC or LPA, enhanced its PDE enzymatic activity (Fig. 5C). PA also has been mechanistically linked to activation of mammalian target of rapamycin (mTOR) signaling (22), which may regulate lipolytic activity as well (23). Adipose tissue of Adn-Lpin1−/− mice or fld mice, which also exhibit adipose tissue PA accumulation (5), showed increased phosphorylation of mTOR (Ser2448) and its downstream target p70 S6 kinase (S6K) compared with WT littermates (Fig. 5D). Phosphorylation of S6K was also increased in 3T3-L1 adipocytes treated with lipin 1 shRNA (Fig. 5D). Importantly, treatment with an mTOR inhibitor (Torin) attenuated the observed increase in PDE activity in 3T3-L1 adipocytes after lipin 1 knockdown (Fig. 5E). These data are consistent with a model in which lipin 1 regulates PDE activity by controlling PA levels in adipocytes through mechanisms involving mTOR and possibly direct allosteric activation of PDE4.
Relationship Between Adipose Tissue Lipin 1 Expression and Lipolysis in Obese Human Subjects.
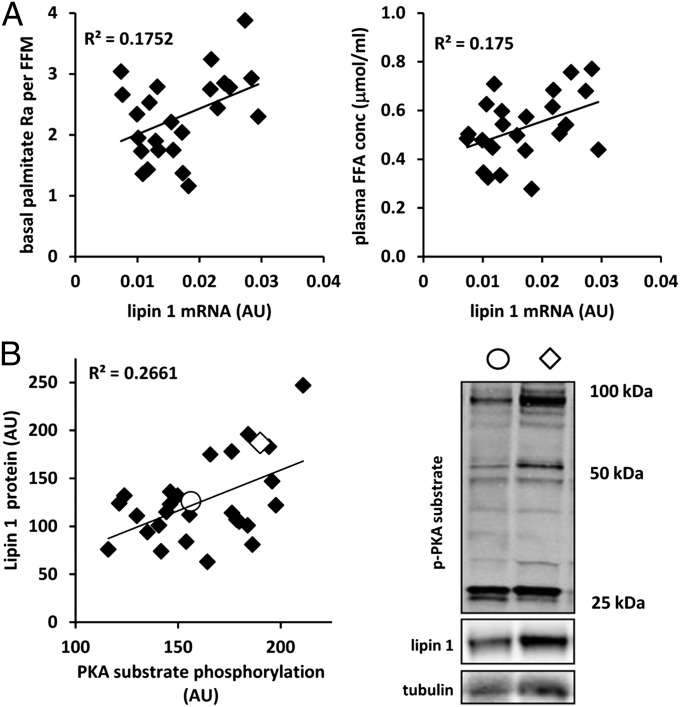
The relationships between lipin 1 expression in adipose tissue and lipolytic rates and circulating free fatty acid levels were assessed in 26 obese human subjects (mean body mass index, 35.8 ± 5.0 kg/m2). As predicted, lipin 1 mRNA expression in abdominal s.c. adipose tissue was directly related to a measure of basal adipose tissue lipolytic rate (palmitate rate of appearance into plasma; P = 0.045), as well as to plasma free fatty acid concentration (P = 0.047) (Fig. 6A). Lipin 1 protein abundance was significantly correlated with PKA substrate phosphorylation in s.c. adipose tissue of 28 obese subjects (P < 0.01) (Fig. 6B). Although a cause-and-effect relationship cannot be ascertained, the correlations among these factors are consistent with a physiological role of lipin 1 in regulating adipose tissue lipolytic activity through PKA signaling in humans.
Fig. 6.
Lipin 1 expression is related to a lipolytic rates and phospho-PKA substrate abundance in obese subjects. (A) Graphs showing the relationship between the basal palmitate rate of appearance (normalized to fat-free mass) (Left) and plasma free fatty acid concentration and s.c. adipose tissue lipin 1 expression (Right) in 26 obese human subjects. (B, Left) Graph showing the relationship between s.c. adipose tissue lipin 1 protein abundance and PKA substrate phosphorylation (quantification of the three most abundant bands corrected for tubulin abundance) in 28 obese subjects as assessed by Western blot analysis. (B, Right) Representative Western blots for phospho-PKA, lipin 1, and tubulin for two subjects, indicated by open circles and diamonds on the graph.
Discussion
In this work, we have developed and characterized adipose tissue lipid metabolism in mice with adipocyte-specific lipin 1 hypomorphism. These studies led us to define an unexpected regulatory role for lipin 1 in modulating PDE activity through control of PA concentration and mTOR signaling. The regulated control of PDE by lipin 1 is interesting in light of previous work showing that acute treatment with β-adrenergic agonists or activators of PKA signaling increase PAP activity (24) and promote lipin 1 dephosphorylation and trafficking to its active site at microsomal membranes (16). Lipin 1 may serve to control the opposing pathways of TG synthesis and degradation. When PA is abundant, TG synthesis is favored, and lipolysis is inhibited. Our data also suggest that this mechanism is relevant to human physiology, given the significant correlations between adipose tissue lipin 1 expression and lipolytic rates and PKA signaling in obese human subjects.
Previous work on PA-mediated regulation of intracellular signaling events, including allosteric activation of mTOR (22) and PDE4 (20, 25), linked the activity of PLD and DAG kinase enzymes to controlling this “signaling pool” of PA. An important finding of the present study is that the pool of PA dephosphorylated by lipin 1 that is likely destined for phosphoglycerolipid synthesis can regulate mTOR and PDE4 activity as well. It should be noted that PA was recently found to suppress formation of the mTORC2 complex (26); thus, further work is needed to determine whether mTORC1 is specifically activated by this pool of PA. We also acknowledge that we cannot rule out the possibility that alterations in other lipid species upstream or downstream of PA in these pathways are mediating these signaling events. Our observation that Torin inhibits PDE activity is consistent with previous work showing that inhibition of mTOR promotes lipolysis in 3T3-L1 adipocytes (23). Protein kinase B/Akt regulates PDE3B activity via direct phosphorylation, and a parallel link between PDE4 and mTOR is possible.
The mouse model that we describe herein has allowed us to specifically define the metabolic functions of lipin 1-mediated PAP activity in adipocytes. Although markedly reduced, DAG and TG synthesis was still measurable. It is likely that the remaining PAP activity is mediated by lipin 2, and that this residual PAP activity is sufficient for some glyceride synthesis. Loss of the yeast lipin protein has been shown to result in localization of the remaining PAP activity to the endoplasmic reticulum (27), and by concentrating this activity to the active site, the marginal capacity for DAG synthesis may be preserved. On the other hand, the Δ115-lipin 1 protein retained its ability to interact with and transactivate PPARα. This finding is supported by our previous work, which found that Δ106-lipin 1 coactivated PPARα and repressed NFATc4 (9), collectively suggesting that loss of the N-Lip domain does not affect transcriptional regulatory function with regard to these transcription factors. A caveat to this conclusion is that some partners could be affected by loss of the N-Lip domain or PAP activity of lipin 1. For example, lipin 1-mediated PAP activity is required for inhibition of sterol regulatory element-binding protein 1 in hepatocytes (10). Moreover, whether the endogenously expressed Δ115-lipin 1 is appropriately localized to the nucleus under all physiological conditions cannot be readily determined. Nonetheless, the expression of known lipin 1 target genes was not affected in adipose tissue of the Adn-Lpin1−/− mice, in contrast to the effect of knockdown of lipin 1 in adipocytes (9). Thus, we seem to have serendipitously generated a model for distinguishing the transcriptional regulatory and enzymatic functions of lipin 1 in intact mice.
In conclusion, the generation of mice lacking lipin 1-mediated PAP activity demonstrates an important role for lipin-mediated control of PA concentration in the regulation of adipocyte PDE activity. It remains to be determined whether these mechanisms are also important to the regulation of PDE activity in other cell types, which could have a variety of applications for controlling PKA signaling and cellular function.
Experimental Procedures
Lipin 1-Deficient Mouse Models.
Mice harboring a Lpin1 allele with exons 3 and 4 of the Lpin1 gene flanked by LoxP sites in a C57BL6 background have been described previously (5). To generate mice with the Lpin1 gene selectively inactivated only in adipocytes or hepatocytes, the Lpin1 floxed (fl/fl) mice were mated with adiponectin-Cre (AdnCre) (28) or albumin-Cre (AlbCre) (29) transgenic mice (both in the C57BL6 background), respectively. Mice constitutively deficient in lipin 1 (fld mice) were compared with WT (+/+) littermate control mice (Balb/cByJ strain). The experiments were done using 6- to 8-wk-old male mice. All animal experiments were approved by Washington University School of Medicine’s Animal Studies Committee.
Studies in Human Subjects.
Twenty-eight obese (mean body mass index, 35.8 ± 5.0 kg/m2) men and women participated in this study. All subjects provided written informed consent before participation. The study was approved by Washington University School of Medicine’s Human Research Protection Office. The experiments are described in detail in SI Experimental Procedures.
Statistical Analysis.
Statistical comparisons were made using ANOVA or the t test. The relationships between tissue gene expression and lipolytic rates, fatty acid concentration, or PKA activity were determined by Pearson correlation coefficient analyses. All data are presented as mean ± SE, with a statistically significant difference defined as P < 0.05.
Supplementary Material
Acknowledgments
We acknowledge Drs. Dequan Zhou and Elizabeth Brunt for technical assistance. This work was supported by National Institutes of Health Grants R01 DK78187 (to B.N.F.), R01 GM50388 and P20 RR-21954 (to A.J.M.), and R01 DK37948 (to S.K.); Nutrition Obesity Research Center Grant P30 DK56341; Digestive Diseases Research Core Center Grant P30 DK52574, Diabetes Research Core Grant P60 DK020579, and Institute for Clinical and Translational Research Grant UL1 RR024992 at Washington University School of Medicine; and Center for Research in Obesity and Cardiovascular Disease at the University of Kentucky Grant P20RR021954. H.R. is an American Heart Association Postdoctoral Fellow. T.E.H. is supported by American Diabetes Association Junior Faculty Grant 7-11-JF-21. X.S. is supported by a Scientist Development Grant 835140N from the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213493110/-/DCSupplemental.
References
- 1.Brindley DN, Pilquil C, Sariahmetoglu M, Reue K. Phosphatidate degradation: Phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochim Biophys Acta. 2009;1791(9):956–961. doi: 10.1016/j.bbalip.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris TE, Finck BN. Dual-function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011;22(6):226–233. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281(14):9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294(5548):1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 5.Nadra K, et al. Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 2008;22(12):1647–1661. doi: 10.1101/gad.1638008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet. 2010;375(9733):2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finck BN, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4(3):199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Koh YK, et al. Lipin1 is a key factor for the maturation and maintenance of adipocytes in the regulatory network with CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma 2. J Biol Chem. 2008;283(50):34896–34906. doi: 10.1074/jbc.M804007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HB, et al. Lipin 1 represses NFATc4 transcriptional activity in adipocytes to inhibit secretion of inflammatory factors. Mol Cell Biol. 2010;30(12):3126–3139. doi: 10.1128/MCB.01671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson TR, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146(3):408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Péterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27(1):121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 12.Reue K, Xu P, Wang XP, Slavin BG. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J Lipid Res. 2000;41(7):1067–1076. [PubMed] [Google Scholar]
- 13.Phan J, Péterfy M, Reue K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J Biol Chem. 2004;279(28):29558–29564. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- 14.Ren H, et al. A phosphatidic acid binding/nuclear localization motif determines lipin1 function in lipid metabolism and adipogenesis. Mol Biol Cell. 2010;21(18):3171–3181. doi: 10.1091/mbc.E10-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eguchi J, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13(3):249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris TE, et al. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J Biol Chem. 2007;282(1):277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- 17.Hall AM, et al. Dynamic and differential regulation of proteins that coat lipid droplets in fatty liver dystrophic mice. J Lipid Res. 2010;51(3):554–563. doi: 10.1194/jlr.M000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langner CA, et al. The fatty liver dystrophy (fld) mutation: A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J Biol Chem. 1989;264(14):7994–8003. [PubMed] [Google Scholar]
- 19.Choi YH, et al. Alterations in regulation of energy homeostasis in cyclic nucleotide phosphodiesterase 3B-null mice. J Clin Invest. 2006;116(12):3240–3251. doi: 10.1172/JCI24867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norambuena A, et al. Phosphatidic acid induces ligand-independent epidermal growth factor receptor endocytic traffic through PDE4 activation. Mol Biol Cell. 2010;21(16):2916–2929. doi: 10.1091/mbc.E10-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Némoz G, Sette C, Conti M. Selective activation of rolipram-sensitive, cAMP-specific phosphodiesterase isoforms by phosphatidic acid. Mol Pharmacol. 1997;51(2):242–249. doi: 10.1124/mol.51.2.242. [DOI] [PubMed] [Google Scholar]
- 22.Yoon MS, Sun Y, Arauz E, Jiang Y, Chen J. Phosphatidic acid activates mammalian target of rapamycin complex 1 (mTORC1) kinase by displacing FK506 binding protein 38 (FKBP38) and exerting an allosteric effect. J Biol Chem. 2011;286(34):29568–29574. doi: 10.1074/jbc.M111.262816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soliman GA, Acosta-Jaquez HA, Fingar DC. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45(12):1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moller F, Wong KH, Green P. Control of fat cell phosphohydrolase by lipolytic agents. Can J Biochem. 1981;59(1):9–15. doi: 10.1139/o81-002. [DOI] [PubMed] [Google Scholar]
- 25.Zakaroff-Girard A, El Bawab S, Némoz G, Lagarde M, Prigent AF. Relationships between phosphatidic acid and cyclic nucleotide phosphodiesterases in activated human blood mononuclear cells. J Leukoc Biol. 1999;65(3):381–390. doi: 10.1002/jlb.65.3.381. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, et al. Glycerolipid signals alter mTOR complex 2 (mTORC2) to diminish insulin signaling. Proc Natl Acad Sci USA. 2012;109(5):1667–1672. doi: 10.1073/pnas.1110730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi HS, et al. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J Biol Chem. 2012;287(14):11290–11301. doi: 10.1074/jbc.M112.346023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ZV, Deng Y, Wang QA, Sun K, Scherer PE. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology. 2010;151(6):2933–2939. doi: 10.1210/en.2010-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26(2):149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.