Abstract
Quantitative analyses of protein concentrations, modifications and activities in their native environments are playing an increasingly vital role in unraveling the general principles underlying signal transduction pathways. The prevalent bacterial two-component systems (TCSs) use a central phosphotransfer for signaling; however, in vivo characterization of the kinase and phosphatase activities of TCS proteins is often limited by traditional transcriptional reporter assays and complicated by simultaneous actions of multiple TCS activities. Here, we report a strategy that combines concentration-dependent phosphorylation profiling and mathematical modeling to characterize the cellular activities of the archetype Escherichia coli PhoR/PhoB system. Phosphorylation of the response regulator (RR) PhoB has been found to be dependent on the total concentrations of PhoB/PhoR and saturated at high concentrations. The relationship between RR phosphorylation and total concentrations has been defined by the modeling of the kinase and phosphatase reactions and quantified to derive the biochemical parameters of the PhoR/PhoB system in vivo. In a further test of this approach on a PhoB mutant, PhoBF20D, it proved highly effective in exploring the mechanistic differences of TCSs that are not revealed by traditional reporter assays. Measurement of biochemical parameters for PhoBF20D led to the discovery that a weaker interaction between the histidine sensor kinase and RR could result in a higher and nonrobust phosphorylation due to diminished phosphatase activities.
Keywords: in vivo phosphorylation, two-component signal transduction
Two-component systems (TCSs), one of the predominant signaling schemes in bacteria, connect input stimuli and output responses with a core phosphotransfer between a histidine sensor kinase (HSK) and a cognate response regulator (RR) (1, 2). The signaling pathway is often simply described as a series of steps that include autophosphorylation of HSKs, phosphotransfer to cognate RRs, and output modulation, usually via transcription regulation, mediated by phosphorylated RRs. Far from a simple on/off switch, phosphorylation levels of many TCS proteins are under sophisticated control by multiple enzymatic activities. One of the fundamental questions in TCS studies is what percentage of protein molecules are phosphorylated in the presence or absence of the stimuli, but the exact phosphorylation levels have not been well quantified in vivo. Without the quantification of phosphorylation, it is extremely difficult to characterize TCS kinase and phosphatase activities in their native cellular environments. This has been identified as one of the key questions outstanding in TCS research (3, 4).
RR phosphorylation levels are commonly inferred from transcriptional reporter activities even though gene transcription is downstream of RR phosphorylation and often complicated by additional regulatory factors. A change in transcription could result from alteration of RR phosphorylation levels as well as effects of additional unidentified regulators that control the promoters of interest. More importantly, transcription measurements are inadequate to report the actual RR phosphorylation levels. A lack of reporter activity does not necessarily reflect an absence of phosphorylation, whereas saturation of reporter activity may originate from reasons other than phosphorylation saturation, such as saturated binding of RR∼P to promoters. Direct characterization of TCS activities in vivo could avoid potential misinterpretations from reporter assays.
Direct measurement of TCS protein phosphorylation has been traditionally performed in cells radiolabeled by inorganic phosphate 32PO4 (3). However, this method does not provide the absolute phosphorylation level because only a fraction of phosphoryl groups become radiolabeled and the exact fraction is difficult to quantify. Recently a Phos-tag acrylamide method has been developed to separate phosphorylated and unphosphorylated RR proteins based on their mobility differences in gels containing the phosphoryl group chelator, Phos-tag (5). Phosphorylation levels of RRs have been successfully followed (5–7), but it is still challenging to characterize all of the TCS activities in vivo because HSKs often possess multiple activities, including the autokinase, phosphotransferase, and phosphatase activities.
Comprehensive characterization of the system relies on correct choices of kinetic models accounting for the essential activities that influence the output RR phosphorylation level. The dependence of reaction rates on protein concentrations has been routinely used to distinguish kinetic models and derive biochemical parameters of kinase and phosphatase activities in vitro. However, the effects of protein amount on cellular phosphorylation levels are rarely quantitatively explored in vivo to obtain biochemical parameters of TCSs. One potential concern for such quantitative studies is that the output responses of signaling systems could be robust to concentration variations of signaling proteins to overcome the intrinsic stochasticity in gene expression (4, 8, 9). However, not all responses of bacterial signaling systems display robustness to variations in protein levels. It has been shown that the steady-state output of the Escherichia coli chemotaxis system is dependent on concentrations of chemotaxis proteins (10). Even for a few TCSs in which concentration robustness has been modeled and demonstrated, only within a certain range of concentrations do the output responses become relatively insensitive to TCS protein levels (4, 11, 12). For a wider range of protein levels, the steady-state level of phosphorylated RR (RR∼P) is described as a function of total RR concentrations defined by kinetic parameters unique to individual TCSs (11–13). Therefore, robust or not, the relationship of RR∼P and total RR amount still provides rich information for in vivo characterization of these unique kinetic parameters.
In this study, we developed a strategy to define the concentration-dependent profile of RR phosphorylation in vivo for the E. coli PhoR/PhoB system and it allowed us to quantitatively describe both kinase and phosphatase activities. The PhoR/PhoB system regulates the assimilation of phosphorus in response to environmental phosphate (Pi) concentrations (14, 15). Under Pi-depleted conditions, the HSK PhoR is relieved from inhibition by Pi transport proteins and induces the phosphorylation of PhoB, which regulates the expression of genes responsible for assimilation of alternative phosphorus resources. Most of these genes are also differently regulated by the stress σ factor RpoS that is activated under Pi-depleted conditions as well (16, 17). Therefore, transcription reporter activities do not accurately mirror the phosphorylation levels of PhoB. Moreover, the requirement of phosphate starvation before 32P labeling precludes use of this radiolabeling method for the PhoR/PhoB system that is sensitive to phosphate concentrations. Instead, we were able to use a modified protocol for Phos-tag gel analyses to quantify the exact phosphorylation extent of PhoB proteins under different expression levels. This enabled us to distinguish between two different kinetic models that explain robustness of TCSs. The concentration-dependent phosphorylation profiling successfully revealed the otherwise nonapparent activity differences between PhoB alleles and demonstrated the prominent role of phosphatases in determining the TCS output. The approach presented here can be broadly applied to other TCSs for in vivo biochemical characterization.
Results
Quantification of PhoB and PhoR Protein Levels.
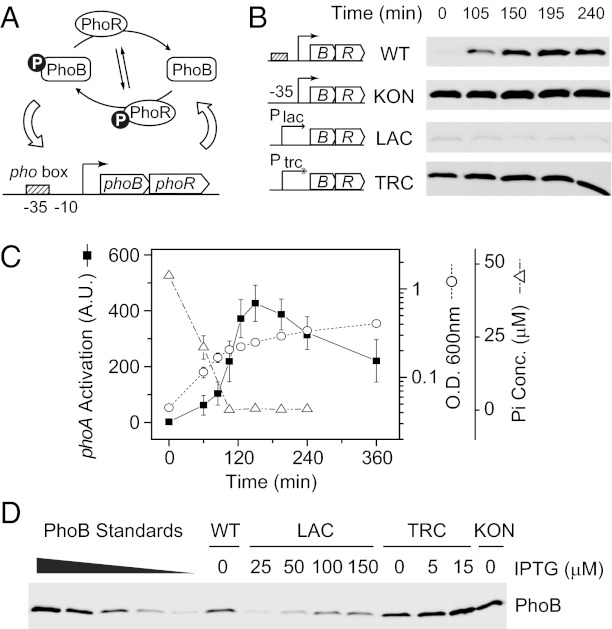
The E. coli PhoR/PhoB system is a typical autoregulated TCS. PhoR modulates the phosphorylation of PhoB, and phosphorylated PhoB regulates the expression of the phoBR operon (Fig. 1A). Thus, both the PhoB level and the expression of PhoB-regulated phoA gene encoding an alkaline phosphatase (AP) could serve as reporters for the activity of the wild-type system. Analysis confirmed that, once the phosphate concentration in the media became limited, the PhoB protein level increased along with the AP activity (Fig. 1 B and C). However, the PhoB protein level remained constant during sustained growth in Pi-limited media, different from the decreasing trend of the AP activity.
Fig. 1.
Quantification of PhoB expression. (A) Autoregulation of the PhoR/PhoB system. PhoR autophosphorylates and modulates the phosphorylation of PhoB with its phosphotransferase and phosphatase activities. Phosphorylated PhoB binds to the pho box and activates the transcription of phoBphoR. (B) Time-dependent expression of PhoB upon phosphate starvation. Nonautoregulated strains KON (RU1617), LAC (RU1616), TRC (RU1618), and autoregulated wild type (BW25113), were grown in Mops media starting with 50 μM Pi. Aliquots were taken at indicated times and analyzed by Western blot with anti-PhoB antisera. (C) Phosphate starvation responses of wild type. Phosphate concentration (open triangles), OD600 (open circles), and PhoB-regulated phoA activation (solid squares) were monitored during the growth of E . coli BW25113. Decrease of the Pi concentration below the detection limit accompanies the change of growth rate and the rise of AP activity. Error bars represent SDs from at least three independent experiments and unseen error bars are smaller than the symbols. (D) Quantitative Western analyses of PhoB expression. Indicated E. coli strains were grown under different IPTG concentrations and assayed for PhoB expression. Pure PhoB proteins were added to lysates of ΔphoBR cells as standards. The displayed Western blot shows only a representative subset of samples; the rest of the quantification is shown in Table S1.
To obtain different constitutive expression levels of PhoB and PhoR for direct phosphorylation profiling, autoregulation was disrupted by replacing the PhoB-binding Pho box of the phoB promoter with a consensus −35 sequence or substituting the entire phoB promoter with an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible lac or trc promoter. The resulting KON, LAC, and TRC strains displayed constant expression levels of PhoB independent of Pi limitation (Fig. 1B). A range of expression levels were achieved by varying IPTG concentrations and protein levels were quantified by Western blots with pure protein as standards (Fig. 1D, Fig. S1, and Table S1). For the wild-type strain under Pi-limited conditions, the histidine kinase PhoR was expressed at a concentration of 0.30 ± 0.10 pmol per 0.3 OD600 equivalent of cells, a much lower quantity than the PhoB level at 2.79 ± 0.46 pmol. As phoB and phoR were maintained in the same operon for all these strains, this PhoB/PhoR ratio remained equally high at different protein concentrations (Table S1). This allowed investigation of stimuli-dependent PhoB phosphorylation without concerns about very unnatural PhoB/PhoR ratios.
Suppression of PhoB Phosphorylation by PhoR at Pi-Replete Conditions.
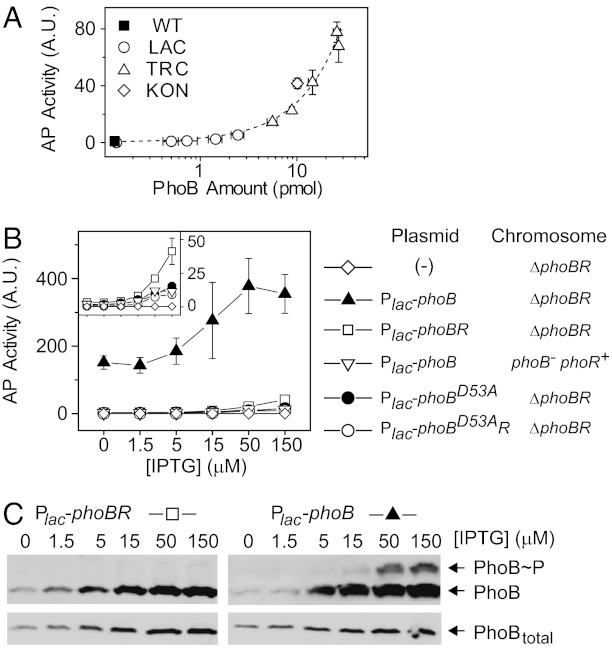
Under Pi-replete conditions, transcription activation of phoA remained modestly low across a wide range of PhoB levels, even though the basal AP activity did increase with increased PhoB and PhoR concentrations (Fig. 2A). To uncover the mechanism for PhoB activation at high Pi conditions, we constructed a series of plasmids with only phoB or both phoBR genes behind a lac promoter. The coexpressed phoBR from the plasmid displayed a similar dependence of basal activity on protein levels as strains with phoBR expressed from the chromosome (Fig. S2A). Despite its basal reporter activity (Fig. 2B), no significant phosphorylation was observed in the absence of stimuli (Fig. 2C).
Fig. 2.
Suppression of PhoB phosphorylation by PhoR at Pi-replete conditions. (A) Dependence of basal AP activities on PhoB expression levels. PhoB and PhoR were expressed from the chromosome under different IPTG concentrations using the indicated strains. The dashed line indicates a linear trend line. (B) Effect of PhoR on basal AP activity. Indicated IPTG concentrations were used to achieve different expression levels. Strains included RU1621 (ΔphoBR) carrying pTRM11 (Plac-phoB), pRG226 (Plac-phoBR), pRG305 (Plac-phoBD53AphoR), pJZG137 (Plac-phoBD53A) or no plasmid, and RU1631 (ΔphoB) carrying pTRM11. All results are shown as the mean and SD of at least three independent experiments, and unseen error bars are smaller than symbols. (C) Effect of PhoR on phosphorylation suppression. Strains with PhoR expression (RU1621/pRG226; Left) or without PhoR (RU1621/pTRM11; Right) were grown under Pi-replete conditions. Samples were analyzed with Phos-tag gels for PhoB phosphorylation (Upper) or normal SDS gels for total PhoB levels (Lower). Phos-tag retards the migration of PhoB∼P relative to unphosphorylated PhoB.
Consistent with earlier reports (15, 18), a much higher activity was observed if PhoB was expressed in the absence of PhoR and the activity was further elevated with higher PhoB concentrations (Fig. 2B). The high activity is dependent on the conserved Asp-53 residue because the unphosphorylatable allele, PhoBD53A, displayed only a low AP activity (Fig. 2B). Correspondingly, significant phosphorylation of PhoB was detected at high PhoB expression levels (Fig. 2C). This provides direct evidence that PhoB is phosphorylated in the absence of PhoR, presumably by acetyl phosphate or other noncognate HSKs, such as CreC (19, 20). When PhoR was coexpressed with PhoB or expressed at a low level from the chromosome, AP activities were suppressed (Fig. 2B) and PhoB phosphorylation was not apparent despite similar PhoB levels (Fig. 2C and Fig. S2). Similar to other TCSs (21–23), the phosphatase activity of the HSK PhoR appears to play a significant role in suppression of PhoB phosphorylation from stimuli-independent sources. The slight difference of AP activities between strains with chromosomally expressed and coexpressed PhoR may originate from different ratios of PhoR to PhoB or other signaling proteins in the pathway.
Characterization of in Vivo PhoB Phosphorylation at Pi-Depleted Conditions.
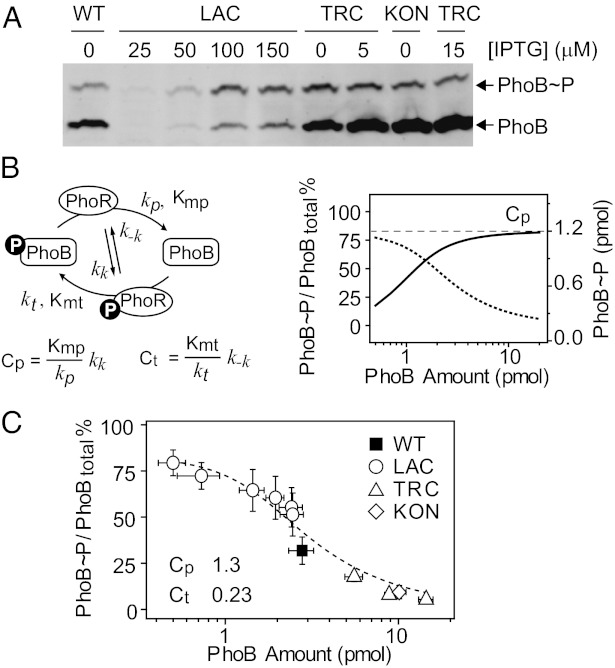
The Pi concentration became depleted ∼1.8 h after inoculation (Fig. 1C), and PhoB phosphorylation started to emerge and increased gradually (Fig. S3). Phosphorylation levels remained steady after 2.5 h of growth; thus, a 3-h growth time was chosen for all subsequent analyses to ensure that the system reached the steady state. Steady-state levels of PhoB phosphorylation were clearly dependent on protein concentration, but phosphorylation levels saturated at high protein concentrations. Further increase of total PhoB/PhoR expression only resulted in elevated levels of unphosphorylated PhoB (Fig. 3A).
Fig. 3.
In vivo phosphorylation of PhoB at Pi-depleted conditions. (A) Saturation of PhoB phosphorylation analyzed with Phos-tag gels. Indicated strains and IPTG concentration were used to achieve different expression levels. Additional samples and standards are shown in Fig. S5. (B) Model of the PhoR/PhoB phosphorylation cycle. (Left) Four reactions were modeled, including autophosphorylation and dephosphorylation of PhoR, and PhoR-mediated phosphotransfer and dephosphorylation of PhoB. Cp and Ct constants define the relationship between PhoB∼P and total PhoB concentration. (Right) Steady-state PhoB∼P level (solid line) saturates at the value of Cp (dashed line), giving continuously decreased PhoB∼P fractions (dotted line). (C) Dependence of PhoB∼P percentages on total PhoB levels. Percentages of phosphorylation were quantified with PhoB∼P standards. The dashed line indicates the best fit of all data points. Error bars are SDs of at least three independent experiments, and unseen error bars are smaller than symbols.
To fit the concentration-dependent phosphorylation saturation with minimal sets of parameters, we adopted a simple kinetic model that has been successful in describing the behaviors of other TCSs (11, 12). The model uses two Michaelis–Menten kinetic reactions to describe PhoR-catalyzed phosphotransfer and dephosphorylation, respectively (Fig. 3B). The steady-state solution of the model suggests that the relationship between phosphorylated RR and total RR concentration is decided by two composite constants, Cp and Ct, which are inversely proportional to the enzyme efficiency, defined by kcat/Km, for these two Michaelis reactions. The phosphorylated PhoB concentration is predicted to increase with total PhoB concentration and saturate at the level of Cp (Fig. 3B and Fig. S4). The phosphorylation saturation has been suggested as the basis of the system robustness at high protein levels (4, 11).
As quantified by PhoB∼P standards, phosphorylated PhoB saturated at ∼1.2 pmol when total PhoB concentration exceeded 2∼3 pmol per 0.3 OD600 equivalent of cells (Fig. S5). Because Phos-tag gel analyses usually give a more accurate quantification of phosphorylation fractions, percentages of PhoB∼P were measured and fitted against the total PhoB concentrations to yield a Cp of 1.3 ± 0.1 pmol and a Ct of 0.23 ± 0.09 pmol per 0.3 OD600 equivalent of cells (Fig. 3C). Given an estimated value of 109 cells per 1 OD and an approximate size of 10−15 L per cell, these values correspond to a Cp of 4 μM and a Ct of 0.8 μM, respectively.
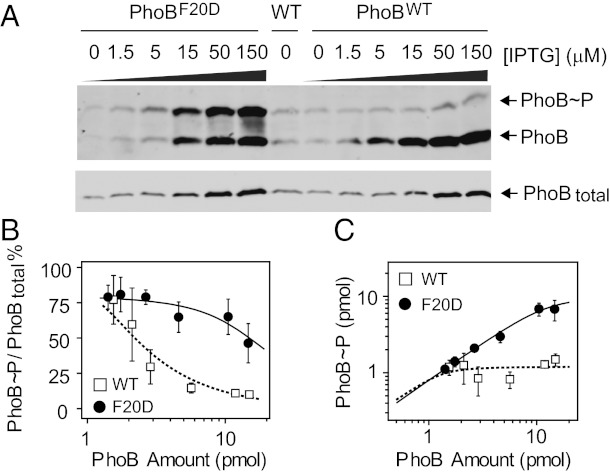
Measurement of in vivo phosphorylation parameters Cp and Ct provides insights into intrinsic TCS phosphorylation reactions and enables exploration of the mechanism behind different in vivo behaviors of TCS mutants. A F20D mutation in PhoB protein has been previously suggested to disrupt an alternative dimer conformation that may play a role in PhoB deactivation (24, 25). AP activity assay of the PhoBF20D mutant did not reveal any significant difference from wild type at the native PhoB expression level (24). Correspondingly, there is little difference in phosphorylation levels between PhoBF20D and PhoBWT at low expression levels (Fig. 4). However, at higher levels of PhoB and PhoR proteins, phosphorylation of PhoBF20D continues to increase, which is in stark contrast to the robust phosphorylation level of PhoBWT (Fig. 4 A and C). Simple incorporation of inactive dimer formation into the model could not explain the elevated phosphorylation if identical Cp and Ct were used (Fig. S6). Therefore, the F20D mutation must have additional effects on central phosphorylation reactions that change the robust behavior of the PhoR/PhoB system. Fitting the phosphorylation percentages of PhoBF20D gave a Cp of 10 pmol and a Ct of 2.3 pmol per 0.3 OD of cells, both of which are 8–10 times greater than corresponding values of wild-type PhoB (Fig. 3B). Because the F20D mutation in PhoB is unlikely to alter the PhoR autophosphorylation parameters kk and k-k, greater Cp and Ct indicate lower enzyme efficiencies for both phosphotransfer and dephosphorylation reactions.
Fig. 4.
In vivo comparison of PhoBWT and PhoBF20D. (A) Phosphorylation of PhoBWT and PhoBF20D at Pi-depleted conditions. Strain RU1621 carrying pRG298 (Plac-phoBF20DphoR) or pRG226 (Plac-phoBWTphoR), and the wild-type strain BW25113 (WT) were analyzed with Phos-tag gels (Upper) and normal SDS gels (Lower). (B and C) Comparison of phosphorylation percentages (B) and levels (C) of PhoBWT and PhoBF20D. Phosphorylation percentages were quantified for plasmid-expressed PhoBWT (open squares) and PhoBF20D (solid circles), respectively. Phosphorylation levels were calculated as the product of phosphorylation percentages and total PhoB amount. The solid line indicates the best fit of the PhoBF20D data with a Cp of 10 ± 1.8 pmol and a Ct of 2.3 ± 0.8 pmol. The dotted line is a simulated curve for PhoBWT with a Cp of 1.2 pmol and a Ct of 0.1 pmol. Error bars are SDs from three independent experiments, and unseen error bars are smaller than symbols.
PhoB Dephosphorylation and Phosphotransfer in Vitro.
To confirm the prediction based on Cp and Ct values of wild-type PhoB and the F20D substituted protein, dephosphorylation and phosphotransfer reactions were individually performed in vitro with the cytoplasmic fragment of PhoR, PhoRcyt (Fig. 5). Consistent with a lower enzyme efficiency observed in vivo, PhoRcyt dephosphorylated PhoBF20D more slowly than PhoBWT (Fig. 5 A and B). Phosphotransfer from phosphorylated PhoRcyt was quick for both PhoBF20D and PhoBWT (Fig. 5D). Unphosphorylated PhoB became almost completely phosphorylated 20 s after addition of PhoRcyt, whereas PhoBF20D required more than 1 min for complete phosphorylation. This is also consistent with a lower enzyme efficiency for phosphotransfer to PhoBF20D.
Fig. 5.
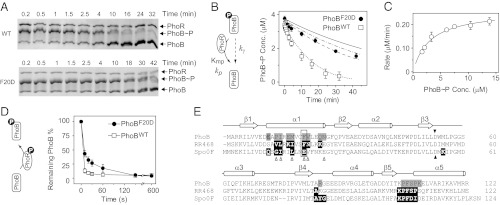
In vitro comparison of PhoBWT and PhoBF20D. (A) Dephosphorylation of PhoBWT (Upper) and PhoBF20D (Lower) by PhoRcyt. Phosphorylated PhoB proteins were incubated at room temperature with 0.5 μM PhoRcyt and 3 mM ADP to initiate dephosphorylation (SI Text). Initial concentrations of PhoB∼P were as follows: PhoBWT∼P, 3.4 μM; PhoBF20D∼P, 3.7 μM. (B) Time course of PhoB dephosphorylation by PhoRcyt. Concentrations of phosphorylated PhoBWT (open squares) and PhoBF20D (solid circles) were calculated from phosphorylation percentages and total PhoB amount. The solid line represents the autodephosphorylation curve calculated from a single exponential decay with the initial concentration of PhoBF20D∼P and experimentally determined decay constant kγ. The dotted line is an ODE simulation of PhoBWT dephosphorylation with following parameters: Km, 3 μM; kp, 0.0087 s−1; kγ, 2.6 × 10−4 s−1. The dissociation rate constant of the PhoB∼P and PhoR complex was assumed to be 0.1 s−1 and its variation was found to have little effect on simulated curves as long as Km was maintained at the same value (SI Text). The dashed and dash-dotted lines represent ODE simulations of PhoBF20D dephosphorylation with different sets of phosphatase parameters: dashed line, Km, 25 μM, kp, 0.0087 s−1; dash dotted line, Km, 3 μM; kp, 0.001 s−1. (C) Dependence of initial dephosphorylation rates on PhoB∼P concentrations. Initial rates were calculated from data within the first 5–10 min of dephosphorylation. The data were fitted to a rectangular hyperbola (solid line) with the Km at 3.0 ± 0.4 μM and Vmax at 0.26 ± 0.01 μM·min−1. kp was derived from dividing the Vmax by the PhoR concentration at 0.5 μM. (D) Phosphotransfer to PhoBWT (open squares) and PhoBF20D (solid circles). Approximately 3 μM of PhoR∼P was incubated with 2 μM of PhoB to initiate phosphotransfer. The remaining unphosphorylated PhoB amount was quantified to compare the extent of phosphotransfer. Error bars are SDs from two to three independent experiments, and unseen error bars are smaller than symbols. (E) Sequence alignment of PhoB with RR468 and Spo0F. Structures of HSK–RR or HPt–RR complexes have been solved for RR468 and Spo0F (27, 30). Secondary structural elements are shown above the alignment. The dark triangles highlight the conserved phosphorylation site, and the open triangles mark residues that are suggested to be important for HSK–RR interaction specificity (28). The gray shaded residues are involved in formation of the alternative PhoB dimer (24), and the black shaded residues are the RR residues that contact the HSK or HPt proteins.
We focused our analyses on dephosphorylation to explore the mechanism behind the increased Cp value of PhoBF20D. Initial rates of PhoBWT dephosphorylation were measured at different PhoB concentrations and fitted well with a rectangular hyperbola equation to give a Km of 3.0 ± 0.4 μM and a kp of 0.0087 s−1 (Fig. 5C). However, it is difficult to repeat the same analysis on PhoBF20D because the PhoR-assisted dephosphorylation of PhoBF20D is extremely slow and the autodephosphorylation of PhoB proteins is no longer negligible (Fig. 5B). Autodephosphorylation of PhoBF20D∼P followed similar kinetics as PhoBWT∼P with a decay constant kγ of 2.4 ± 0.23 × 10−4 s−1 (Fig. S7). PhoB autodephosphorylation was then included into the modeling of PhoR dephosphorylation with ordinary differential equation (ODE)-based simulations. The simulated curve with experimentally determined Km, kp, and kγ agreed well with the dephosphorylation data of PhoBWT (Fig. 5B). Given a larger Cp value for PhoBF20D than for PhoBWT, PhoBF20D may have a larger Km, smaller kp, or both. Correspondingly, we simulated PhoBF20D dephosphorylation with different combinations of Km and kp, including two extreme cases with either a Km of 25 μM or a kp of 0.001 s−1, all of which could give a Cp value about eight times larger than that of PhoBWT. The curve simulated with the Km of 25 μM appeared the best fit to the experimental data (Fig. 5B), implying that the increased Km, rather than a decreased kp, contributes greatly to the high Cp value of PhoBF20D. As Km usually reflects the binding affinity between the enzyme and substrates, a larger Km suggests a weaker interaction between PhoR and phosphorylated PhoB. Interestingly, the F20 residue is located in a region that has been suggested to participate in HSK–RR interactions (Fig. 5E).
Discussion
The TCS is one of the fundamental signaling modules that control information flow in prokaryotes and lower eukaryotes via protein phosphorylation. Mechanistic studies of TCSs have greatly benefited from biochemical analyses in vitro, yet such studies still face great challenges due to the lack of quantitative characterization of the central phosphorylation/dephosphorylation reactions. Neither the cellular protein amount nor the biochemical parameters have been extensively measured in vivo for most TCSs. In this study, we developed a strategy that combines the quantification of concentration-dependent phosphorylation profiles with the simple modeling of TCS reactions to characterize in vivo behaviors of the archetype E. coli PhoR/PhoB system.
Concentration-dependent phosphorylation profiles are highly effective in revealing differences in TCS behaviors that are otherwise masked at a single expression level. It is common practice to compare TCS behaviors at limited expression levels, usually close to the wild-type concentration of TCS proteins. PhoBF20D showed similar phosphorylation levels as PhoBWT near the wild-type concentration despite apparent differences at high expression or in vitro, demonstrating that lack of phenotypic differences at a specific protein level does not necessarily imply absence of differences across all protein levels. Thus, caution should be taken when interpreting data from assays performed at a single or limited range of protein levels. Conversely, observation of biochemical differences in vitro does not guarantee these differences to be manifested in vivo at certain natural expression levels. The balance of kinase and phosphatase activities can yield similar net results of phosphorylation, masking the mechanistic differences and providing some tolerance for the system to withstand mutational variations.
In contrast to direct phosphorylation measurements, traditional transcription reporter assays only give an indirect readout several steps downstream of RR phosphorylation. Many factors specific to individual reporters could potentially interfere with assessment of RR phosphorylation levels. For instance, expression of either phoA or phoB could report the activity of the system but they display different time-dependent behaviors. A lower level of phoA activation was observed at 240 min than that at 150 min, whereas PhoB expression appeared to be the same. Lengthened growth in Pi-limited media elicits stress responses and results in accumulation of the stress σ factor RpoS that represses transcription of phoA and phoB (16, 17). The difference in phoA and phoB expression profiles may reflect different regulation by RpoS. Interpretation of reporter activities is intrinsically complicated by such additional regulatory schemes that are largely unknown for most RR-regulated genes.
Phos-tag analyses provided an explicit quantification of the absolute cellular PhoB phosphorylation levels. One important observation is that significant fractions of PhoB proteins remain unphosphorylated across all expression levels, even at fully activated states. This can be accounted for by the currently used model (11), but it does not agree with the prediction made by a kinetic model in which RR is anticipated to reach complete phosphorylation when the total RR concentration is below a threshold value (8). As both models predict phosphorylation saturation and robustness, transcription reporter assays would not be possible to tell whether an RR is fully phosphorylated or not, whereas the phosphorylation quantification can distinguish between these two models. Compared with the model that predicts complete RR phosphorylation, the currently used model considers additional reactions that remove phosphoryl groups from the HSK, such as autodephosphorylation of HSK or reverse phosphotransfer to ADP, which may not be negligible for the PhoR/PhoB system. Apparently the success of modeling relies on the particular TCS fulfilling the model assumptions (see more discussions in SI Text). Many TCS activities displayed distinct differences, for instance, the rate of RR autodephosphorylation has been shown to range over six orders of magnitude (26). So great differences are expected for the vast number of TCSs; thus successful modeling requires careful examination of individual TCS behaviors.
Combined with mathematic modeling, quantification of phosphorylation profiles allows determination of the Cp and Ct values that define the relationship between RR∼P and total RR concentrations. Although it is still not possible to resolve every single kinetic parameter for each reaction, Cp and Ct provide substantial insights into the cellular characteristics of TCS systems. Traditionally, higher phosphorylation observed for one RR protein than another at high concentrations may be attributed to either a high phosphotransferase activity or a low phosphatase activity. Model fitting of PhoBF20D and PhoBWT phosphorylation profiles gave higher values of Cp and Ct for PhoBF20D and revealed lower enzyme efficiencies for both phosphotransfer and dephosphorylation reactions. Furthermore, the kinetic parameters derived from in vivo determination of Cp and Ct agreed well with in vitro measurements of PhoBF20D dephosphorylation. The Cp and Ct constants appear to capture the central properties of the steady-state behavior of the PhoR/PhoB system and they may represent valuable parameters for in vivo characterization of other TCSs.
Dephosphorylation analyses in vitro suggest that the higher Cp of PhoBF20D is likely the result of a higher Km instead of a diminished catalytic rate constant kp. Consistently, the F20 residue is located far from the catalytic phosphorylation site (Fig. S6) and the F20D substitution does not cause significant alterations in kinetics of autodephosphorylation or phosphorylation by small-molecule phosphodonors (24). Instead, the F20 residue is in the middle of the α1 helix that has been shown to interact with the DHp domain of HSKs (Fig. 5E) (27, 28). Replacing the aromatic Phe residue with the charged Asp residue likely reduces the affinity between PhoR and PhoB∼P, giving a higher Km, hence a higher Cp. This mutation could exert a similar effect on the affinity between PhoR∼P and unphosphorylated PhoB for the phosphotransfer reaction, accounting for the similar fold of increase on Ct. Even though the phosphotransferase and phosphatase activities appear similarly reduced as indicated by the Cp and Ct values, a weaker PhoR–PhoB interaction for PhoBF20D ultimately leads to a higher phosphorylation level at high protein concentrations. This counterintuitive observation illustrates the prominent role of phosphatase activity in modulating the TCS output response. It has been shown in both modeling and experimental analyses that response to stimuli mediated by a noncognate HSK and RR pair relies on the emergence of phosphatase activity, which requires strong HSK–RR interaction (29). Additionally, interaction strength could be potentially adjusted by stimuli to achieve regulation of the phosphatase activity.
Phosphatase activity is clearly essential for the concentration robustness of TCS phosphorylation. The robustness results from the saturation of RR phosphorylation at the value of Cp. The Cp value of the PhoR/PhoB system is only 4 μM compared with the total PhoB concentration of ∼9 μM under conditions of activation. Consequently, more than one-half of the wild-type PhoB proteins remain unphosphorylated due to the phosphatase activity of PhoR. Decreasing the phosphatase activity in PhoBF20D gives a high Cp value and greatly raises the threshold concentration for saturation. For typical TCSs, as long as the total RR concentration remains within the saturating range, variations of RR concentration cause little change in RR phosphorylation. It appears that the activated wild-type expression levels of the PhoR/PhoB, PhoQ/PhoP, and EnvZ/OmpR systems are all within, or close to, the saturating range (11, 12). For a different system, with proteins expressed below saturating concentrations, phosphorylation robustness may not be anticipated.
In summary, our analyses offer a straightforward strategy to investigate in vivo biochemistry of TCS proteins. Traditional transcription reporter assays could suffer from potential complication by additional regulators while the common practice of examination at limited protein expression levels may lead to misinterpretation. Our strategy provides direct measurements of both protein and phosphorylation levels that are amenable for modeling to define essential features of TCS circuits. It may prove particularly useful in complex pathways, such as branched or phosphorelay systems where phosphorylation of individual or intermediate components is difficult to probe by reporters.
Materials and Methods
Strains and Growth Conditions.
The strains and plasmids used in this study are listed in Table S2. Bacteria were grown in 3-(N-morpholino)propanesulfonic acid (Mops) minimal media containing either 2 mM (Pi-replete) or 50 μM (Pi-depleted) KH2PO4. Detailed growth conditions are described in SI Text.
Analyses of PhoB Phosphorylation in Vivo.
PhoB phosphorylation was analyzed by Phos-tag acrylamide SDS/PAGE as described before (5) with a faster and more gentle cell lysis method. Bacteria cells equivalent to 0.3 OD600∙mL per pellet were immediately resuspended in 55 μL of 1× BugBuster reagent (Novagen) in 50 mM Tris, 100 mM NaCl, pH 7.4, with 0.1% (vol/vol) Lysonase reagent (Novagen). Sufficient lysis was achieved by repeated pipetting up and down for ∼10 s followed by addition of 18 μL of 4× SDS loading buffer. Samples were immediately transferred and kept on ice for <10 min until all samples were prepared. For time-course experiments for which total sample preparation in short time was not possible, denatured samples were flash frozen in dry ice/ethanol. Known amounts of unphosphorylated PhoB and PhoB phosphorylated by phosphoramidate were mixed to create standards. PhoB and PhoB∼P were separated on 10% (wt/vol) acrylamide gels containing 25 μM Phos-tag acrylamide and 50 μM MnCl2, followed by EDTA treatment and Western blotting as described before (5).
Quantitative Western Blot Analyses.
Blots probed with the indicated primary antisera, and HRP- or Cy5-conjugated secondary antibodies were visualized by either chemiluminescence or fluorescence imaging with a FluoChem Q (Alpha Innotech). Blot images were analyzed with ImageJ (National Institutes of Health) as described previously (5). Protein amounts were calculated based on band intensities, and standard curves generated from pure proteins (Fig. S1). For quantification of Phos-tag gels, intensities of both phosphorylated and unphosphorylated PhoB bands were used to calculate the fraction of PhoB∼P. Standard curves of PhoB∼P fractions were plotted to determine the PhoB∼P fractions in unknown samples (Fig. S5).
Detailed procedures for data fitting, in vitro phosphotransfer, and phosphatase assays are described in SI Text.
Supplementary Material
Acknowledgments
We thank Ti Wu and Jizong Gao for assistance with protein purification and plasmid construction. This work was supported by National Institutes of Health Grant R37GM047958.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214587110/-/DCSupplemental.
References
- 1.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scharf BE. Summary of useful methods for two-component system research. Curr Opin Microbiol. 2010;13(2):246–252. doi: 10.1016/j.mib.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Goulian M. Two-component signaling circuit structure and properties. Curr Opin Microbiol. 2010;13(2):184–189. doi: 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem. 2008;376(1):73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolfe MD, et al. 2011. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem 286(12):10147–10154.
- 7.Wayne KJ, Li S, Kazmierczak KM, Tsui HC, Winkler ME. Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol Microbiol. 2012;86(3):645–660. doi: 10.1111/mmi.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinar G, Milo R, Martínez MR, Alon U. Input output robustness in simple bacterial signaling systems. Proc Natl Acad Sci USA. 2007;104(50):19931–19935. doi: 10.1073/pnas.0706792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinar G, Feinberg M. Structural sources of robustness in biochemical reaction networks. Science. 2010;327(5971):1389–1391. doi: 10.1126/science.1183372. [DOI] [PubMed] [Google Scholar]
- 10.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397(6715):168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 11.Batchelor E, Goulian M. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc Natl Acad Sci USA. 2003;100(2):691–696. doi: 10.1073/pnas.0234782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyashiro T, Goulian M. High stimulus unmasks positive feedback in an autoregulated bacterial signaling circuit. Proc Natl Acad Sci USA. 2008;105(45):17457–17462. doi: 10.1073/pnas.0807278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitrophanov AY, Hadley TJ, Groisman EA. Positive autoregulation shapes response timing and intensity in two-component signal transduction systems. J Mol Biol. 2010;401(4):671–680. doi: 10.1016/j.jmb.2010.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh YJ, Wanner BL. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol. 2010;13(2):198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanner BL. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella. Washington, DC: American Society for Microbiology; 1996. pp. 1357–1381. [Google Scholar]
- 16.Taschner NP, Yagil E, Spira B. A differential effect of sigmaS on the expression of the PHO regulon genes of Escherichia coli. Microbiology. 2004;150(Pt 9):2985–2992. doi: 10.1099/mic.0.27124-0. [DOI] [PubMed] [Google Scholar]
- 17.Lamarche MG, Wanner BL, Crépin S, Harel J. The phosphate regulon and bacterial virulence: A regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev. 2008;32(3):461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 18.Haldimann A, Daniels LL, Wanner BL. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol. 1998;180(5):1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wanner BL. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174(7):2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wanner BL, Wilmes-Riesenberg MR. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J Bacteriol. 1992;174(7):2124–2130. doi: 10.1128/jb.174.7.2124-2130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 22.Siryaporn A, Goulian M. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol. 2008;70(2):494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groban ES, Clarke EJ, Salis HM, Miller SM, Voigt CA. Kinetic buffering of cross talk between bacterial two-component sensors. J Mol Biol. 2009;390(3):380–393. doi: 10.1016/j.jmb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mack TR, Gao R, Stock AM. Probing the roles of the two different dimers mediated by the receiver domain of the response regulator PhoB. J Mol Biol. 2009;389(2):349–364. doi: 10.1016/j.jmb.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bachhawat P, Swapna GV, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure. 2005;13(9):1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pazy Y, et al. Matching biochemical reaction kinetics to the timescales of life: Structural determinants that influence the autodephosphorylation rate of response regulator proteins. J Mol Biol. 2009;392(5):1205–1220. doi: 10.1016/j.jmb.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139(2):325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Skerker JM, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133(6):1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siryaporn A, Perchuk BS, Laub MT, Goulian M. Evolving a robust signal transduction pathway from weak cross-talk. Mol Syst Biol. 2010;6:452. doi: 10.1038/msb.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zapf J, Sen U, Madhusudan , Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8(8):851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.