Abstract
Visible phenotypes based on locomotion and posture have played a critical role in understanding the molecular basis of behavior and development in Caenorhabditis elegans and other model organisms. However, it is not known whether these human-defined features capture the most important aspects of behavior for phenotypic comparison or whether they are sufficient to discover new behaviors. Here we show that four basic shapes, or eigenworms, previously described for wild-type worms, also capture mutant shapes, and that this representation can be used to build a dictionary of repetitive behavioral motifs in an unbiased way. By measuring the distance between each individual's behavior and the elements in the motif dictionary, we create a fingerprint that can be used to compare mutants to wild type and to each other. This analysis has revealed phenotypes not previously detected by real-time observation and has allowed clustering of mutants into related groups. Behavioral motifs provide a compact and intuitive representation of behavioral phenotypes.
Keywords: phenotyping, imaging, ethology, nematode
The study of unconstrained spontaneous behavior is the core of ethology, and it has also made significant contributions to behavioral genetics in model organisms. A powerful approach has been the careful expert observation of mutants to identify those with visible locomotor phenotypes, as demonstrated for many model organisms (1–6). However, as with most manually scored experiments, subjectivity can reduce reproducibility, whereas subtle quantitative changes or those that happen on very short or long time-scales are likely to be missed. Furthermore, manual observations are not scalable, and this has led to a widening gap between our ability to sequence and manipulate genomes and our ability to assess the effects of genetic variation and mutation on behavior.
Several recent reports describe systems that begin to address this gap by automatically recording and quantifying spontaneous behavior in animals ranging from worms (7–15) to flies (16–19), fish (20, 21), and mice (22, 23). The advantage of these approaches is that they provide a means to quantify movement parameters such as velocity precisely and in some cases to automatically detect predefined behaviors based on a manually annotated training data set. This automated analysis eliminates some of the problems of a purely manual approach, but it still relies on preselected behavioral parameters that may not be optimal for phenotypic comparisons and precludes the discovery of new behaviors that have not already been observed by eye. An alternative approach is to use unsupervised learning, which attempts to use the inherent structure of a data set to identify informative patterns; to do this, we first needed to extract worm postures from movie data and have as compact and complete a representation of worm behavior as possible.
Results and Discussion
Using eight inexpensive USB microscope-based trackers we recorded high-resolution movie data of freely crawling worms covering 307 different mutant strains with a total of 7,708 individual worms. Worms were transferred to the surface of agar plates seeded with Escherichia coli OP50 and were allowed to habituate for 30 min before being recorded for 15 min at 25 frames per second. To automatically extract worm posture, the outline of the worm was determined after thresholding, and the skeleton was found by tracing the midline connecting the two points of highest curvature on the outline, which correspond to the worm’s head and tail. The head position was determined automatically using the distribution of pixel brightness and the magnitude of lateral motion of the head and tail (Fig. 1A). Skeleton coordinates were converted to a position- and orientation-independent representation by taking 48 tangent angles evenly distributed from head to tail and subtracting off the mean angle (24).
Fig. 1.
The same four basic shapes, or eigenworms, capture both wild-type and mutant postures. (A) Worms crawling freely on a bacterial lawn on an agar pad can be segmented and accurately skeletonized (outline and midline; color indicates curvature). The green dot indicates the worm’s head, and the red dot indicates the vulval side. (B) The four wild-type eigenworms are shown as thick red lines. Eigenworms derived from 307 mutant strains (gray lines) are similar to the wild type. (C) By projecting worm shapes onto the four-dimensional basis formed by the eigenworms, a sequence of behavior can be compactly represented as a four-channel time series. The images above the time series show the worm posture at the times indicated by the red vertical lines. The blue dots indicate the worm’s head. (D) The rmsd between the raw worm shape from the skeletonization and the worm shape reconstructed using just the four eigenworms for 7,008 individuals. As more eigenworms (1–4) are used, the fit improves. The rmsd distribution for the wild-type data alone is shown in purple on the left. The fit to all of the mutant data are comparable, as can be seen more clearly in the Inset where the distributions have been rescaled.
It has been shown that the space of shapes explored by Caenorhabditis elegans during spontaneous behavior on agar without bacterial food is only four-dimensional (24). In other words, just four fundamental shapes, or eigenworms, can be added together in different proportions to reconstruct any worm posture. Because the four eigenworms (Fig. 1B; Fig. S1) provide an essentially complete description of worm posture, each frame in a movie of worm behavior can be represented as just four numbers, the amplitudes along each dimension when the shape is projected onto the eigenworms (Fig. 1C).
To use this representation as a common basis in behavioral genetics experiments, it must also capture mutant worm shapes, even though many mutants adopt postures that appear very different from those of wild-type animals. Even in wild type, locomotion is known to depend on environment; in particular, the presence of a bacterial food lawn significantly affects many aspects of locomotion (25, 26). Nonetheless, using data from wild-type (N2) worms tracked on food, we found that four eigenworms were sufficient to capture 93 ± 3% (mean ± SD) of the variance of worm shapes (Fig. 1D). Likewise, when we projected behavioral data from 307 mutant strains (n = 7,708) onto the wild-type–derived eigenworms and used the four amplitudes to reconstruct the skeleton angles, the fit of the mutant data was comparable to wild type—92 ± 4% of the variance, including mutant shapes, is captured by the wild-type eigenworms (Fig. 1D). Even eigenworms derived directly from mutant data (Fig. 1B) show a remarkable reproducibility and similarity to the wild-type–derived eigenworms.
The fact that the wild-type–derived eigenworms also capture mutant postures may reflect fundamental constraints on worm behavior, with even highly uncoordinated mutants exploring different regions of essentially the same shape space. It is not clear what constrains worm behavior, but the mutant strains that are least-well fit by the wild-type eigenworms suggest some possibilities (Fig. S2). Among the worst-fit mutants are lon-2(e67) which is longer than wild type, suggesting a role for body mechanics, and unc-4(gk705) and unc-34(e566), which affect synaptic specificity (27, 28), suggesting that neural circuit architecture also plays a role. Though these mutants may give insight into behavioral constraints, they are not alone sufficient to escape them completely: 86 ± 6% of the variance of unc-2(gk366) shapes (the worst-fit mutant in terms of rmsd) is still captured by the wild-type eigenworms.
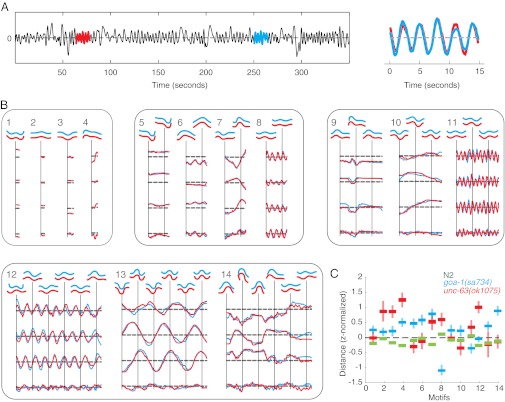
Having established a compact and common basis for representing worm shapes, we next turn to the problem of automatic behavior detection. To avoid subjectivity, we used what might be considered a minimal definition of stereotyped behavior and simply searched for the most repetitive subsequence of a given length in a movie (Fig. 2A); in the amplitude representation, this is equivalent to time series motif-finding (29), and so we call these subsequences behavioral motifs. We used the minimum rmsd to define the best match and used either a brute force search or the MK algorithm (30) to find exact motifs.
Fig. 2.
Unsupervised discovery of behavioral motifs. Repetitive subsequences are identified by discovering time-series motifs, which are the best-matching subsequences of a given length. In the sample time-series shown in A, the best-matching subsequences are shown in red and blue and overlaid on the Right. (B) Fourteen sample motifs ranging from 1.6 to 32 s (40–800 frames) representing diverse but repetitive behaviors. (C) A quantitative phenotypic profile is generated by finding the distance between movies and each element of the sample motif dictionary shown in B. Phenotypic profiles are shown for N2 wild type (green), a hyperactive mutant goa-1(sa734) (blue) (50, 51), and an uncoordinated mutant unc-63(ok1075) (1, 36). For each strain, the lines show the mean distance from each motif ± the SE for a population of worms. goa-1 is significantly closer to the two relatively high-frequency bouts of forward locomotion in motifs 8 and 11 than either N2 or unc-63, consistent with its hyperactivity; likewise, unc-63 is further from the flat posture of motif 2 because it is uncoordinated with a tendency to have higher body curvature.
We constructed a dictionary of 2,223 behavioral motifs by searching for motifs in movies sampled across all mutant strains (SI Methods). Each motif is a behavior performed by a single individual at two different times. The dictionary contains a wide range of behaviors, including short motifs essentially consisting a single posture (Fig. 2B, motifs 1–3) as well as long and sometimes seemingly irregular behaviors that are nonetheless almost perfectly repeated (Fig. 2B, motif 14). Bouts of forward locomotion are relatively common but can have widely different frequencies and amplitudes (Fig. 2B, motifs 11–13). Finally, some motifs contain subtle behaviors that may not be picked in a manual inspection for stereotyped behaviors such as motif 9, which contains a pause interrupted by a small head bend.
We next used the motif dictionary to construct a quantitative phenotypic fingerprint for each recording. The fingerprint for each individual is a vector of distances between that worm’s behavior and each dictionary element (illustrated for three strains and 14 motifs in Fig. 2C). The distance between the fingerprints averaged over a strain provides a quantitative measure of their phenotypic dissimilarity. More specifically, for each element in the motif dictionary, we identified the best matching subsequence in the movie of interest and used the rmsd as the distance between a motif and a time series (31). To compare hundreds of strains, we found the distance between all of the motifs in the dictionary and each of the movies in the database. We then used a minimum redundancy maximum relevance (mRMR) criterion (32) to select 700 motifs for clustering. Reducing the number of motifs saves computational time, but the method is robust to exactly how features are selected: a random selection of 700 motifs also performs well. The results do not depend sensitively on the number of motifs. Qualitatively similar results are found using 300 motifs. Affinity propagation (33) was used to cluster mutants into phenotypically related groups using inverse Mahalanobis distance between strains as the similarity measure. We then resampled individuals from each group with replacement, recalculated the distance, repeated the clustering 100 times, and determined the frequency with which strains were in the same cluster. We kept only the most frequent 10% of connections, and illustrated these as edges in a phenotypic similarity network (Fig. 3; see Fig. S3 for a version with node labels).
Fig. 3.
Phenotypic association network. Nodes are mutant strains, and edges show phenotypic connections. Edge transparency indicates the frequency with which two strains cluster together after resampling from the data with replacement (frequently clustering strains are connected by dark edges). The network layout is determined using spring embedding with edge weights determined by the inverse phenotypic distance. Color-coding indicates either known phenotypic classes or molecular pathway families. (Inset) Network around N2 with increased transparency and smaller node labels for clarity. The DEG/ENaC mutants discussed in Fig. 5 are shown with a red rectangle.
The nodes in Fig. 3 are colored by phenotypic or molecular class. For a complete list of the strains and their corresponding class designation, see Dataset S1. There are four broad groups of mutants in the network: (i) on the lower left there is a cluster primarily of monoamine related genes (e.g., receptors, putative monoamine transporters, and enzymes involved in monoamine synthesis) and some transient receptor potential (TRP) ion channels; (ii) on the lower right is a cluster of neuropeptides and G protein-coupled receptors; (iii) on the upper left is a loosely connected uncoordinated cluster; and (iv) on the upper right is a cluster of strains with nearly normal locomotion, including several mutant groups and the wild-type N2 strain itself. For the last group, it is important to emphasize that some of the mutants in the cluster with N2 are still significantly different from N2, as we discuss in more detail below.
To assess the clustering outcome in more detail, we compared the proximity of strains with predicted associations at increasing scales: loss-of-function alleles of the same gene, mutations affecting different genes but disrupting the same molecular complex, and at the largest scale, genes affecting a common molecular pathway. There were several loss-of-function deletion, nonsense, or splice-site mutant alleles of the same gene in the network, and these clustered significantly closer to each other than the network average (gene pairs are indicated by dashed red lines in Fig. 3): they were separated by an average of 1.6 edges, significantly less than the average network distance of 3.7 (P = 6 × 10−5, Wilcoxon rank-sum test). These genes, with the corresponding network distances are egl-21(n476) and egl-21(n611) (one edge); ocr-4(tm2173) and ocr-4(vs137) (two edges); trp-2(sy691) and trp-2(gk298) (two edges); unc-10(md1117) and unc-10(e102) (two edges); unc-89(e1460) and unc-89(st85) (one edge); and trpa-2(ok3189), trpa-2(tm3085), and trpa-2(tm3092) (the ok3189 allele is two edges from the other alleles which are one edge from each other). The unc-108(n501) and unc-108(n777) (one edge) alleles are both dominant activating mutations in a Rab small GTPase (34) and are therefore also expected to result in a similar phenotype. Although there are two loss-of-function alleles of unc-4, these were not included in the analysis because one of them (gk705) is not part of the main network. There are also two pairs of genes that form subunits of the same complex: unc-38 and unc-63 encode subunits of the same acetylcholine receptor (35, 36), and unc-79 and unc-80 encode subunits of the NALCN neuronal sodium leak channel (37, 38). Both pairs of mutants cluster together in the network as expected (Fig. 3, Upper Left). syg-1 is required for specifying synaptic specificity and acts as a receptor for syg-2; however, they are separated by three edges in the network, just under the network average of 3.7.
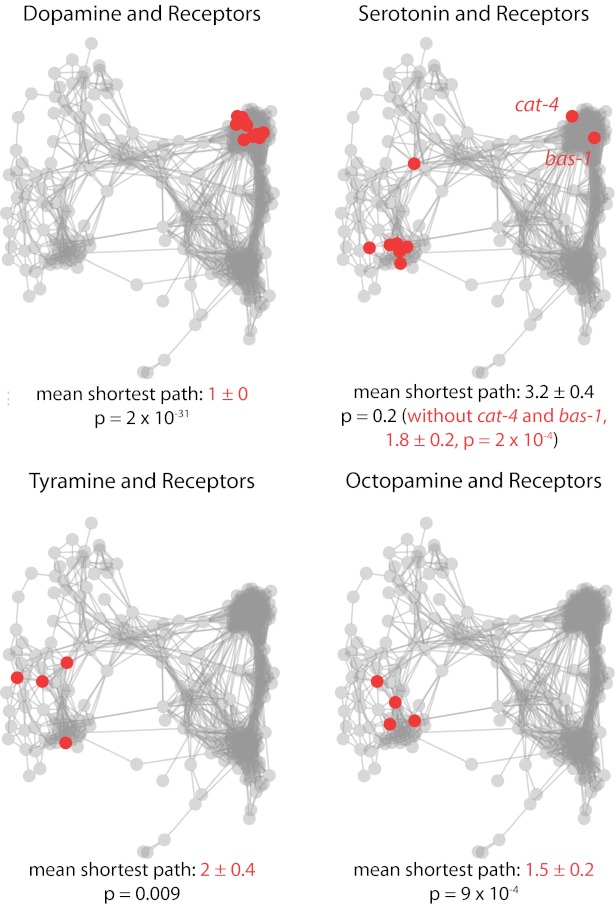
We performed a similar analysis for genes in common molecular pathways. Fig. 4 highlights four examples of monoamine signaling (see Fig. S4 for acetylcholine receptors and pathways regulating synaptic release, insulin signaling, Go/Gq signaling, and mechanosensation). Most of the monoamine mutants are expected to have relatively subtle behavioral phenotypes, but they still form significantly tighter clusters than the network overall. The exception appears to be for serotonin; however, the outlying pair of genes cat-4 and bas-1 encode molecules required for both serotonin and dopamine biosynthesis—indeed, they cluster tightly with other dopamine-related genes. If we consider only the genes involved exclusively in serotonin signaling, we find that these too are significantly more tightly clustered than the overall network (Fig. 4).
Fig. 4.
Genes involved in monoamine pathways cluster together. In each panel, genes in the indicated class of monoamine signaling are highlighted in red. The mean ± SE of the shortest path connecting each pathway member is listed below the network. Cases where the intragroup distance is significantly smaller than the network overall based on a Wilcoxon rank-sum test are highlighted in red. In the case of serotonin, the results are also shown without cat-4 and bas-1 because they encode molecules required for both serotonin and dopamine biosynthesis. Included genes are as follows: dopamine and receptors: cat-2(e1112), dop-1(vs101), dop-1(vs100); dop-2(vs105), dop-1(vs100); dop-2(vs105); dop-3(vs106), dop-1(vs100); dop-3(vs106), dop-2(vs105), dop-2(vs105); dop-3(vs106), dop-3(vs106), dop-4(tm1392), bas-1(ad446), cat-4(e1141). Serotonin and receptors: bas-1(ad446), cat-4(e1141), ser-1(ok345), ser-4(ok512), ser-5(tm2654), ser-7(tm1325), tph-1(mg280). Tyramine and receptors: tdc-1(n3419), tyra-2(tm1846), tyra-3(ok325), ser-2(pk1357). Octopamine and receptors: octr-1(ok371), tdc-1(n3419), ser-6(tm2146), tbh-1(n3247).
To determine how the different components of the algorithm contribute to this clustering result, we repeated the analysis with different versions of some subroutines. In each case, nothing was changed about the analysis procedure except the specified substitution. Instead of the inverse Mahalanobis distance we used the inverse Euclidean distance between strains as the phenotypic similarity measure and found that the resulting network (Fig. S5) showed fewer of the predicted associations discussed above and had lower modularity (proportional to the number of within-group connections minus the number of connections expected for a random network of the same degree) (39). However, when we used 700 randomly selected motifs from the 2,223 element dictionary instead of mRMR and repeated the clustering, the resulting phenotypic network was of similar quality to the original in terms of its modularity and the significant association of molecular pathway components, although it has slightly fewer expected allelic and molecular complex connections (Fig. S5B). When short behaviors with the same lengths as the motifs in the dictionary are selected at random and used for clustering, we find a similar-quality network with three fewer expected associations (Fig. S5C). This finding suggests that selecting a large-enough number of behavioral sequences is almost as good for phenotypic profiling, and that stereotyped behaviors, at least according to our minimal definition, are not specifically required to usefully compare mutants.
In addition to identifying broad categories and analyzing pathways, we want to generate specific hypotheses about functional interactions based on phenotypic similarity. To demonstrate the potential of this approach, we considered two degenerin/epithelial Na+ (DEG/ENaC) channels present in the N2-like cluster: asic-2(ok289) (n = 19) and acd-5(ok2657) (n = 20). Neither of these genes has a known function, nor do the deletion strains have a previously reported phenotype. We used the mRMR criterion to find the two most-distinguishing behavioral motifs with respect to N2 (Fig. 5). Qualitatively, in both cases we find one motif that represents a bout of forward locomotion and another that is a pause in a curved shape. The DEG/ENaC channel mutants are, on average, further from the forward locomotion and closer to the curved pause. Both differences are statistically significant based on a Hotelling T2 test with permutation (40, 41) [asic-2(ok289) vs. N2, P = 0.0019; acd-5(ok2657) vs. N2, P = 9 × 10−6]. Comparing the two DEG/ENaC mutants to each other (Fig. 5C), two motifs are selected but there is no significant difference between their distances to the motifs (P = 0.796, Hotelling T2 with permutation). In other words, these two mutants were found to be different from N2 yet they were not distinguishable from each other using the same procedure. It should be noted that this is not true for all mutants in the N2-like group; for example, two other DEG/ENaC channels near each other in the same cluster are still distinguishable using the same approach (Fig. S6). Given that DEG/ENaC channels are known to form heteromeric complexes (42), we arrive at the hypothesis that these channels share a similar function and may even operate in the same channel complex in some cells.
Fig. 5.
Maximally distinguishing behavioral motifs. For each of the indicated comparisons (A–C) the two most-distinguishing behavioral motifs from the dictionary are found using mRMR. The plots on the left show the z-normalized distance between the compared strains and the motif (mean ± SE). The motif amplitudes and the corresponding worm postures are shown in gray. The colored lines show the mean-matching motifs from each of the compared strains. For example, acd-5(ok2657) matches the first motif in A more closely than N2 on average, and this is visible in the amplitude overlay.
Phenotypic profiling using automatically extracted behavioral motifs can reveal abnormal phenotypes in mutants that were not apparent from manual observation. Furthermore, phenotypic associations can sharpen hypotheses of gene function, especially when combined with other information, such as sequence similarity, and can therefore help guide functional experiments. Although the discovery phase is unsupervised, the end result is still an intuitive summary of a phenotype in terms of a small number of short behaviors. Because the assumptions underlying behavioral motif extraction are minimal, we expect our method to apply generally to many model organisms. In particular, because zebrafish larvae are of fixed length and generate body movements in two dimensions, little modification would be required to adapt the approach described here. Although a representation based on skeleton angles would not be optimal for Drosophila larvae, a representation based directly on outline curvature could make Drosophila larva locomotion and in vitro cell motility (43) amenable to unsupervised motif discovery as well. Motif-derived phenotypes are related to functional classes but are derived completely independently from other data; we therefore expect them to provide complementary information that may be usefully combined with proteomic and transcriptomic data in the future.
Methods
Strains.
All strains were maintained at 22 °C as previously described (44). A complete strain list is included in Dataset S1.
Tracking.
Single-worm tracking was performed as previously described (45, 46). Briefly, single worms were transferred to agar plates seeded with 20 μL of OP50 bacteria using a platinum wire and allowed to habituate for 30 min. Then each worm was tracked for 15 min at 25 frames per second using one of eight trackers consisting of a USB microscope mounted on a motorized stage. The camera was moved beneath the crawling worm, which was kept stationary to avoid mechanically stimulating the worm. Each tracker was controlled by a computer running home-built software written in Java (www.mrc-lmb.cam.ac.uk/wormtracker/).
Segmentation and Skeletonization.
All analysis software described below was written in MATLAB. Each movie frame was segmented using the Otsu threshold (47), and the worm was taken to be the largest connected component in the resulting image. The curvature of the outline of this connected component was determined, and the two points of highest curvature were taken to be the head and the tail. The skeleton was found by tracing the midline of the outline between these two points. The skeleton was divided into 49 equally spaced points, which were used to define 48 tangent angles. The mean of these angles was then subtracted.
Eigenworm Representation.
Eigenworms were derived from at least 1 h of pooled crawling data for each strain as previously described (24). The skeleton angles in each frame were then projected onto the wild-type–derived eigenworms, and these amplitudes were used for all further behavior analysis.
Motif Dictionary.
Motifs were discovered as previously described† with the following modifications. Motifs were discovered using distances across all four eigenworm amplitudes simultaneously, ensuring an essentially one-to-one correspondence between a time-series motif and a segment of worm behavior. Furthermore, no normalization was performed on the candidate subsequences to preserve the amplitude offsets and magnitudes that are essential for maintaining the times-series behavior mapping. Nine different length motifs from 1.6 to 32 s (40–800 frames) were discovered in each of 1,542 movies (each 15 min long) sampled from all mutant strains, resulting in an initial motif index with 13,878 entries. Motifs were discovered on data downsampled by a factor of four. The 20% worst matching motifs of each length were dropped, and from the remaining 80%, the 20% of each length with the highest variance across all dimensions were kept, leaving 2,223 motifs in the pruned dictionary. Keeping high variance elements ensures that the pruned dictionary would not contain only pauses, which tend to be the most perfectly repeated behaviors. However, it should be emphasized that the remaining set does contain pauses at different postures, which can be important distinguishing features. The full-resolution versions of each remaining motif were then recovered and used in subsequent analysis.
A feature vector for each individual was constructed by finding the best-matching subsequence between each amplitude time-series and each motif in the dictionary. The rmsd between the motif and its best match in the amplitude time-series is taken as the distance between that motif and the time series. The set of distances from each motif defines the feature vector for each individual worm. The feature matrix consisting of the full set of feature vectors was further reduced to 700 features using mRMR (32) to find the 700 most-informative motifs. The resulting feature matrix was z-normalized to prevent outlying motifs from dominating the dissimilarity measure and allow subtle but potentially important differences from common motifs to contribute. Pair-wise Mahalanobis distances were calculated between all of the strains. Because the number of features is significantly greater than the number of individuals in each group, the Mahalanobis distance is calculated using a shrinkage estimate of the covariance matrix (47) for each strain.
Clustering.
Clustering was performed using affinity propagation (33) with the inverse Mahalanobis distance as the similarity measure and the median similarity as the clustering preference factor. Individual worms were then resampled with replacement from each strain, and the clustering was repeated 100 times. The clustering frequency was determined, and we kept only the most frequent 10% of connections and illustrated these as edges in a phenotypic similarity network. Cytoscape (48) was used to display the network. The nodes are arranged using a force-directed layout with inverse Mahalanobis distance as the edge weight so that more similar highly interconnected strains are nearer to each other (49).
Supplementary Material
Acknowledgments
The authors acknowledge Eamonn Keogh, Abdullah Al Mueen, Hanchuan Peng, Kevin Murphy, Andrew Duchi, and Suchi Saria for sharing code; Roland Schwarz and Greg Stephens for useful discussions and sharing code; and Robyn Branicky for critically reading the manuscript. Some strains were provided by National Institutes of Health (NIH) Chemical Genomics Center Office of Research Infrastructure Program P40 OD010440. Support was also provided by Medical Research Council Grant MC-A022-5PB91, NIH Grant DA018341, and a long-term fellowship from the Human Frontier Science Program (to A.E.X.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211447110/-/DCSupplemental.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sokolowski MB. Drosophila: Genetics meets behaviour. Nat Rev Genet. 2001;2(11):879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- 3.Granato M, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- 4.Vitaterna MH, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264(5159):719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hrabé de Angelis MH, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25(4):444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 6.Nolan PM, et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet. 2000;25(4):440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 7.Baek JH, Cosman P, Feng Z, Silver J, Schafer WR. Using machine vision to analyze and classify Caenorhabditis elegans behavioral phenotypes quantitatively. J Neurosci Methods. 2002;118(1):9–21. doi: 10.1016/s0165-0270(02)00117-6. [DOI] [PubMed] [Google Scholar]
- 8.Geng W, Cosman P, Baek JH, Berry CC, Schafer WR. Quantitative classification and natural clustering of Caenorhabditis elegans behavioral phenotypes. Genetics. 2003;165(3):1117–1126. doi: 10.1093/genetics/165.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsibidis GD, Tavernarakis N. Nemo: A computational tool for analyzing nematode locomotion. BMC Neurosci. 2007;8:86. doi: 10.1186/1471-2202-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. The Parallel Worm Tracker: A platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE. 2008;3(5):e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sznitman J, Purohit PK, Krajacic P, Lamitina T, Arratia PE. Material properties of Caenorhabditis elegans swimming at low Reynolds number. Biophys J. 2010;98(4):617–626. doi: 10.1016/j.bpj.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krajacic P, Shen X, Purohit PK, Arratia P, Lamitina T. Biomechanical profiling of Caenorhabditis elegans motility. Genetics. 2012;191(3):1015–1021. doi: 10.1534/genetics.112.141176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swierczek NA, Giles AC, Rankin CH, Kerr RA. High-throughput behavioral analysis in C. elegans. Nat Methods. 2011;8(7):592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh R, Mohammadi A, Kruglyak L, Ryu WS. Multiparameter behavioral profiling reveals distinct thermal response regimes in Caenorhabditis elegans. BMC Biol. 2012;10(1):85. doi: 10.1186/1741-7007-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, et al. Profiling a Caenorhabditis elegans behavioral parametric dataset with a supervised K-means clustering algorithm identifies genetic networks regulating locomotion. J Neurosci Methods. 2011;197(2):315–323. doi: 10.1016/j.jneumeth.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branson K, Robie AA, Bender J, Perona P, Dickinson MH. High-throughput ethomics in large groups of Drosophila. Nat Methods. 2009;6(6):451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dankert H, Wang L, Hoopfer ED, Anderson DJ, Perona P. Automated monitoring and analysis of social behavior in Drosophila. Nat Methods. 2009;6(4):297–303. doi: 10.1038/nmeth.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo L, et al. Navigational decision making in Drosophila thermotaxis. J Neurosci. 2010;30(12):4261–4272. doi: 10.1523/JNEUROSCI.4090-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Marin A, Stephens GJ, Louis M. Active sampling and decision making in Drosophila chemotaxis. Nat Commun. 2011;2:441. doi: 10.1038/ncomms1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rihel J, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327(5963):348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokel D, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6(3):231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jhuang H, et al. Automated home-cage behavioural phenotyping of mice. Nature Communications. 2010;1:1–9. doi: 10.1038/ncomms1064. [DOI] [PubMed] [Google Scholar]
- 23.de Chaumont F, et al. Computerized video analysis of social interactions in mice. Nat Methods. 2012;9(4):410–417. doi: 10.1038/nmeth.1924. [DOI] [PubMed] [Google Scholar]
- 24.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLOS Comput Biol. 2008;4(4):e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26(3):619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 26.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102(9):3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White JG, Southgate E, Thomson JN. Mutations in the Caenorhabditis elegans unc-4 gene alter the synaptic input to ventral cord motor neurons. Nature. 1992;355(6363):838–841. doi: 10.1038/355838a0. [DOI] [PubMed] [Google Scholar]
- 28.Yu TW, Hao JC, Lim W, Tessier-Lavigne M, Bargmann CI. Shared receptors in axon guidance: SAX-3/Robo signals via UNC-34/enabled and a Netrin-independent UNC-40/DCC function. Nat Neurosci. 2002;5(11):1147–1154. doi: 10.1038/nn956. [DOI] [PubMed] [Google Scholar]
- 29.Lin J, Keogh E, Lonardi S, Patel P. Proceedings of the Eighth Association for Computing Machinery Special Interest Group on Knowledge Discovery and Data Mining International Conference on Knowledge Discovery and Data Mining. New York: Assoc Comp Machinery; 2002. Finding motifs in time series; pp. 53–68. [Google Scholar]
- 30.Mueen A, Keogh E, Zhu Q, Cash S, Westover B. Exact discovery of time series motifs. In Society for Industrial and Applied Mathematics International Conference on Data Mining. 2009. Available at www.siam.org/proceedings/datamining/2009/dm09_045_mueena.pdf. [DOI] [PMC free article] [PubMed]
- 31.Ye L, Keogh E. In 15th Conference on Knowledge Discovery and Data Mining. 2009. Time series shapelets: a new primitive for data mining. (ACM SIGKDD) (Assoc Comp Machinery, New York), pp 947–956. [Google Scholar]
- 32.Peng H, Long F, Ding C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 33.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315(5814):972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 34.Park EC, Horvitz HR. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics. 1986;113(4):821–852. doi: 10.1093/genetics/113.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleming JT, et al. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J Neurosci. 1997;17(15):5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culetto E, et al. The Caenorhabditis elegans unc-63 gene encodes a levamisole-sensitive nicotinic acetylcholine receptor alpha subunit. J Biol Chem. 2004;279(41):42476–42483. doi: 10.1074/jbc.M404370200. [DOI] [PubMed] [Google Scholar]
- 37.Sedensky MM, Meneely PM. Genetic analysis of halothane sensitivity in Caenorhabditis elegans. Science. 1987;236(4804):952–954. doi: 10.1126/science.3576211. [DOI] [PubMed] [Google Scholar]
- 38.Yeh E, et al. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 2008;6(3):e55. doi: 10.1371/journal.pbio.0060055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70(6 Pt 2):066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 40.Good PI. Resampling Methods a Practical Guide to Data Analysis. Boston: Birkhäuser; 2006. [Google Scholar]
- 41.Tsai C-A, Chen JJ. Multivariate analysis of variance test for gene set analysis. Bioinformatics. 2009;25(7):897–903. doi: 10.1093/bioinformatics/btp098. [DOI] [PubMed] [Google Scholar]
- 42.Arnadóttir J, Chalfie M. Eukaryotic mechanosensitive channels. Annu Rev Biophys. 2010;39:111–137. doi: 10.1146/annurev.biophys.37.032807.125836. [DOI] [PubMed] [Google Scholar]
- 43.Driscoll MK, et al. Cell shape dynamics: From waves to migration. PLOS Comput Biol. 2012;8(3):e1002392. doi: 10.1371/journal.pcbi.1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yemini E, Kerr RA, Schafer WR. 2011. Preparation of samples for single-worm tracking. Cold Spring Harb Protoc 2011, 10.1101/pdb.prot066993.
- 45.Yemini E, Kerr RA, Schafer WR. 2011. Illumination for worm tracking and behavioral imaging. Cold Spring Harb Protoc 2011, 10.1101/pdb.prot067009.
- 46.Nobuyuki O. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–66. [Google Scholar]
- 47.Schäfer J, Strimmer K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat Appl Genet Mol Biol. 2005;4:e32. doi: 10.2202/1544-6115.1175. [DOI] [PubMed] [Google Scholar]
- 48.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamada T, Kawai S. An algorithm for drawing general undirected graphs. Inf Process Lett. 1989;31(1):7–15. [Google Scholar]
- 50.Mendel JE, et al. Participation of the protein Go in multiple aspects of behavior in C. elegans. Science. 1995;267(5204):1652–1655. doi: 10.1126/science.7886455. [DOI] [PubMed] [Google Scholar]
- 51.Ségalat L, Elkes DA, Kaplan JM. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science. 1995;267(5204):1648–1651. doi: 10.1126/science.7886454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.