Abstract
The assembly of static supramolecular structures is a culminating event of developmental programs. One such structure, the proteinaceous shell (called the coat) that surrounds spores of the bacterium Bacillus subtilis, is composed of about 70 different proteins and represents one of the most durable biological structures known. The coat is built atop a basement layer that contains an ATPase (SpoIVA) that forms a platform required for coat assembly. Here, we show that SpoIVA belongs to the translation factors class of P-loop GTPases and has evolutionarily lost the ability to bind GTP; instead, it uses ATP hydrolysis to drive its self-assembly into static filaments. We demonstrate that ATP hydrolysis is required by every subunit for incorporation into the growing polymer by inducing a conformational change that drives polymerization of a nucleotide-free filament. SpoIVA therefore differs from other self-organizing polymers (dynamic cytoskeletal structures and static intermediate filaments) in that it uses ATP hydrolysis to self-assemble, not disassemble, into a static polymer. We further show that polymerization requires a critical concentration that we propose is only achieved once SpoIVA is recruited to the surface of the developing spore, thereby ensuring that SpoIVA polymerization only occurs at the correct subcellular location during spore morphogenesis.
Keywords: actin, cytoskeleton, sporulation, spoVM, tubulin
The assembly of static structures represents an important developmental end point that can contribute to the characteristic morphology of an organism. Unlike dynamic structures, such as those made of actin and tubulin, that are frequently assembled and disassembled to suit the needs of a cell at a particular time, large static structures, such as eggshells, flagella, and teeth, are built to remain intact for longer periods of time (1–4). Among the most durable structures in biology is the proteinaceous shell, called the coat, that surrounds the dormant endospores of Gram-positive bacteria and self-assembles to create a structure that may last for many years (5–9). Indeed, this structure contributes to the remarkable resilience of major pathogens like Bacillus anthracis and Clostridium difficile against environmental onslaughts (10, 11).
Spore formation (sporulation) in the rod-shaped bacterium Bacillus subtilis initiates when the cell senses the imminent deprivation of nutrients (12–15). The bacterium responds to this starvation condition by dividing asymmetrically and elaborating a spherical internal organelle, called the forespore, that is enveloped by a double membrane and contains a copy of the genetic material (Fig. 1A). The outer cell (the “mother cell”) nurtures the forespore as it matures into a largely dormant cell; at that point, the mother cell lyses and releases the now mature spore into the environment. Part of this nurturing consists of the deposition of some 70 different proteins produced in the mother cell onto the surface of the forespore in a highly coordinated manner that eventually will form the coat, the outermost feature of mature B. subtilis spores (6, 16–19).
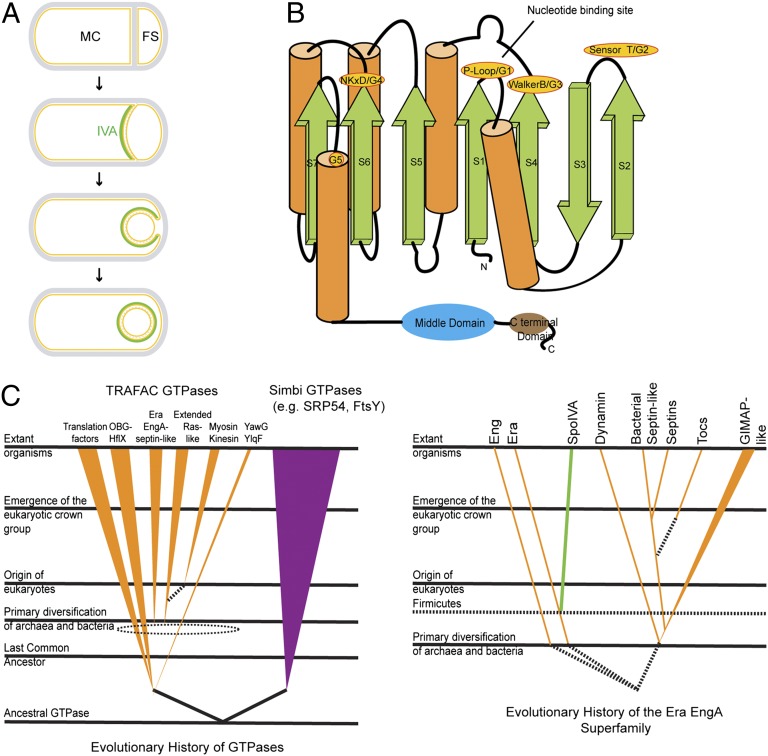
Fig. 1.
ATPase IVA belongs to the TRAFAC class of P-loop GTPases. (A) Schematic representation of sporulation in B. subtilis. Asymmetrical division results in a larger mother cell (MC) and a smaller forespore (FS) that are held together by a cell wall that surrounds the sporulating cell (gray). Membranes are depicted in yellow. At the onset of engulfment, IVA (green) localizes to the engulfing membrane [anchored to the membrane surface by SpoVM (not depicted for clarity)] and tracks the engulfing membrane until it eventually surrounds the forespore (Bottom). (B) Topological representation of IVA. The active site motifs are labeled in yellow, whose numbering (G1–G5) corresponds to the “idealized” GTPase (37). The “middle domain” and the C-terminal domain are shown as cartoons (blue and brown, respectively). The colors of the strands, helices, and domains correspond to colors in Fig. S1. (C) Evolutionary history of GTPase families and the Era/EngA superfamily. Several relative temporal epochs separated by the major evolutionary transitions that mark their boundaries are shown. The filled colored lines and triangles indicate the maximum depth to which the GTPase lineages can be traced with respect to these temporal epochs. The broken lines indicate uncertainty in terms of the exact point of origin of a lineage. The broken-line ellipses bundle groups of lineages from within which a new lineage potentially could have emerged.
Formation of the complex coat is absolutely dependent on the assembly of a platform, called the basement layer, on top of which the coat assembles (20) and whose structural component is composed of a protein called SpoIVA (pronounced “Spo-four-A”; hereafter called IVA) (16, 21, 22) (Fig. 1A). IVA is anchored to the surface of the forespore by a small amphipathic protein that dictates the correct subcellular location of IVA (22–26), and its encasement around the forespore depends on a soluble protein in the mother cell (27). Previously, we reported that IVA binds and hydrolyzes ATP in vitro and that disruption of a “Walker A” motif in IVA, required for ATP binding, disrupted sporulation efficiency of cells producing the variant protein in vivo (28). We proposed that ATP hydrolysis, not simply ATP binding, was required for polymerization of IVA at the surface of the forespore. Here, we used a bioinformatics approach to reveal that based on predicted secondary structure, the ATPase IVA belongs to a large class of P-loop GTPases, called the translation factor (TRAFAC) GTPases, that contain proteins involved in translation, signaling, protein transport, and membrane remodeling found in all domains of life. Unlike other nucleotide binding cytoskeletal proteins, we show that IVA assembles into largely static polymers in vitro that do not display dynamic assembly and disassembly. By exploiting the similarity of IVA to TRAFAC GTPases, we made targeted disruptions in IVA that specifically impaired its ATPase activity without abrogating its ability to bind ATP. Such variants of IVA were unable to polymerize in vitro and failed to promote coat assembly in vivo. We show that ATP binding to IVA results in a conformational change in the protein and that subsequent hydrolysis of the bound ATP results in an additional conformational change that drives IVA polymerization into rigid filaments. Interestingly, the conformational change resulting from ATP hydrolysis when the protein was below its threshold concentration for polymerization was a functional intermediate that was capable of polymerization once the threshold concentration for polymerization was exceeded. We propose that proper localization of IVA to the forespore surface, where its local concentration increases, ensures that IVA only polymerizes at the correct subcellular location. IVA and its related orthologs therefore appear to represent a distinct evolutionary solution that arose when the firmicutes first emerged, in which ATP hydrolysis was used to drive the assembly of extremely long-lived structures.
Results
IVA Is a Unique Member of the TRAFAC Class of P-Loop GTPases.
In an effort to identify the basis for IVA’s ATP-dependent polymerization, we first performed computational analyses of its amino acid sequence and its predicted secondary structure. Sequence searches with the BLAST program showed that orthologs of IVA occur only in firmicutes and always in a single orthologous copy per genome. Sequence profile searches with the PSI-BLAST program (29) recovered the Era GTPases (e = 10−25, iteration 2), rather than any type of P-loop ATPase, as the best hit for the N-terminal region of IVA (from Met1 to approximately Val238). We confirmed the specific relationship of the IVA N-terminal region to the Era GTPases using profile-profile comparisons with the HHpred program (30), in which a profile generated using the IVA alignment recovered the Era GTPases as the clear best match (P = 10−17, probability of 90%). This indicated that IVA belonged to the translation factors (TRAFAC) clade of GTPases, which, along with the sSignal recognition particle, MinD, BioD (SIMIBI) clade, comprises the entire superclass of P-loop GTPases that are found in all three domains of life (31) (Fig. 1C, Left). The TRAFAC GTPases include not only translation factors and ribosome-associated proteins but proteins involved in signal transduction and vesicular dynamics, such as those of the extended Ras family; proteins involved in cytokinesis (eukaryotic septins); and GTPases of immunity-associated protein 2 (GIMAP2s) and dynamins involved in membrane remodeling (32, 33). Interestingly, the TRAFAC clade of the GTPase superfamily also includes a number of ATPases of the myosin and kinesin families of motor proteins (31, 34). Like other TRAFAC GTPases, the predicted secondary structure of the N-terminal domain of IVA consisted of seven mostly parallel β-strands that form a central β-sheet flanked on either side by α-helices (Fig. 1B). The predicted nucleotide binding site of IVA harbors a previously described Walker A motif (GxxxxGKS) between the first β-strand and the α-helix (28) that is usually implicated in binding the γ-phosphoryl group of a nucleoside triphosphate (35). In addition, the constructed topology diagram predicted a Walker B motif (hhhhDxxG, where h is a hydrophobic residue) at the end of the fourth β-strand, which contains an aspartate residue (position 97 of IVA; Fig. S1A) that typically binds a water-bridged Mg2+ ion (35, 36) and is absolutely conserved among IVA orthologs. Finally, TRAFAC GTPases are distinguished by a conserved “sensor” threonine or serine residue between the second and third β-strands that participates in coordinating the Mg2+ ion (37). The computed topology diagram of IVA displays a highly conserved threonine at position 70 that may correspond to this residue (Fig. 1B and Fig. S1A). Disruption of the Walker A motif of TRAFAC GTPases typically abolishes GTP binding, whereas disruption of either the Walker B motif or the sensor T generally abrogates GTP hydrolysis specifically, and nucleotide binding tends to be retained (35). A further hallmark of P-loop GTPases is a conserved [NT]KxD motif at the end of the sixth core strand that confers specificity for binding GTP over other nucleoside triphosphates (37). In IVA orthologs, this motif is modified, with the conserved lysine being substituted with a serine or threonine (position 189, a serine, in B. subtilis IVA), consistent with its previously demonstrated inability to bind GTP (28). In an effort to restore potential GTP binding to IVA, we substituted S189 of IVA with lysine. Although B. subtilis cells harboring IVAS189K as the only copy of IVA were unable to sporulate, immunoblot analysis of cell extracts revealed that the resulting protein was present at very low levels (Fig. S2), suggesting that the cell did not tolerate this substitution. Because the protein appeared to be largely unstable in vivo, we did not pursue this avenue of research further.
Beyond the N-terminal GTPase-related domain, based on secondary structure prediction and sequence conservation patterns, we were able to identify two additional C-terminal globular domains (Fig. 1B and Fig. S1A). The predicted secondary structure of the IVA middle domain, which immediately follows the ATPase domain, is composed of two symmetrical units with three-stranded sheets. Consistent with this, profile-profile searches of this domain with the HHpred program recovered the CheX/FliN family of phosphoesterases and flagellar proteins with a comparable secondary structure (P = 10−5, probability of 75%) as the best hits. However, the catalytic residues of the CheX phosphoesterase are not conserved in the IVA middle domain, suggesting that it is likely a structural domain that does not harbor any enzymatic activity. The predicted secondary structure of the extreme C-terminal domain displayed three conserved helices followed by a β-hairpin. This progression, along with profile-profile comparisons, suggested a potential winged-helix-turn-helix (wHTH)–like structure. However, the conservation patterns of typical known DNA binding versions of the wHTH domain were absent in the C-terminal region of IVA. Instead, it showed a highly conserved hydrophobic pattern at the extreme C terminus, which has been shown to be absolutely essential for IVA function (22) and harbors a region that interacts with a small amphipathic protein that anchors IVA to the surface of the forespore (23). IVA was seen as being well conserved across the firmicutes, transcending the deep phylogenetic branches of Bacillales, Clostridia, Moorella, and Thermoanaerobacterium but not outside of firmicutes. This conservation pattern suggests that IVA emerged close to the stem of the firmicute tree via a duplication and rapid divergence from the ancestral Era GTPase that is conserved throughout bacteria (Fig. 1C, Right). Taken together, we conclude that IVA is a relatively ancient protein that arose when sporulation first evolved and that the protein consists of an N-terminal ATPase domain, a middle structural domain, and a hydrophobic C-terminal domain that participates in anchoring IVA to the outer forespore membrane.
Walker B Motif and Sensor T of IVA Are Required for ATP Hydrolysis But Not ATP Binding.
To test if the predicted Walker B motif and sensor T of IVA are required for proper function of IVA in vivo, we disrupted either motif by site-directed mutagenesis and measured the sporulation efficiency of cells harboring the mutant allele of IVA (genotypes are listed in Table S1). Disruption of the Walker B motif of IVA by substituting Asp97 with alanine almost completely abolished the sporulation efficiency in cells harboring IVAD97A (hereafter called IVAB*) as the only copy of IVA (Table S2, strain D). Substitution of the predicted sensor T at position 70 with Ala resulted in a ∼500-fold decrease in sporulation efficiency in cells harboring IVAT70A as the only copy of IVA. We noticed that IVA of B. subtilis harbors a less conserved threonine at position 71, immediately adjacent to the predicted (and well-conserved) sensor T at position 70 (Fig. S1A). Substitution of both threonine residues with alanine resulted in a five-log decrease in sporulation efficiency (Table S2, strain E), whereas substitution of the less conserved Thr71 alone with alanine did not abrogate sporulation (Table S2, strain G). We therefore concluded that Thr70 is primarily required for IVA function in vivo, whereas the adjacent Thr71, although not necessary for IVA function, may partially compensate for disruption of the well-conserved Thr70; consequently, we used the IVAT70A,T71A (hereafter called IVAT*) double mutant allele of IVA in all further experiments in which we wished to disrupt the sensor T. Immunoblot analysis of sporulating extracts of B. subtilis indicated that disruption of the Walker B motif or the sensor T did not appreciably reduce the steady-state levels of the protein compared with the WT protein produced from an ectopic chromosomal locus (thr, Fig. S3).
Next, we wished to examine if disruption of either the putative Walker B motif or sensor T specifically abrogated the ATPase activity of IVA without affecting ATP binding. We therefore produced in and purified from Escherichia coli WT IVA, IVAB*, or IVAT* harboring an N-terminal six-histidinyl tag. We then measured the affinity of IVA for ATP by exploiting the ability of dry nitrocellulose membranes to separate free radiolabeled ligands from protein–ligand complexes by capillary diffusion (38, 39). Incubating varying amounts of purified protein with a limiting amount of radiolabeled nucleotide produced the saturation binding curve in Fig. S1C, which indicates that half-maximal binding was achieved at 8.0 ± 1.5 μM. As a negative control, purified IVA harboring a previously described K30A substitution that disrupted the Walker A motif (28) did not appreciably bind the nucleotide (A*, Fig. S1C). Purified IVAB* and IVAT* achieved half-maximal binding of ATP at 6.1 ± 1.8 μM and 11.8 ± 3.2 μM, respectively, suggesting that disruption of either the Walker B motif or the sensor T did not abrogate the ATP binding capacity of IVA (the modest increase and decrease in affinity for B* and T*, respectively, were within the errors of measurement). We then investigated the ability of these variants to hydrolyze the bound nucleotide by incubating the purified proteins with varying concentrations of [α-32P]-ATP and directly measuring the production of ADP by separating the products of the reaction by TLC. The saturation curve in Fig. S1D indicates that incubating WT IVA with ATP resulted in ATP hydrolysis with a turnover rate of about 1.0 pmol⋅min⋅pmol of IVA at Vmax. In contrast, both IVAB* and IVAT* hydrolyzed ATP at levels similar to that of IVAA* (which was largely unable to bind ATP). We therefore conclude that disruption of the Walker B motif or the sensor T of IVA specifically abrogates ATP hydrolysis while leaving ATP binding largely unaffected.
ATP Hydrolysis Drives the Stable Polymerization of IVA.
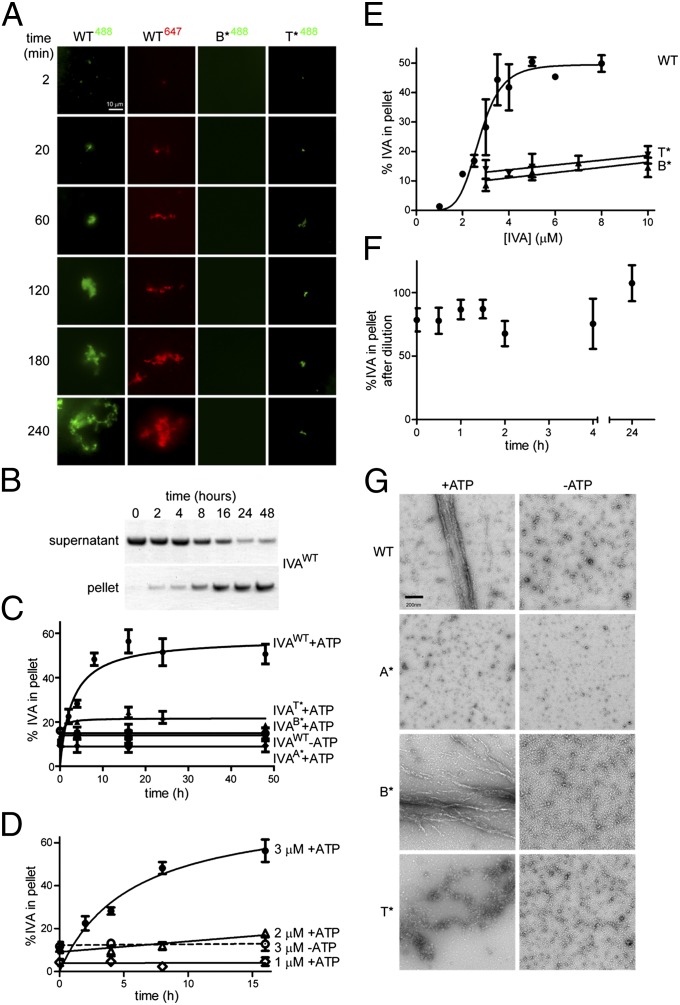
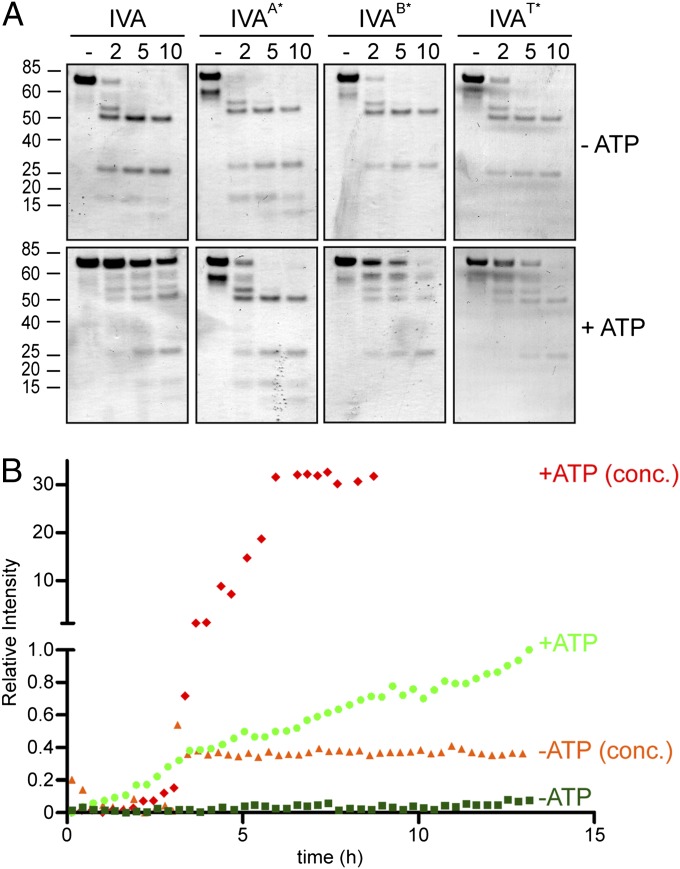
Previously, we had proposed that the polymerization of IVA may require not simply ATP binding but ATP hydrolysis as well (28). To test this hypothesis rigorously, we established two assays that measured polymerization over time of WT IVA or either IVA variant that was capable of binding but not hydrolyzing ATP. For the first assay, we covalently linked the fluorescent molecule Alexa 488 to purified IVA, incubated the labeled protein in buffered solution in the presence of ATP, and monitored the accumulation of insoluble (polymerized) IVA over time on the surface of a glass coverslip using total internal reflection fluorescence (TIRF) microscopy without any further manipulation of the sample. Unlike standard epifluorescence microscopy, which detects the fluorescence emitted from the entire depth of an illuminated sample, the use of TIRF microscopy allowed us to detect only the labeled material that accumulated on the interface between the glass and the aqueous sample, thereby allowing us to monitor selectively the polymerized labeled IVA that had fallen out of solution. At the onset of our observations, we were unable to detect any accumulated signal, but 20 min after the reaction started, we began to observe patches of fluorescent material that occupied a larger surface area over time as it increased in size roughly radially outward (WT488, Fig. 2A). Similarly, when WT IVA was labeled with a different fluorophore, Alexa 647, the labeled protein also accumulated on the surface of the slide over time. In contrast, we were unable to detect any such accumulation of fluorescent label anywhere on the glass slide when we repeated the experiment with labeled IVAB* (B*488, Fig. 2A), and only rarely did we detect small patches of accumulated fluorescent signal when the experiment was performed with labeled IVAT* (T*488, Fig. 2A).
Fig. 2.
ATP hydrolysis drives the self-assembly of static IVA polymers. (A) Purified WT IVA labeled with Alexa 488 (first column) or Alexa 647 (second column) and IVAB* or IVAT* labeled with Alexa 488 (third and fourth columns, respectively) were incubated with ATP, and accumulation of polymerized insoluble material on the surface of the slide over time was monitored using TIRF microscopy. (B) Representative Coomassie-stained polyacrylamide gel of supernatant and pellet fractions of purified IVA incubated in the presence of ATP after centrifugation of aliquots at different indicated time points to monitor polymerization. (C) Kinetic analysis of polymerization of various IVA variants (●, WT; ♦, IVAA*; ■, IVAB*; ▲, IVAT*) in the presence of ATP or WT IVA in the absence of ATP (○) measured by centrifugation. (D) Polymerization of IVA is concentration-dependent. A total of 1 μM, 2 μM, or 3 μM purified IVA was incubated with or without ATP, and polymerization was measured over time by centrifugation. (E) Critical concentration for IVA polymerization. Varying concentrations of purified IVA, IVAB*, or IVAT* were incubated with ATP for 16 h, and polymerization was measured by centrifugation. Data for IVA polymerization were fit using the Hill model. (F) Polymerized IVA is largely static. Purified IVA (3 μM) was incubated for 16 h with ATP. Analysis of a parallel reaction indicated that about 50% of the total IVA had polymerized by this time (this value was set as 100% polymerized at t0). The reaction was then diluted threefold to below the threshold concentration for polymerization (final concentration of 1 μM), and samples of the reaction were centrifuged at the indicated time points to measure the amount of polymerized IVA in the pellet after dilution, normalized against the amount of polymerized IVA before dilution. All symbols represent mean values of three to four independent measurements; error bars represent SEM. (G) Transmission electron micrographs of purified IVA variants after incubation in the presence (Left) or absence (Right) of ATP. (Scale bar: 200 nm.) A*, B*, disruption of Walker A or Walker B motif, respectively; T*, disruption of sensor threonine (Thr).
Although the TIRF assay allowed us to observe polymerization of IVA without extensive manipulation of the sample after polymerization was initiated, the assay did not allow us to quantify and compare the total unpolymerized and polymerized material easily. To measure polymerization of IVA more quantitatively, we exploited the property of polymerized, but not unpolymerized, IVA to accumulate in the pellet fraction after ultracentrifugation. Fig. 2B shows a Coomassie-stained polyacrylamide gel of purified IVA found in either the supernatant or pellet fraction that was taken at various time points after the initiation of IVA polymerization by addition of ATP. Using this assay, we observed the disappearance of IVA in the supernatant and the concomitant appearance of IVA in the pellet fraction over time. Quantifying the intensity of the bands in Fig. 2B revealed that IVA reached a maximal level of polymerization in approximately 8 h and that prolonged incubation (up to 48 h) in the presence of ATP did not result in destabilization of already polymerized material (Fig. 2C). Inclusion of an ATP regeneration system in the reaction resulted in similar polymerization kinetics and absolute level of polymerization (Fig. S4A), suggesting that it was not the depletion of ATP or the accumulation of ADP and phosphate that prevented the quantitative polymerization of IVA in Fig. 2C. As a negative control, IVA did not increasingly accumulate in the pellet fraction over time in the absence of ATP, nor did IVAA*, which is unable to bind ATP, in the presence of ATP. Similarly, IVAB* and IVAT* also did not accumulate in the pellet fraction to significant levels over time (Fig. 2C), although IVAT* did display a higher background level of insolubility than either IVAA* or IVAB* (consistent with our observation that IVAT* occasionally formed small insoluble aggregates visible in the TIRF assay). Next, we varied the concentration of IVA and measured the accumulation of polymerized IVA over time in the pellet fraction after ultracentrifugation. As seen before in Fig. 2C, WT IVA, at a concentration of 3 μM, accumulated in the pellet fraction over time strictly in the presence of ATP (Fig. 2D). However, at lower concentrations of protein (2 μM or 1 μM), we failed to detect any polymerization of the protein even after 16 h of incubation with ATP (Fig. 2D). To determine the critical concentration for polymerization of IVA, we varied the concentration of the protein and measured polymerization by centrifugation after 16 h of incubation in the presence of ATP to generate the sigmoidal curve in Fig. 2E. The data were fit to the Hill model to reveal that the critical concentration for polymerization (the inflection point of the sigmoidal curve) was ∼2.7 μM IVA (the data plotted on a logarithmic scale, wherein the y-axis intercept reveals the critical concentration, are shown in Fig. S5). In contrast, repeating the experiment with either IVAB* or IVAT* failed to generate a saturation curve, even at a protein concentration of 10 μM, indicating that these variants did not display a measurable critical concentration for polymerization. Taken together, we conclude that IVA polymerization requires ATP hydrolysis, not simply ATP binding, and that successful polymerization, measured by centrifugation in the buffer conditions used here, requires a critical protein concentration of about 2.7 μM.
The basement layer of the spore coat is a structure that is presumed to be extremely stable; thus, we have assumed that ATP-dependent polymerization of IVA must form a static structure that does not display the dynamic polymerization and depolymerization exhibited by other nucleotide-dependent cytoskeletal proteins like actin or tubulin. To test formally if IVA polymers are static structures that do not disassemble, we took advantage of the observation that IVA polymerization requires a minimum concentration of protein. We therefore allowed purified IVA (final concentration of 3 μM) to polymerize for 16 h; at that time, 49.8 ± 9.7% of the total protein had polymerized, as measured by centrifugation of a parallel reaction. At this time, the reaction mix was diluted threefold in the same buffer containing ATP; thus, the final concentration of IVA was below the polymerization threshold. At various time points, aliquots were centrifuged and the amount of polymerized IVA in the pellet was measured and reported as a percentage of polymerized IVA before dilution (Fig. 2F). Polymers of cytoskeletal proteins like tubulin, which display dynamic instability, rapidly disassemble on dilution below a threshold polymerization concentration because subsequent reassembly of polymers is prevented (40). In contrast, we found that IVA polymers, once assembled, remained largely polymerized immediately on dilution and that this polymerized material remained stable for even 24 h after dilution. Thus, we conclude that unlike other nucleotide binding cytoskeletal proteins, IVA polymers, once assembled, remain largely static and do not disassemble in the time frames examined here.
To observe the ultrastructure of polymerized IVA, we examined the polymerized material (without centrifugation) by EM. As we previously reported, negative staining revealed that polymerized IVA formed filaments ∼10 nm in diameter but often several tens of microns in length (Fig. 2G). Curiously, unlike actin filaments, which have a similar diameter, IVA filaments appeared rather rigid, often extending even up to 100 μm in length while assuming only a very slight curvature (Fig. S4B). Such structures were not observed in samples incubated in the absence of ATP (Fig. 2G). When purified IVAB* was incubated in the presence of ATP, we rarely observed any higher order structures by EM; however, occasionally, we saw short filaments (several hundred nanometers in length) that did not exceed 1 μm in length (Fig. 2G). Incubation of IVAT* with ATP very rarely produced filaments that exceeded 5 μm in length, but they were often broken and the protein usually aggregated into structures of nonuniform shape (Fig. 2G). We conclude that ATP hydrolysis, and not simply ATP binding, drives the assembly of IVA into long, static, rigid filaments.
Continuous ATP Hydrolysis Is Required for IVA Polymerization.
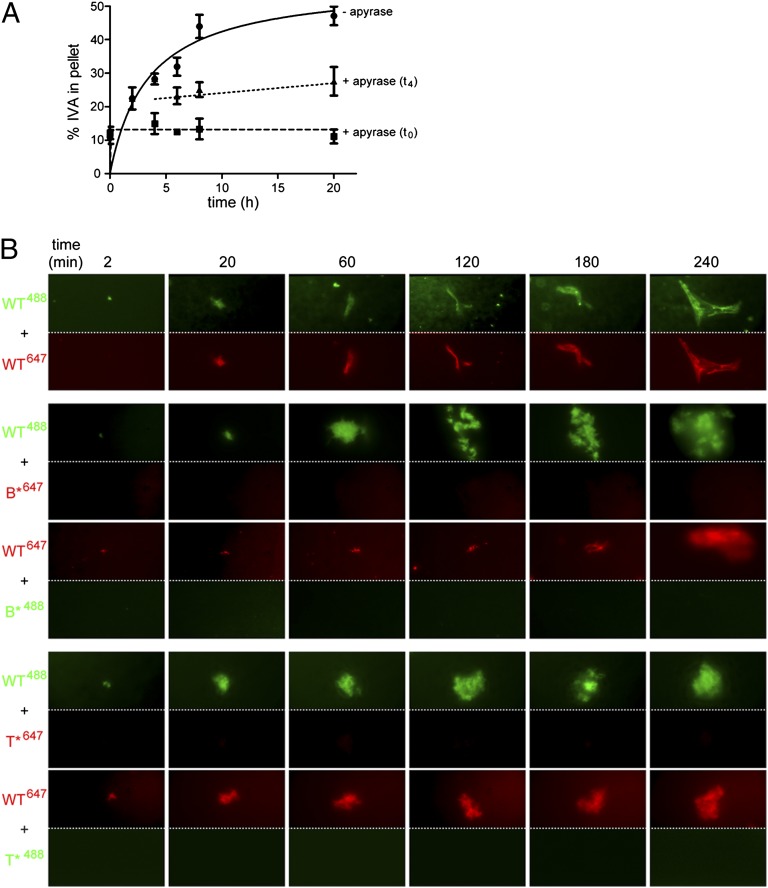
Although the data thus far suggested that ATP hydrolysis was required for robust polymerization of IVA, it was unclear if every IVA monomer incorporated into the growing filament was required to bind and hydrolyze ATP. To test if ATP hydrolysis was continuously required during polymerization, we sought to deplete the pool of ATP at various time points and to monitor if IVA, once assembly was initiated, would continue to polymerize in the absence of ATP; if polymerization would arrest on ATP depletion; or if the already polymerized material would subsequently disassemble. In the presence of ATP, purified IVA polymerized over time, as measured by ultracentrifugation. However, addition of apyrase, an enzyme that catalyzes the hydrolysis of ATP to yield AMP and inorganic phosphate, at the onset of the polymerization reaction prevented the assembly of IVA, suggesting that the enzyme was able to deplete the pool of ATP rapidly in the reaction mix (Fig. 3A). Addition of apyrase 4 h after the initiation of polymerization, when ∼30% of IVA had already polymerized, resulted in the immediate arrest of further polymerization of IVA, suggesting that subsequent assembly of IVA requires the presence of ATP. In addition, IVA that had already assembled did not disassemble on depletion of ATP, further suggesting that the continuous presence of ATP is only required for assembly, but not for the subsequent stability, of IVA polymers.
Fig. 3.
Continued ATP hydrolysis is required for IVA polymerization. (A) Kinetic analysis of IVA polymerization in the presence of ATP as measured by centrifugation in the absence of apyrase (●), when apyrase was added at the beginning of the experiment (■), or when apyrase was added after 4 h of incubation with ATP (▲). Symbols represent mean values of at least three independent measurements; error bars represent SEM. (B) Mixing experiment wherein WT IVA and IVA variants were labeled with a fluorophore [Alexa 488 (green) or Alexa 647 (red)] and incorporation of either variant into the growing polymer over time (indicated above in minutes) was observed using TIRF microscopy. The top and bottom of each panel represent the same field of view taken with the appropriate filter to detect the indicated fluorescent label. First row: WT IVA labeled with Alexa 488 mixed with an equimolar amount of WT IVA labeled with Alexa 647. Second and third rows: WT IVA, labeled with either Alexa 488 or Alexa 647, mixed with an equimolar amount of IVAB*, labeled with either Alexa 647 or Alexa 488, respectively. Fourth and fifth rows: Similar to rows 2 and 3, but WT IVA was mixed with IVAT*.
Next, we wished to examine if individual IVA molecules that do not hydrolyze ATP are able to incorporate into growing IVA polymers in the presence of WT IVA. We therefore performed a mixing experiment in which we combined WT IVA labeled with one color fluorophore with either IVAB* or IVAT* labeled with another fluorophore and measured the incorporation of the mutant protein into WT IVA polymers using TIRF microscopy. As a positive control, we first mixed WT IVA labeled with green fluorophore (Alexa 488) with WT IVA labeled with red fluorophore (Alexa 647). As expected, polymerized material that assumed a similar shape assembled on the glass slide over time when viewed with either a green or red filter (Fig. 3B, first row), indicating that IVA labeled with either fluorophore is capable of incorporating into a growing polymer. When equimolar amounts of WT IVA labeled with green dye were mixed with IVAB* labeled with red dye, increased accumulation of WT IVA labeled was detectable on the glass slide, but we did not observe any incorporation of IVAB* labeled with red dye in the same field of view (Fig. 3B, second row). Similarly, when we mixed WT IVA labeled with red dye with IVAB* labeled with green dye, we observed the accumulation of WT IVA but not IVAB* (Fig. 3B, third row) over time. Similar results were obtained when we repeated the experiment using IVAT* (Fig. 3B, fourth and fifth rows). In total, the data suggest that the continuous presence of ATP is required to drive IVA polymerization, that removal of ATP does not destabilize IVA polymers that have already assembled, and that each molecule of IVA must hydrolyze ATP to incorporate into a growing polymer.
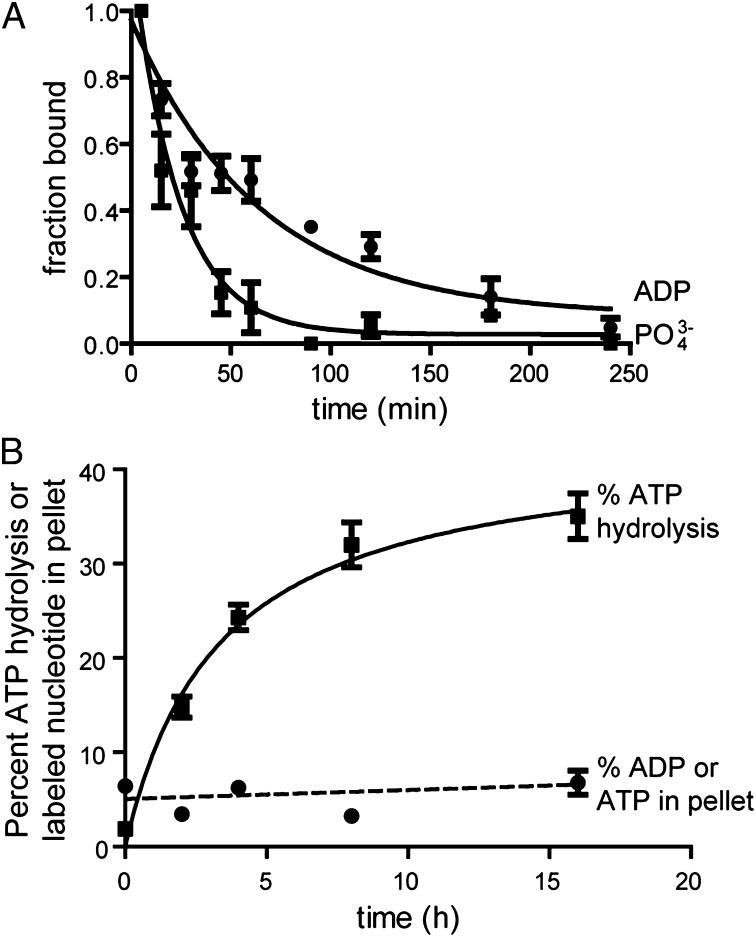
To test if the nucleotide remains associated with IVA after hydrolysis and polymerization, we performed two tests. First, we incubated purified IVA with a limiting amount of ATP labeled with 32P at either the α- or γ-phosphoryl group (to track the fate of ADP or phosphate, respectively). At various time points, the reaction was spotted onto nitrocellulose and the amount of free and bound phosphate or ADP was measured as described above. The results in Fig. 4A show that on hydrolysis, the γ-phosphate was rapidly released. In addition, the resulting ADP was also released by IVA, albeit at a slightly slower rate than the phosphate, suggesting that neither the ADP nor phosphate resulting from hydrolysis remained associated with individual molecules of IVA. To test if ATP or ADP is associated with IVA polymers, we incubated purified IVA with radiolabeled [α-32P]-ATP under polymerizing conditions. At different time points, we measured ATP hydrolysis, IVA polymerization by centrifugation, and the amount of radiolabeled nucleotide associated with the pellet after centrifugation. After 16 h, 44.7 ± 13.7% of total IVA had polymerized and 35 ± 6.4% of the total ATP in the reaction mix had been hydrolyzed (Fig. 4B). However, the amount of radiolabel in the pellet did not increase significantly over background levels as IVA polymerization proceeded (Fig. 4B), suggesting that neither ADP nor ATP was appreciably associated with polymerized IVA. Taken together, we conclude that unlike unstable polymers, IVA molecules release the resulting ADP soon after hydrolysis of the bound ATP and that IVA polymers are largely devoid of bound nucleotide on assembly.
Fig. 4.
IVA releases the bound nucleotide on hydrolysis and polymerization. (A) Kinetic analysis of ADP and phosphate release. Five micromolar purified IVA was incubated with 7 nM [α-32P]-ATP (●) or [γ-32P]-ATP (■) to measure the fate of ADP and phosphate (PO3−4), respectively, and the fraction of bound radiolabel was measured at various time points using the differential radial capillary action of ligand assay and reported as a fraction that initially bound (t = 1 min). (B) Radiolabeled ATP associated with polymerized material was measured by incubating 3 μM purified IVA with [α-32P]-ATP and 4 mM unlabeled ATP. At various time points, aliquots of the polymerization reaction were centrifuged and the amount of radiolabel in the pellet, comprising either ATP or ADP, was measured (●). Aliquots were also removed at various time points, and ATP hydrolysis was measured using TLC (■). Symbols represent mean values of three to eight independent measurements; error bars represent SEM.
ATP Hydrolysis Drives a Conformational Change of IVA That Makes It Polymerization-Competent.
The continued requirement for ATP during IVA polymerization, along with the observation that every molecule of IVA must be able to hydrolyze ATP, led us to wonder if ATP hydrolysis drives a conformational change in IVA that results in polymerization. To avoid measuring conformational changes that would naturally result from protein–protein interactions during polymerization, we took advantage of the observation that IVA is able to hydrolyze ATP but unable to polymerize at lower concentrations (Fig. 2 D and E) and monitored changes in IVA tertiary structure by examining its susceptibility to degradation by limited periods of proteolysis. We therefore incubated 2 μM WT IVA for 4 h in the presence or absence of ATP, digested the sample with trypsin for 0–10 min, separated the products of digestion by gel electrophoresis, and then visualized the products by Coomassie staining of the gel (Fig. 5A). In the absence of ATP, IVA was almost completely degraded even after a 2-min exposure to trypsin; however, after incubation with ATP in nonpolymerizing conditions, IVA was largely resistant to degradation even after 10 min of exposure trypsin, suggesting that the protein had undergone a structural change. As a negative control to ensure that the presence of ATP did not interfere with the proteolytic activity of trypsin, we repeated the experiment with IVAA*. After a 4-h incubation both in the presence and absence of ATP, IVAA*, which is largely unable to bind ATP, displayed an increased sensitivity to proteolysis by trypsin, similar to the pattern shown by WT IVA in the absence of ATP. Next, we measured the sensitivity of IVAB* and IVAT*, which bind but do not hydrolyze ATP, to tryptic digestion. Like WT IVA, both variants were rapidly degraded by trypsin in the absence of ATP. However, unlike IVAA*, IVAB* and IVAT* were more resistant to trypsin after incubation with ATP but less resistant than WT IVA, requiring only about 5 min for almost complete degradation of the protein. Thus, the IVA variants that are able to bind but not hydrolyze ATP displayed an intermediate resistance to trypsin digestion. To ensure that the presence or absence of ATP, even when IVA concentration was below the critical concentration for polymerization, did not affect the oligomerization state of IVA and variants (and thus influence trypsin susceptibility), we examined the elution profile of the purified proteins by size exclusion chromatography (Fig. S4). At a concentration of 2 μM, all IVA variants, in the presence and absence of ATP, eluted as a single peak at the same volume (Fig. S6A), suggesting that the presence or absence of ATP did not affect the oligomerization state of IVA. Interestingly, the elution volume of purified IVA and variants, compared with the elution of size markers of known molecular weight, corresponded to a 202-kDa species. Assuming that IVA behaves as a globular protein, this molecular weight corresponds to ∼3.5 subunits of IVA, suggesting that a single functional unit of IVA, assuming that it behaves as a globular protein, may be a trimer or tetramer. To test if ATP binding alone, above the critical concentration for polymerization, would result in oligomerization of this minimal unit, we examined the elution profile of 4 μM IVAB* and IVAT*, after incubation with ATP for 16 h, by size exclusion chromatography. Immunoblot analysis of the various fractions revealed that even above the critical concentration for polymerization, both variants continued to migrate at an elution volume that corresponded to the single functional unit, indicating that ATP binding alone did not alter the oligomerization state of IVA. Taken together, we conclude that ATP binding results first in a conformational change in the minimal functional unit of IVA (perhaps a trimer or tetramer, based on migration in a size exclusion column) and that ATP hydrolysis drives a further change in conformation of IVA, all without affecting the oligomerization state of the protein.
Fig. 5.
ATP hydrolysis drives a conformational change in IVA to form an intermediate that is able to polymerize once the protein concentration is raised above the threshold. (A) Purified IVA variants (WT, IVAA*, IVAB*, and IVAT* at 2 μM, below the threshold concentration for polymerization) were incubated either in the absence (Upper) or presence (Lower) of ATP at 37 °C for 4 h. The reactions were then exposed to limited proteolysis by trypsin for the times indicated (−, no trypsin; 2, 5, or 10 min); after that time, the proteolysis was stopped by addition of sample buffer and the products were promptly analyzed by Coomassie-stained PAGE. (Left) Mobility of molecular weight markers (kilodaltons) is shown. (B) Purified IVA at a concentration of 4 μM (above the threshold concentration for polymerization) was incubated at 37 °C in the presence (light green) or absence (dark green) of ATP, and polymerization was measured over time by dynamic light scattering. Alternatively, 2 μM (below the threshold concentration) IVA was first incubated in the presence (red) or absence (orange) of ATP for 3 h at 37 °C. The sample was then concentrated by pressure dialysis to achieve a final concentration of 4 μM IVA, and polymerization continued to be monitored by dynamic light scattering. The data were normalized to the maximum level of polymerization achieved by IVA in the presence of ATP at t = 13 h.
Does ATP hydrolysis by IVA below the critical concentration for polymerization result in a polymerization-competent intermediate or a dead-end form of the protein that is unable to polymerize further? To distinguish between these possibilities, we first incubated purified IVA at a concentration of 2 μM in the presence of ATP for 4 h and then increased the concentration of the protein to 4 μM (above the threshold concentration for polymerization) by pressure dialysis and monitored polymerization of the protein using dynamic light scattering. Incubation of 4 μM IVA in the presence, but not absence, of ATP resulted in an increase in the light scattering signal over time, suggesting that the assay properly measured IVA polymerization (Fig. 5B). When we incubated 2 μM IVA in the presence of ATP for 4 h (at which time ATP hydrolysis approaches a plateau; Fig. 4B) and then increased IVA concentration to 4 μM, we observed a sharp increase in the light scattering signal. As a negative control, preincubation of IVA for 4 h in the absence of ATP, followed by increasing the concentration of the protein, failed to result in a steady increase in the signal over time, indicating that the increase in light scattering that we observed in the presence of ATP was due to polymerization and not due to unspecific aggregation of the protein attributable to pressure dialysis. Taken together, the data suggested that premature hydrolysis of ATP, under nonpolymerizing conditions, did not prevent subsequent polymerization of IVA once polymerization conditions were restored. We therefore conclude that ATP hydrolysis by IVA under nonpolymerizing conditions results in a polymerization-competent intermediate that will only polymerize once the threshold concentration is exceeded.
ATP Hydrolysis by IVA Is Required in Vivo for Proper Assembly of the Spore Coat.
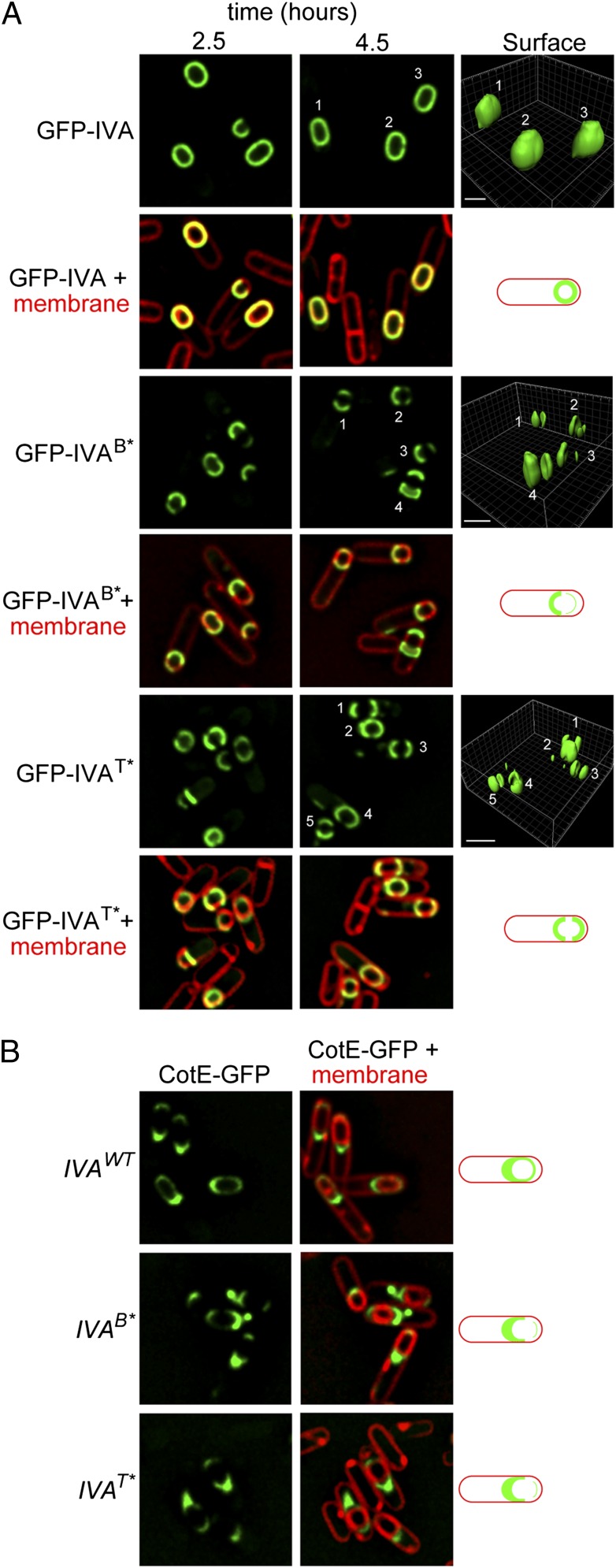
The proper localization and polymerization of IVA on the surface of the forespore is an absolute requirement for the subsequent assembly of the spore coat, which is built atop the platform provided by IVA. To determine if IVA variants that are unable to hydrolyze ATP are defective in vivo in coat assembly, we first examined the subcellular localization of IVA fused to GFP in sporulating cells. As previously reported, GFP-IVA forms a uniform shell around the developing forespore, and in those cells that have not finished engulfment, GFP-IVA tracks the engulfing membrane (16, 22) (Fig. 6A). Whereas GFP-IVA harboring a disruption in the Walker A motif was completely mislocalized in the mother cell cytosol (28), GFP-IVAB* and GFP-IVAT* both properly localized to the surface of the developing forespore. However, unlike WT IVA, GFP-IVAB* and GFP-IVAT* did not form a uniform shell around the forespore, instead localizing as two caps at the mother cell-proximal and mother cell-distal forespore poles (Fig. 6A). This failure to encase the forespore (18, 27) persisted even at a later time point during the sporulation program (4.5 h after initiation), because 91.5% of cells producing GFP-IVAB* (n = 201) and 80% of cells producing GFP-IVAT* (n = 330) failed to form a uniform shell around the forespore, compared with only 7.6% (n = 196) of the cells producing WT IVA that failed to do so. To ensure that free GFP was not liberated from the fusion construct, we examined extracts prepared from these cells by immunoblotting, which revealed that the GFP fusions remained intact (Fig. S3). Quantification of the fluorescence intensity revealed that the levels of GFP-IVAB* and GFP-IVAT* were 80 ± 18% (n = 10) and 113 ± 38% (n = 10), respectively, relative to GFP-IVA, suggesting that both variants were produced at levels similar to the WT protein. To monitor if later coat assembly steps were indeed disrupted, we examined the localization of CotE (a component of the outer layer of the spore coat) fused to GFP (41). In cells harboring WT IVA, CotE-GFP localized as a nonuniform shell on the forespore surface with a biased accumulation on the mother cell-proximal face of the forespore (42). In contrast, in cells expressing either IVAB* or IVAT* as the only allele of IVA, CotE-GFP mainly localized as a single cap on the mother cell-proximal face of the forespore, which was disconnected from any CotE-GFP fluorescence that may have localized at the mother cell-distal pole of the forespore. Quantification of the fluorescence intensity revealed that CotE-GFP was produced at 105 ± 40% (n = 10) and 89 ± 27% (n = 10) in the presence of IVAB* and IVAT*, respectively, relative to the strain harboring WT IVA, suggesting that CotE was produced at a level similar to that of the WT strain. Together with the observation that cells harboring only IVAB* or IVAT* exhibit a severe sporulation defect (Table S2), we conclude that failure of IVA to hydrolyze ATP and polymerize in vivo results in the misassembly of the basement layer of the spore coat and, by extension, the assembly of the entire coat, causing a sporulation defect.
Fig. 6.
Disruption of IVA ATP hydrolysis abrogates coat assembly in vivo. (A) Localization of GFP fused to WT IVA (Top, strain JPC156), IVAB* (Middle, strain JPC174), or IVAT* (Bottom, strain JPC243) in cells 2.5 h (Left) or 4 h (Center) after the induction of sporulation is shown. An overlay of GFP fluorescence and membranes visualized with the fluorescent dye FM4-64 (added to the sporulating culture) is shown below each corresponding panel. (Right) GFP intensity represented as a 3D surface and schematic representation of the localization of GFP fusions are shown. (B) (Left and Center) Localization of CotE-GFP in the presence of WT IVA (Top, strain KR448), IVAB* (Middle, strain JPC138), or IVAT* (Bottom, strain JPC281). (Right) Overlay of GFP fluorescence and membranes and schematic representations of CotE-GFP localizations are shown to the right of corresponding panels. Strain genotypes are listed in Table S1.
Discussion
In this report, we have described a bacterial morphogenetic protein called IVA whose polymerization into rigid, static filaments is driven by a conformational change that requires ATP hydrolysis and not simply ATP binding. Analysis of the primary amino acid sequence of IVA revealed that the predicted secondary structure of the amino terminal domain of the protein resembles the TRAFAC class of P-loop GTPases, which typically function as switches in protein synthesis, signal transduction, and membrane remodeling but not in protein polymerization. Specifically IVA appears to have been derived in firmicutes from a GTPase of the Era family, which binds single-stranded 16S rRNA via its C-terminal KH domain and mediates the assembly of the 30S ribosomal subunit. A comparison of the phyletic distribution of the IVA protein across firmicutes shows a strong concordance with reports of sporulation (43). For example, there is no instance of the occurrence of IVA in the Lactobacillales clade. Similarly, within the Bacillales clade, both Staphylococcus and Listeria, which are known to lack sporulation, also lack an IVA ortholog. The picture gets more complicated in the Clostridiales clade, however, wherein certain lineages with reportedly no endospore formation do display IVA (e.g., Faecalibacterium and Ruminococcus). Similarly, in the more basal halotolerant lineages (e.g., Halothermothrix, Natranaerobius) and the basal thermophilic lineages (e.g., Thermoanaerobacter, Caldanaerobacter), there is no evidence for sporulation, although they do display a bona fide IVA ortholog. These observations, together with the evidence that IVA is the primary structural and temporal determinant of sporulation, have considerable bearing on the evolution of sporulation. First, some of these cases, such as Ruminococcus, have closely related taxa that are sporulating (e.g., Sporobacter), suggesting that they might have lost sporulation recently or possibly display cryptic sporulation. Indeed, some firmicutes, such as C. difficile, exhibit sporulation only on special media, and others, such as Epulopiscium, exhibit it only at night. Although there is little evidence for sporulation in the above-mentioned basal extremophilic firmicutes, they nonetheless exhibit some remarkable features, such as resistance to 120 °C for 45 min (e.g., Caldanaerobacter), which are typically observed only in endospores. IVA might contribute in these organisms to a protective structure that might be mechanistically similar but morphologically distinct from the classic endospore. Indeed, the possibility is raised that such temperature-resistance features in the extremophilic firmicutes eventually gave rise to classic endospores (43, 44).
Although Era and IVA appear to have very distinct functions, it should be noted that several members of the TRAFAC family, including Era, undergo notable conformational changes on binding and hydrolysis of GTP (45, 46). Additionally, members of the Era family have been observed to function without the need for a GTPase-activating protein (GAP) or GDP-exchange factor (GEF), and our in vitro results for ATP hydrolysis by IVA are consistent with a similar GAP- or GEF-free action. However, IVA is distinguished from most other TRAFAC GTPases in binding ATP, and not GTP, which appears to result from the alteration of the classic [NT]KxD motif that confers GTP binding specificity. Most dedicated ATPases among the P-loop NTPases belong to the additional strand conserved glutamate (ASCE) division as opposed to the kinase-GTPase division. The ATPases of the ASCE division use a glutamic acid residue as a base to catalyze the removal of the proton from the “attacking” nucleophile during hydrolysis. The two other exceptional members of the TRAFAC class that bind and hydrolyze ATP instead of GTP are kinesins and myosins, which arose only in eukaryotes. The mechanism of ATP hydrolysis in the case of myosin/kinesin largely mirrors the action of the related GTPases (47). In this mechanism, the proton from the hydrolytic water is ultimately relayed to the γ-phosphate, which, from the viewpoint of catalysis, takes the place of the glutamic acid seen in the ASCE division ATPases. Given that IVA lacks a conserved glutamic acid near the predicted ATPase active site, we propose that it might use a mechanism similar to myosin/kinesin for ATP hydrolysis. However, IVA differs from these proteins in that it uses the hydrolysis of ATP to drive its own polymerization rather than transportation through motor action.
Our model that IVA polymerization requires a conformational change driven by ATP hydrolysis, not simply ATP binding, is based on four observations. First, IVA harbors two motifs (a Walker B motif in the fourth β-strand and a sensor threonine in the third β-strand), conserved in every known IVA ortholog, that are required by TRAFAC GTPases to hydrolyze the bound nucleoside triphosphate. Disruption of either of these motifs did not abrogate ATP binding by IVA but did largely abolish ATP hydrolysis and concomitantly abolished effective polymerization of purified IVA in vitro. Second, IVA variants that simply bind, but do not hydrolyze, ATP were unable to incorporate in vitro into actively growing polymers of WT IVA, suggesting that every molecule of IVA is required to hydrolyze ATP to polymerize. Third, limited proteolysis experiments with purified IVA protein revealed that IVA undergoes a structural change on ATP binding and that ATP hydrolysis drives a further change in conformation that is coincident with polymerization. Failure to hydrolyze the bound ATP prevented the second conformational change. Fourth, IVA variants that did not hydrolyze ATP did not assemble into a uniform shell on the surface of the developing forespore in vivo, failed to promote the proper assembly of subsequently assembling spore coat proteins, and led to a severe sporulation defect.
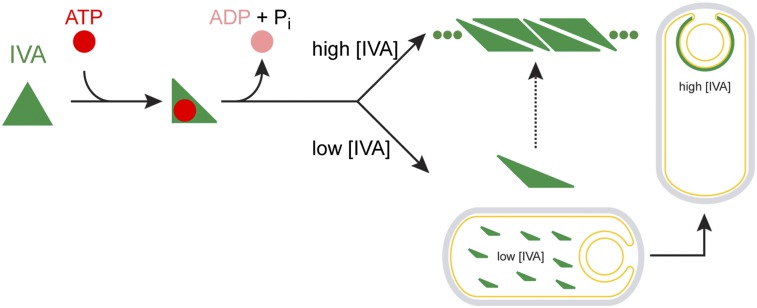
Taken together, Fig. S7 depicts our working model that describes the ATP-dependent polymerization of IVA. We propose that IVA, once synthesized in the mother cell cytosol, binds ATP, which results in the first conformational change in IVA (depicted as an equilateral triangle-to-right triangle change). Migration of this minimal assembly unit of IVA by size exclusion chromatography suggests that the protein may a trimer or tetramer. This is comparable to what has been reported in the related TRAFAC GTPases, such as Era and GIMAP, where nucleotide binding induces conformational change between the GTPase domain and the C-terminal domains or extensions (33, 46). Thereafter, the bound nucleotide is hydrolyzed and the resulting ADP and phosphate molecules are released. Above a certain critical concentration of IVA (2.7 μM IVA in vitro in the buffer conditions reported here), ATP hydrolysis drives the polymerization of IVA into filaments (Fig. S7, upper arrow). At concentrations below the threshold, ATP hydrolysis drives a second conformational change in IVA that may or may not be distinct from the conformation that IVA adopts on polymerization. Under these conditions, rather than representing a terminal off-pathway event in which polymerization cannot subsequently be achieved, our data suggest that this form of IVA is a polymerization-competent intermediate that is able to polymerize once the threshold concentration for polymerization is achieved. It has previously been unclear to us how the premature polymerization of IVA is prevented in the mother cell cytosol, despite the abundance of ATP in this compartment. It is now tempting to speculate that the concentration of IVA in the mother cell is below the in vivo threshold concentration for polymerization and that recruitment of IVA to the forespore surface would raise the local concentration of IVA above the threshold and ensure that IVA polymerization only occurs at the correct subcellular location. Thus, although not ruling out any yet-to-be-identified in vivo factors that may stimulate IVA polymerization, our working model suggests that ATP hydrolysis, combined with an increase in local concentration of IVA (driven by proper localization), is sufficient to drive IVA polymerization specifically at the forespore surface and to prevent spurious polymerization elsewhere.
To our knowledge IVA polymerization is mechanistically unique compared with other well-known instances of biological nanostructure formation. Dynamic cytoskeletal assemblies, such as microtubules (made of tubulin) and thin filaments (made of actin), bind nucleotides to drive their assembly. Subsequent hydrolysis of the bound nucleotide drives disassembly of the polymer, and therefore accomplishes two goals. First, the process becomes reversible, such that the cell can assemble and disassemble a structure according to its changing requirements. Second, energy released by nucleotide hydrolysis, which drives disassembly and therefore contraction of the structure, may be used to perform physical work (e.g., segregation of chromosomes by microtubules). In contrast, structurally analogous contractile tubular structures, such as the type VI secretion system injection apparatus, assemble without the need for free energy from nucleotide hydrolysis. However, they require active nucleotide hydrolysis by an ATPase for disassembling the polymer for recycling the components on injection. On the other hand, largely static cytoskeletal elements, such as intermediate filaments, typically polymerize spontaneously, eschewing the need for a nucleotide altogether. In the case of IVA, we propose that the protein uses the energy released by ATP hydrolysis to drive a massive structural change that results in an irreversible interaction between IVA molecules.
Likewise, IVA polymerization might be distinguished from the formation of higher order structures by bifunctional enzymes to perform a structural or morphogenetic role in a cell. A canonical example is the group of crystallin proteins found in the lens of vertebrate eyes, which, depending on the organism, could be a distinct protein that diverged from an ancestral enzyme or an active enzyme that polymerizes preferentially in the eye to form the lens (48). Similarly, the enzyme CTP synthase (CtpS) of the bacterium Caulobacter crescentus was recently shown to play a morphogenetic role by polymerizing into long filaments that mediate the shape of the comma-shaped Caulobacter cell, in addition to its well-characterized enzymatic activity (49). However, in both cases, the enzymatic functions of these proteins were typically separable from the capacity of these proteins to self-assemble. Thus, whereas disruption of the CTP synthase activity of the CtpS did not abrogate its ability to form filaments that mediate cell shape, disrupting the ATP hydrolysis activity of IVA abolished its ability to polymerize. Moreover, unlike enzymes harboring a particular activity whose tertiary structures have been coopted for cytoskeletal roles, the enzymatic activity of IVA, to date, appears dedicated only to polymerization of the protein. Perhaps this strategy that ties ATP hydrolysis exclusively to the assembly of a static structure represents an evolutionary solution to the problem of assembling a static structure that is meant to display extraordinary resilience to a variety of external insults.
Materials and Methods
Sequence Analysis.
Sequence profile searches were performed using the PSI-BLAST program, which was run against the nonredundant protein database of National Center for Biotechnology Information to identify further homologs. The multiple sequence alignment of IVA was built using the Kalign programs, followed by manual adjustments on the basis of profile-profile and structural alignments. Secondary structures were predicted using the JPred program. The HHpred program was used for profile-profile comparisons. Phylogenetic analysis was conducted using an approximately maximum-likelihood method implemented in the FastTree 2.1 program under default parameters.
Strain Construction, Cell Growth, and General Methods.
Strains are otherwise congenic derivatives of B. subtilis PY79 (50). Additional details are provided in SI Materials and Methods.
His6-IVA Purification.
His6-IVA and derivatives were overproduced in and purified from E. coli harboring plasmid pKR145 (and derivatives thereof). The detailed protocol is provided in SI Materials and Methods.
ATP Binding and Hydrolysis Assays.
ATP binding was measured using a capillary diffusion assay on dry nitrocellulose (differential radial capillary action of ligand assay) (39). The detailed protocol is provided in SI Materials and Methods.
In Vitro Polymerization Assays.
Detailed protocols for centrifugation, TIRF microscopy, EM, and dynamic light scattering assays are provided in SI Materials and Methods.
Limited Proteolysis.
Two micromolar His6-IVA was incubated with trypsin at 25 μg⋅mL−1 (Sigma) in buffer A containing 10 mM MgCl2 at 37 °C. At the times indicated, 10 μL was removed, proteolysis was stopped by addition of sample buffer, and the products were separated by PAGE and analyzed by Coomassie staining. Where indicated, 4 mM ATP was preincubated with proteins for 4 h before the addition of trypsin.
Epifluorescence Microscopy.
Cells were visualized as described previously (51). The detailed protocol is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Antonina Roll Mecak, Erin Goley, Vincent Lee, Adrian Ferré-D’Amaré, Sue Wickner, Mike Maurizi, Susan Gottesman, and members of our laboratories for discussion and comments on the manuscript; James Sellers for advice and in whose laboratory the TIRF experiments were performed; and Ulrich Baxa of the Electron Microscopy Laboratory (Frederick National Laboratory for Cancer Research) for assistance with EM. This work was funded by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (K.S.R. and J-P.C.); National Heart, Lung, and Blood Institute (A.N.); and National Library of Medicine (V.A. and L.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 400 (volume 110, number 2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210554110/-/DCSupplemental.
References
- 1.Richman JM, Handrigan GR. Reptilian tooth development. Genesis. 2011;49(4):247–260. doi: 10.1002/dvg.20721. [DOI] [PubMed] [Google Scholar]
- 2.Waring GL. Morphogenesis of the eggshell in Drosophila. Int Rev Cytol. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- 3.Qin H. Regulation of intraflagellar transport and ciliogenesis by small G proteins. Int Rev Cell Mol Biol. 2012;293:149–168. doi: 10.1016/B978-0-12-394304-0.00010-5. [DOI] [PubMed] [Google Scholar]
- 4.Erhardt M, Namba K, Hughes KT. Bacterial nanomachines: The flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2(11):a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15(4):172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 7.Cano RJ, Borucki MK. Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science. 1995;268(5213):1060–1064. doi: 10.1126/science.7538699. [DOI] [PubMed] [Google Scholar]
- 8.Jacotot H, Virat B. La longévité des spores de B. anthracis (premier vaccin de Pasteur) Ann Inst Pasteur (Paris) 1954;87(2):215–217. [PubMed] [Google Scholar]
- 9.Vreeland RH, Rosenzweig WD, Powers DW. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature. 2000;407(6806):897–900. doi: 10.1038/35038060. [DOI] [PubMed] [Google Scholar]
- 10.Giorno R, et al. Morphogenesis of the Bacillus anthracis spore. J Bacteriol. 2007;189(3):691–705. doi: 10.1128/JB.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Permpoonpattana P, et al. Surface layers of Clostridium difficile endospores. J Bacteriol. 2011;193(23):6461–6470. doi: 10.1128/JB.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 13.Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1(2):117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 14.Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7(6):579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36(1):131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driks A, Roels S, Beall B, Moran CP, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8(2):234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 17.Driks A. The bacillus spore coat. Phytopathology. 2004;94(11):1249–1251. doi: 10.1094/PHYTO.2004.94.11.1249. [DOI] [PubMed] [Google Scholar]
- 18.McKenney PT, Eichenberger P. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol. 2012;83(2):245–260. doi: 10.1111/j.1365-2958.2011.07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebmeier SE, Tan IS, Clapham KR, Ramamurthi KS. Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol Microbiol. 2012;84(4):682–696. doi: 10.1111/j.1365-2958.2012.08052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenney PT, et al. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol. 2010;20(10):934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174(2):575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price KD, Losick R. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol. 1999;181(3):781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62(6):1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 24.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323(5919):1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2(4):a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramamurthi KS. Protein localization by recognition of membrane curvature. Curr Opin Microbiol. 2010;13(6):753–757. doi: 10.1016/j.mib.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang KH, et al. The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol. 2009;74(3):634–649. doi: 10.1111/j.1365-2958.2009.06886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell. 2008;31(3):406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 32.Blombach F, Brouns SJ, van der Oost J. Assembling the archaeal ribosome: Roles for translation-factor-related GTPases. Biochem Soc Trans. 2011;39(1):45–50. doi: 10.1042/BST0390045. [DOI] [PubMed] [Google Scholar]
- 33.Schwefel D, et al. Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2) Proc Natl Acad Sci USA. 2010;107(47):20299–20304. doi: 10.1073/pnas.1010322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Delius M, Leigh DA. Walking molecules. Chem Soc Rev. 2011;40(7):3656–3676. doi: 10.1039/c1cs15005g. [DOI] [PubMed] [Google Scholar]
- 35.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21(11):2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 38.Donaldson GP, Roelofs KG, Luo Y, Sintim HO, Lee VT. A rapid assay for affinity and kinetics of molecular interactions with nucleic acids. Nucleic Acids Res. 2012;40(7):e48. doi: 10.1093/nar/gkr1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108(37):15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 41.Zheng LB, Donovan WP, Fitz-James PC, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2(8):1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- 42.Webb CD, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177(20):5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onyenwoke RU, Brill JA, Farahi K, Wiegel J. Sporulation genes in members of the low G+C Gram-type-positive phylogenetic branch (Firmicutes) Arch Microbiol. 2004;182(2-3):182–192. doi: 10.1007/s00203-004-0696-y. [DOI] [PubMed] [Google Scholar]
- 44.Holt JG, editor. Bergey’s Manual of Determinative Bacteriology. 9th Ed. Baltimore: Williams & Wilkins; 1994. [Google Scholar]
- 45.Sharma MR, et al. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell. 2005;18(3):319–329. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 46.Tu C, et al. Structure of ERA in complex with the 3′ end of 16S rRNA: Implications for ribosome biogenesis. Proc Natl Acad Sci USA. 2009;106(35):14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heiner Linke AM, editor. Controlled Nanoscale Motion: Nobel Symposium 131. 1st Ed. New York: Springer; 2007. [Google Scholar]
- 48.Wistow GJ, Piatigorsky J. Lens crystallins: The evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 49.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12(8):739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12(1):1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 51.Eswaramoorthy P, et al. Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis. MBio. 2011;2(6):e00257–e00211. doi: 10.1128/mBio.00257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]