Abstract
Homologous spectrin-like repeats can mediate specific protein interaction. The underlying mechanism is poorly understood. Dystrophin contains 24 spectrin-like repeats. However, only repeats 16 and 17 (R16/17) are required for anchoring neuronal NOS (nNOS) to the sarcolemma. Through an adeno-associated virus-based in vivo binding assay, we found that membrane expression of correctly phased R16/17 was sufficient to recruit nNOS to the sarcolemma in mouse muscle. Utrophin R15/16 is homologous to dystrophin R16/17. Substitution of dystrophin R16/17 microdomains with the corresponding regions of utrophin R15/16 suggests that the nNOS binding site is located in a 10-residue fragment in dystrophin R17 α1 helix. Interestingly, swapping this microdomain back into utrophin did not convey the nNOS binding activity. To identify other structural features that are required for nNOS interaction, we replaced an individual α-helix of dystrophin R16/17 with an equivalent α-helix from another dystrophin repeat. In vitro study with yeast two-hybrid suggests that most α-helices of R16/17, except for the R17 α1 helix, were dispensable for nNOS interaction. Surprisingly, in vivo binding assay showed that α2 and α3 helices of both R16 and R17 were essential for nNOS binding in muscle. We concluded that a microdomain in the α1 helix of dystrophin R17 binds to nNOS in a way uniquely defined by two pairs of the flanking helices. Our results provide an explanation for how structurally similar spectrin-like repeats in dystrophin display selective interaction with nNOS. The results also open new therapeutic avenues to restore defective nNOS homeostasis in dystrophin-null Duchenne muscular dystrophy.
Keywords: Becker muscular dystrophy, BMD, DMD, gene therapy, microdystrophin
Spectrin-type repeat (STR) is a common structural element in a variety of proteins, especially cytoskeletal proteins. STR is composed of 106–122 amino acids folded in a triple α-helical unit. STR exists either as a single-copy or tandem repeats. STR-containing proteins play a fundamental role in maintaining the cytoskeletal architecture and organizing protein complexes (1, 2). Dystrophin is a vital STR-containing protein in striated muscles that links the cytoskeleton with the extracellular matrix and, hence, preserves sarcolemmal integrity during muscle contraction. Besides mechanical support, dystrophin also scaffolds neuronal nitric oxide synthase (nNOS) and several other signaling proteins to the sarcolemma.
Absence of dystrophin results in Duchenne muscular dystrophy (DMD), an X-linked lethal muscle disease (3). Although increased membrane fragility has been considered as a primary pathogenic mechanism of DMD, accumulated evidence suggests that the loss of sarcolemmal nNOS also contributes to the dystrophic process (4–7). A clear understanding of how nNOS is localized to the membrane may thus offer insight to our understanding of the disease and open new therapeutic avenues.
Dystrophin has four functional domains including the N-terminal (NT), middle rod, cysteine-rich (CR), and C-terminal domains. The middle rod domain contains 24 STRs and four interspersed hinges. It was initially thought that nNOS indirectly binds to the dystrophin C-terminal domain via syntrophin (8, 9). Surprisingly, later studies show that merely restoring syntrophin to the membrane cannot anchor nNOS (10–12). Through systemic structure-function analysis, we recently found that dystrophin STRs 16 and 17 (R16/17), rather than the C-terminal domain, are required for sarcolemmal distribution of nNOS (4). Basically, dystrophins that contain R16/17 show membrane expression of nNOS but those without R16/17 do not. Our findings raise an important question about why only R16/17 but not other structurally similar dystrophin STRs interact with nNOS. Here we dissected molecular attributes of dystrophin R16/17 that are responsible for nNOS binding. We found that membrane localized R16/17 was the minimal unit for dystrophin–nNOS interaction. We also found that a 10-residue microdomain in the α1 helix of dystrophin R17 contains the nNOS binding site. To our surprise, in vitro yeast two-hybrid assay failed to predict some critical structural features essential for dystrophin-nNOS interaction in muscle. Using an adeno-associated virus (AAV)-based in vivo nNOS-binding assay, we demonstrated that the last two α-helices (α2 and α3 helices) of both R16 and R17 were required to anchor nNOS to the sarcolemma although they were dispensable for nNOS binding in vitro.
Results
Membrane Expression of Dystrophin R16/17 Alone Is Sufficient to Target nNOS to the Sarcolemma.
Our previous studies suggest that dystrophin R16/17 is necessary for membrane-associated nNOS expression (4, 7). However, our smallest nNOS binding dystrophin (ΔR2-R15/ΔR18-R23/ΔC) also carries the NT and CR domains, H1, H4, R1, and R24 (Fig. S1 and Table S1) (4). To determine whether these regions contributed to dystrophin–nNOS interaction, we examined in vivo nNOS binding in constructs carrying additional deletions. Removing R1 and R24 did not compromise sarcolemmal nNOS expression in dystrophin-null mdx muscle. Further deletion of the NT domain and H1 or H4 and the CR domain did not alter nNOS membrane localization either (Fig. S1). These results suggest that dystrophin R16/17 can recruit nNOS to the sarcolemma independent of other dystrophin domains.
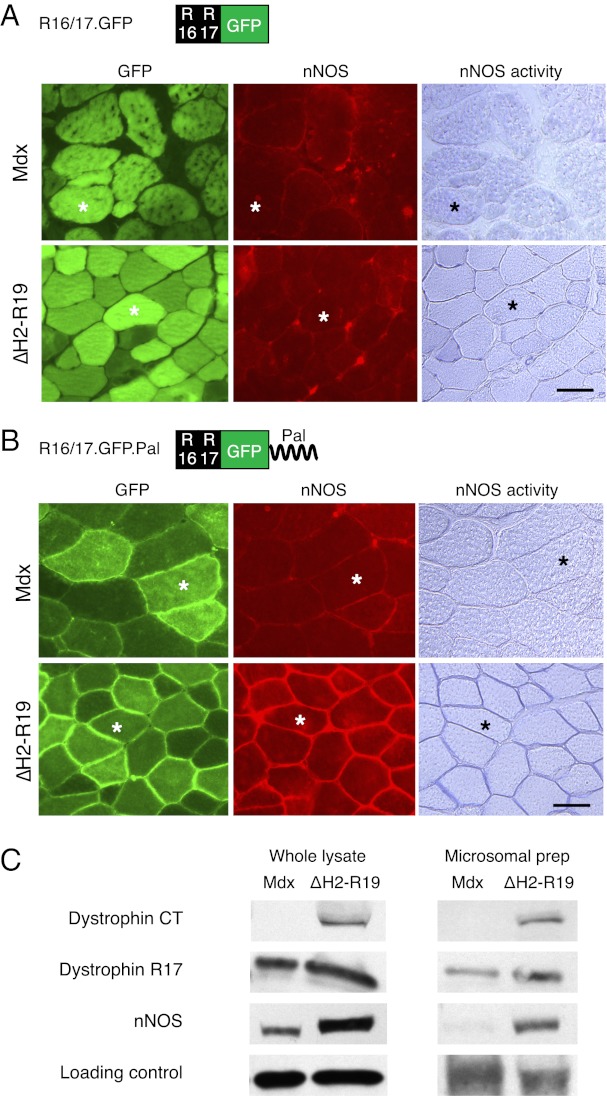
Next, we tested whether a stripped-down construct of only dystrophin R16/17 can localize nNOS to the sarcolemma. To facilitate detection, we fused a GFP tag to dystrophin R16/17 (R16/17.GFP) (Fig. 1A and Table S1). Robust expression of R16/17.GFP was observed in mdx muscle but nNOS was not detected at the sarcolemma (Fig. 1A). Loss of dystrophin results in the disassociation of syntrophin from the membrane. Syntrophin is also required for sarcolemmal nNOS localization (14, 15). To more stringently test the R16/17.GFP construct, we introduced it to skeletal muscle specific ∆H2-R19 minidystrophin transgenic mdx mice (Fig. 1A) (4). The ∆H2-R19 minidystrophin gene does not restore nNOS to the membrane but it anchors syntrophin to the sarcolemma (4, 10, 16). The R16/17.GFP AAV virus successfully transduced transgenic mdx muscle. However, the virus still did not restore nNOS to the sarcolemma (Fig. 1A). We noticed that R16/17.GFP expression was limited to the sarcoplasm only. We reasoned that failure to localize nNOS to the sarcolemma might be because of the lack of membrane targeting of R16/17.GFP. To address this theory, we attached a palmitoylation membrane targeting sequence to the C terminus of R16/17.GFP and generated R16/17.GFP.Pal (Fig. 1B and Table S1) (17). Compared with R16/17.GFP, palmitoylated dystrophin R16/17 was clearly enriched at the sarcolemma (Fig. 1B). Importantly, membrane-associated nNOS was detected in R16/17.GFP.Pal-treated ∆H2-R19 transgenic mdx mice (Fig. 1 B and C). Collectively, our data suggest that R16/17 is the only dystrophin component required for sarcolemmal nNOS targeting (4).
Fig. 1.
Membrane targeting of dystrophin R16/17 restores sarcolemmal nNOS expression in ΔH2-R19 transgenic but not parental mdx mice. (A) Morphological evaluation of nNOS expression following R16/17.GFP.Pal AAV virus infection. (Scale bar, 50 μm.) (B) Morphological evaluation of nNOS expression following R16/17.GFP.Pal AAV virus infection. (Left) GFP expression; (Center) nNOS immunofluorescence staining; (Right) in situ nNOS activity staining. Asterisks indicate the same myofiber in serial sections. (Scale bar, 50 μm.) (C) Western blot evaluation of nNOS expression following R16/17.GFP.Pal AAV virus infection. (Left) Representative results from whole muscle lysate; (Right) representative results from microsomal preparation. α-Tubulin was used as the loading control for whole muscle lysate. α1-Na+/K+ ATPase was used as the loading control for microsomal preparation.
Dystrophin R17 α1 Helix Contains the nNOS-Binding Domain.
Utrophin is an autosomal paralog of dystrophin. Utrophin R15/16 is homologous to dystrophin R16/17. However, utrophin R15/16 cannot bring nNOS to the sarcolemma (13). To test whether dystrophin R16/17 can restore sarcolemmal nNOS in a foreign context, we engineered a chimeric microutrophin gene in which utrophin R15/16 was replaced by dystrophin R16/17 (Fig. S2 and Table S1). Modified microutrophin effectively restored sarcolemmal nNOS expression in utrophin/dystrophin double knockout (u-dko) mouse muscle (Fig. S2). These results reiterate that dystrophin R16/17 bind nNOS in a context-independent manner.
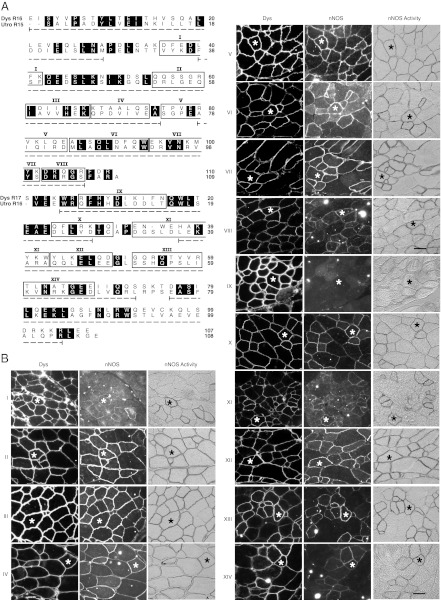
To identify the nNOS-binding domain in dystrophin R16/17, we generated 14 chimerical microdystrophin constructs. In these constructs, a microdomain of dystrophin R16/17 was substituted by the corresponding sequence from utrophin R15/16 (Fig. 2A and Table S1). Each construct was named after the matching microdomain (I to XIV). Following AAV gene transfer to mdx muscle, sarcolemmal nNOS expression was examined. The pattern was not altered in 13 constructs (Fig. 2B). The only exception is construct IX, in which a 10-residue microdomain in the first half of dystrophin R17 α1 helix was replaced. Membrane-associated nNOS expression was completely abolished in muscles treated with this construct (Fig. 2B). These results suggest that the 10-residue microdomain in construct IX contains the nNOS-binding site (Fig. 2B).
Fig. 2.
Microdomain substitution study reveals the nNOS binding site in dystrophin R17 α1 helix. Individual microdomain in dystrophin R16/17 was replaced by the corresponding microdomain of utrophin R15/16 in the ΔR2-R15/ΔR18-R23/ΔC microdystrophin gene. The modified microgene was delivered to mdx muscle by AAV. (A) Amino acid sequence alignment of dystrophin R16/17 and utrophin R15/16. Identical residues are shaded in black. The α-helices are marked by the underlying dotted line. Microdomains are boxed and numbered from I to XIV. (B) Representative photomicrographs of dystrophin and nNOS immunostaining, and nNOS activity staining. Asterisks indicate the same myofiber in serial sections. (Scale bar, 50 μm.)
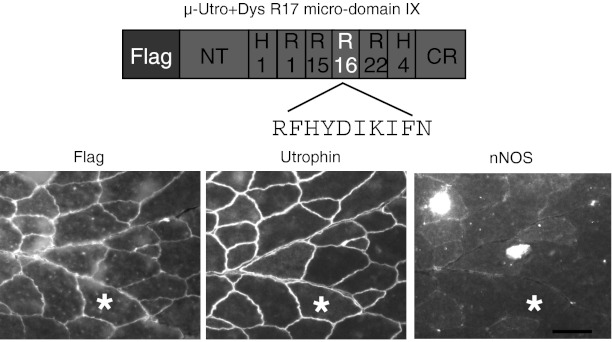
To further establish dystrophin R17 α1 helix microdomain IX as the nNOS-binding domain, we engineered this microdomain into the microutrophin gene. Specifically, we replaced the corresponding sequence in utrophin R16 with that of dystrophin R17 (Table S1). Despite strong expression, dystrophin R17 microdomain IX did not anchor nNOS to the sarcolemma in the context of utrophin (Fig. 3). We deduced that in addition to dystrophin R17 microdomain IX, other yet undefined structural features of dystrophin R16/17 are also needed for sarcolemmal nNOS localization.
Fig. 3.
Dystrophin nNOS binding domain dose not recruit nNOS to the sarcolemma in microutrophin. Schematic outline of the chimerical microutrophin construct (μ-Utro +Dys R17 microdomain IX). The microdomain IX of utrophin R16 was replaced by the corresponding microdomain of dystrophin R17 in the ΔR2-14/ΔR17-21/ΔC microutrophin gene. Modified microutrophin was delivered to utrophin/dystrophin double null mouse muscle. Shown are the representative Flag, utrophin, and nNOS immunostaining photomicrographs. Asterisks indicate the same myofiber in the serial sections. (Scale bar, 50 μm.)
Sarcolemmal nNOS Binding Requires Five Correctly Phased α-Helices, Including α2 and α3 Helices of Dystrophin R16 and all Three α-Helices of Dystrophin R17.
The linker between adjacent STRs has been implicated in protein–protein interaction (18, 19). Therefore, we first tested whether the junction between dystrophin R16 and R17 was involved in nNOS binding. We generated four linker mutants (mutants 1–4). However, none of the mutants altered nNOS membrane localization (Tables S1 and S2). These results suggest that the linker between R16 and R17 is not required for nNOS binding.
To decipher other regions that may contribute to nNOS binding, we decided to re-examine the whole STR. The nNOS-binding domain is located in dystrophin R17 (Fig. 2); hence, replacing this STR will destroy nNOS interaction. For this reason, we focused our attention on dystrophin R16. Individual replacement of eight microdomains of dystrophin R16 with the corresponding microdomains of utrophin R15 had minimal impact on nNOS binding (Fig. 2). This finding seems to suggest that dystrophin R16 and utrophin R15 may be exchangeable. To determine the contribution of dystrophin R16 in its entirety, we generated another chimeric microdystrophin (μ-Dys+Utro R15) in which dystrophin R16 was replaced by utrophin R15 (Table S1). Surprisingly, modified microdystrophin only yielded very faint sarcolemmal nNOS staining (Fig. S3A). On microsomal preparation Western blot, modified microdystrophin did not localize nNOS to the sarcolemma (Fig. S3B). These results suggest that dystrophin R16 may tolerate single microdomain substitution but not whole STR exchange by homological utrophin R15.
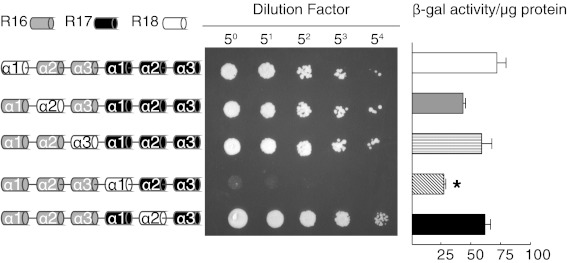
The α-helix is the basic structural unit of STR. Each STR contains three α-helices. To determine contribution of individual α-helix on nNOS binding, we screened a series of α-helix substitution constructs by yeast two-hybrid (Fig. 4). In these constructs, one of the α-helices of dystrophin R16/17 was replaced by the corresponding α-helix from dystrophin R18. Interaction with nNOS was not disrupted in most cases, except when R17 α1 helix was replaced (Fig. 4).
Fig. 4.
In vitro interaction between dystrophin R16/17 and the nNOS PDZ domain depends on R17 α1 helix. (Left) Schematic outlines of the activation constructs used in yeast two-hybrid assay. The binding construct carries the nNOS PDZ domain. (Center) From dot assay results. (Right) Quantitative yeast two-hybrid assay measured by β-galactosidase activity. Mean ± SE of mean. Asterisk, significantly different from other groups by one-way ANOVA (P < 0.05).
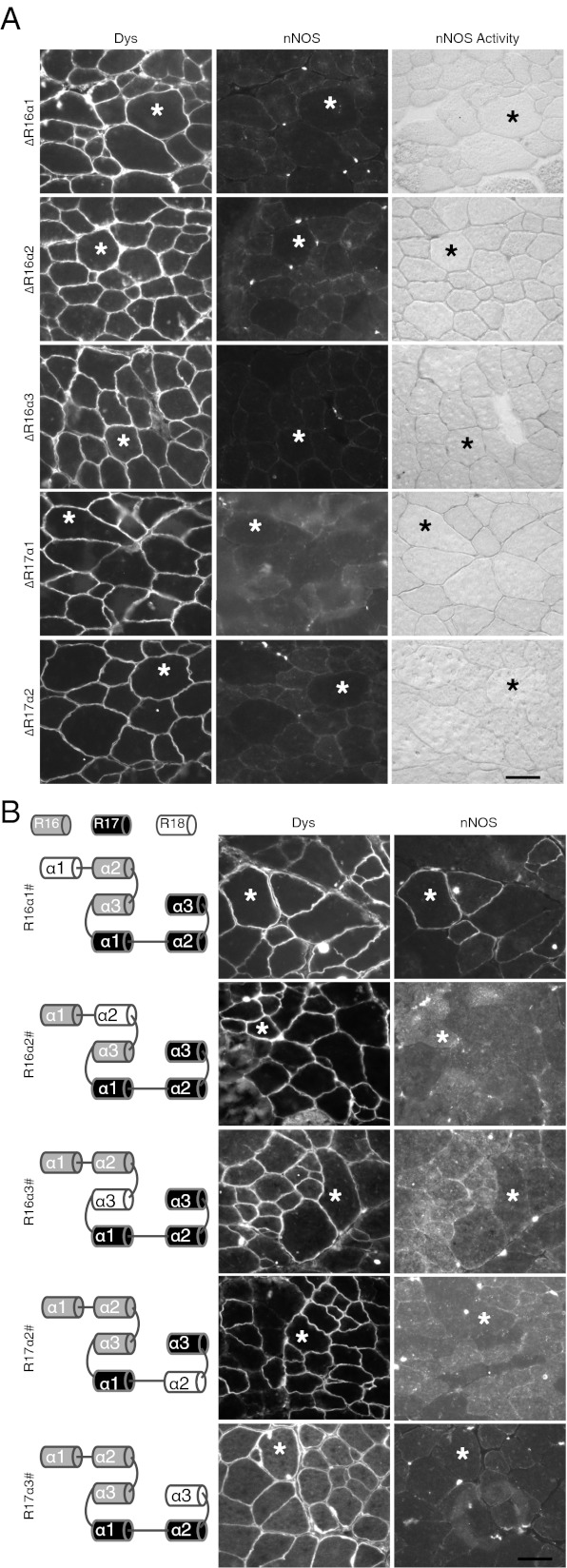
Considering the possibility that in vitro assay may fail to predict protein interaction in vivo, we then performed the in vivo binding assay using AAV gene transfer. We first examined the impact of single α-helix deletion. Interestingly, nNOS binding was abolished in all of the deletion constructs we examined (Fig. 5A and Table S1). This finding suggests that either every α-helix is required, or more likely, single α-helix deletion has shifted the normal phasing of the entire STR and hence disrupted 3D structure of the binding motif. To further determine the importance of each α-helix, we generated α-helix substitution microdystrophin constructs. In these constructs, one α-helix (or multiple α-helices) in dystrophin R16/17 was replaced by the corresponding α-helix (or helices) from another dystrophin STR (Fig. 5B, and Tables S1 and S2). This design allows the modified constructs to maintain normal α-helix phasing. As expected, substitution of R17 α1 helix destroyed nNOS binding (Table S2). Interestingly, replacement of other α-helices also abolished nNOS binding (Table S2). Single helix substitution of the remaining five α-helices revealed more striking results. Although R16 α1 helix replacement did not affect nNOS binding, swapping the α2 or α3 helix of either R16 or R17 eliminated dystrophin–nNOS interaction (Fig. 5B and Table S2). Collectively, our in vivo data suggest that α2 and α3 helices of both R16 and R17 are essential for membrane localization of nNOS in muscle.
Fig. 5.
In vivo interaction between dystrophin R16/17 and nNOS requires correct α-helix phasing and all but R16 α1 helices. (A) Representative photomicrographs of dystrophin and nNOS immunostaining and nNOS activity staining in mdx muscle infected by AAV viruses carrying a single α-helix deletion. The deleted α-helix is marked on the left side of each panel. (B) Representative photomicrographs of dystrophin and nNOS immunostaining in mdx muscle infected by AAV viruses carrying single α-helix substitution. The α-helix replaced in each construct is depicted in the cartoon drawing (Left). The “#” refers to the referred α-helix that is substituted by the corresponding α-helix of dystrophin R18. Asterisks indicate the same myofiber in serial sections. (Scale bar, 50 μm.)
Discussion
In this study, we investigated molecular mechanisms underlying dystrophin R16/17-mediated nNOS sarcolemmal localization. Because dystrophin STRs have never been successfully crystallized (20), we decided to take an in vivo biochemical approach to study how dystrophin recruits nNOS to the sarcolemma. Specifically, we generated more than 48 different dystrophin and utrophin constructs to express various sequence changes that might be involved in dystrophin–nNOS interaction. These constructs were packaged in muscle tropic AAV viruses and delivered to limb muscles of mdx, u-dko, and ∆H2-R19 minidystrophin transgenic mdx mice. nNOS expression was examined by immunofluorescence staining, in situ enzymatic activity assay, and microsomal preparation Western blot. Positive nNOS binding was defined as the detection of nNOS on the sarcolemma. We found that membrane bound dystrophin R16/17 anchored nNOS to the sarcolemma in the presence of syntrophin. We further showed that dystrophin R17 α1 helix carried the nNOS-binding microdomain. Finally, we demonstrated that the function of the nNOS binding microdomain not only required correct phasing of all α-helices in R16/17 but also depended on the structural environment formed by four surrounding helices.
STR is a highly conserved structural module consisting of a triple helical bundle. Interestingly, some paired STRs have evolved unique properties to mediate specific protein–protein interaction while still maintaining their tertiary conformation. The molecular basis for functional specialization of STR is poorly understood. The crystal structure of a ligand-bound STR has only been resolved in one case (19). Ipsaro and colleagues recently deciphered the atomic structure of spectrin R14/15 in complex with its binding partner ankyrin (19). The authors find that a negatively charged patch in the α3 helix of spectrin R14 interacts with a positively charged patch in ankyrin. They also show that the linker region between spectrin R14 and R15, and the loop between the α2 and α3 helices of spectrin R15, are important for binding (19). The authors propose that: (i) a large tilting between spectrin R14 and R15 brings the linker region and spectrin R15 α2/α3 loop close to each other to form the docking interface, and (ii) ankyrn binding occurs through patch electrostatic interaction (19). Our results herein revealed a different interaction mode. Specifically, we found that nNOS recognition was likely accomplished via a 10-residue microdomain in dystrophin R17 α1 helix (Fig. 2). This microdomain is highly conserved through evolution, suggesting it may represent an essential structural feature (20). Based on the fact that dystrophin R17 α1 helix alone supported nNOS binding in vitro in yeast two-hybrid assay (Fig. 4), we hypothesize that the 10-residue motif contains the authentic nNOS binding site. In contrast to the negatively charged patch in spectrin R14 α3 helix reported by Ipsaro et al., the nNOS binding microdomain we identified consists of amino acids of various electrostatic properties. This finding suggests that dystrophin R16/17 may bind to nNOS through a mechanism different from what was shown for spectrin–ankyrin interaction. Future elucidation of this binding mechanism with X-ray crystallography and NMR may shed new light on our understanding of other STR-mediated protein interaction.
Another intriguing aspect of dystrophin R16/17-nNOS interaction is the striking difference between in vitro and in vivo assay results. Yeast two-hybrid revealed dystrophin R17 α1 helix is the only component needed for nNOS binding. The requirement for other α-helices was appreciated only when the binding assay was performed in vivo. Because the α1 helix of dystrophin R17 independently recruited nNOS in vitro, we reasoned that α2 and α3 helices of dystrophin R16 and R17 may not directly participate in the binding. Rather, these helices may function to stabilize R16/17 in a specific configuration to facilitate in vivo nNOS binding. Considering the fact that such information can only be obtained from studies performed in muscle, our results highlight the importance of in vivo biochemical approach in studying protein interaction.
The rod domain of dystrophin was initially considered as a flexible spacer that separates more important functional domains at the N and C termini. However, recent studies suggest that some STRs in the rod domain actually play a more active role in a plethora of cellular functions via interaction with membrane phospholipids, cytoskeletal proteins, and signaling proteins (21). Of particular interest is the ability of dystrophin R16/17 to compartmentalize nNOS to the sarcolemma (4). Failure to do so causes functional ischemia and muscle fatigue, hence more severe muscle disease (4, 22). Although our previous studies explained why nNOS is delocalized from the membrane in patients carrying deletion mutations involving dystrophin R16/17, they cannot justify cases in which R16/17 is intact yet nNOS is lost from the sarcolemma (23–25). The results from single α-helix deletion/substitution experiments suggest that in-frame deletion in other regions of dystrophin may disrupt nNOS interaction by altering α-helix phasing.
Last but not least, our findings also reveal several new therapeutic opportunities to treat DMD. Utrophin overexpression has been considered as a promising therapy for DMD. Unfortunately, utrophin cannot bind nNOS (13). The unique dystrophin R16/17-containing microutrophin gene described herein may thus improve utrophin-based gene therapy. Another very exciting possibility is to use membrane-targeted R16/17 as a supplementary therapy to restore sarcolemmal nNOS expression in situations in which nNOS binding activity is lost in muscle because of deletions affecting dystrophin R16/17 coding region (such as in some Becker muscular dystrophy patients or in DMD patients treated with exon 42–45 skipping).
Methods
AAV-Mediated in Vivo nNOS Binding.
All animal experiments were approved by the University of Missouri Institutional Animal Care and Use Committee. Modified microdystrophins/utrophins were packaged in Y445F AAV-6 vector. The 1010 viral particles were injected to the tibialis anterior muscle to young adult mice. Microgene expression and nNOS expression were examined 5 wk later by immunofluorescence staining, in situ nNOS activity assay, and Western blot (whole-muscle lysate and microsomal preparation) (4). Details of each assay are provided in SI Methods.
In Vitro nNOS Binding Assay with Yeast Two-Hybrid.
The assay was performed as elaborated in SI Methods. The binding construct carried the nNOS PDZ domain. The activation constructs express various α-helix substituted dystrophin R16/17.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant AR-49419 (to D.D.), and grants from the Muscular Dystrophy Association (to D.D.), Jessey’s Journey-The Foundation for Cell and Gene Therapy (to D.D.), and the University of Missouri Research Council (to Y.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 387.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211431109/-/DCSupplemental.
References
- 1.Djinovic-Carugo K, Gautel M, Ylänne J, Young P. The spectrin repeat: A structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513(1):119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- 2.Le Rumeur E, Hubert JF, Winder SJ. A new twist to coiled coil. FEBS Lett. 2012;586(17):2717–2722. doi: 10.1016/j.febslet.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel LM. 2004 William Allan Award address. Cloning of the DMD gene. Am J Hum Genet. 2005;76(2):205–214. doi: 10.1086/428143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai Y, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest. 2009;119(3):624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sander M, et al. Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 2000;97(25):13818–13823. doi: 10.1073/pnas.250379497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas GD, et al. Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proc Natl Acad Sci USA. 1998;95(25):15090–15095. doi: 10.1073/pnas.95.25.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Yue Y, Lai Y, Hakim CH, Duan D. Nitrosative stress elicited by nNOSµ delocalization inhibits muscle force in dystrophin-null mice. J Pathol. 2011;223(1):88–98. doi: 10.1002/path.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284(5415):812–815. [PubMed] [Google Scholar]
- 9.Tochio H, Zhang Q, Mandal P, Li M, Zhang M. Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide. Nat Struct Biol. 1999;6(5):417–421. doi: 10.1038/8216. [DOI] [PubMed] [Google Scholar]
- 10.Lai Y, et al. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat Biotechnol. 2005;23(11):1435–1439. doi: 10.1038/nbt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue Y, Liu M, Duan D. C-terminal-truncated microdystrophin recruits dystrobrevin and syntrophin to the dystrophin-associated glycoprotein complex and reduces muscular dystrophy in symptomatic utrophin/dystrophin double-knockout mice. Mol Ther. 2006;14(1):79–87. doi: 10.1016/j.ymthe.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Judge LM, Haraguchiln M, Chamberlain JS. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J Cell Sci. 2006;119(Pt 8):1537–1546. doi: 10.1242/jcs.02857. [DOI] [PubMed] [Google Scholar]
- 13.Li D, et al. Sarcolemmal nNOS anchoring reveals a qualitative difference between dystrophin and utrophin. J Cell Sci. 2010;123(Pt 12):2008–2013. doi: 10.1242/jcs.064808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams ME, et al. Absence of alpha-syntrophin leads to structurally aberrant neuromuscular synapses deficient in utrophin. J Cell Biol. 2000;150(6):1385–1398. doi: 10.1083/jcb.150.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kameya S, et al. alpha1-syntrophin gene disruption results in the absence of neuronal-type nitric-oxide synthase at the sarcolemma but does not induce muscle degeneration. J Biol Chem. 1999;274(4):2193–2200. doi: 10.1074/jbc.274.4.2193. [DOI] [PubMed] [Google Scholar]
- 16.Harper SQ, et al. Modular flexibility of dystrophin: Implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8(3):253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 17.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63(1):133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 18.Stabach PR, et al. The structure of the ankyrin-binding site of beta-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties. Blood. 2009;113(22):5377–5384. doi: 10.1182/blood-2008-10-184291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ipsaro JJ, Mondragón A. Structural basis for spectrin recognition by ankyrin. Blood. 2010;115(20):4093–4101. doi: 10.1182/blood-2009-11-255604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legrand B, Giudice E, Nicolas A, Delalande O, Le Rumeur E. Computational study of the human dystrophin repeats: interaction properties and molecular dynamics. PLoS ONE. 2011;6(8):e23819. doi: 10.1371/journal.pone.0023819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Rumeur E, Winder SJ, Hubert JF. Dystrophin: More than just the sum of its parts. Biochim Biophys Acta. 2010;1804(9):1713–1722. doi: 10.1016/j.bbapap.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi YM, et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 2008;456(7221):511–515. doi: 10.1038/nature07414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chao DS, et al. Selective loss of sarcolemmal nitric oxide synthase in Becker muscular dystrophy. J Exp Med. 1996;184(2):609–618. doi: 10.1084/jem.184.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells KE, et al. Relocalization of neuronal nitric oxide synthase (nNOS) as a marker for complete restoration of the dystrophin associated protein complex in skeletal muscle. Neuromuscul Disord. 2003;13(1):21–31. doi: 10.1016/s0960-8966(02)00191-8. [DOI] [PubMed] [Google Scholar]
- 25.Torelli S, et al. Absence of neuronal nitric oxide synthase (nNOS) as a pathological marker for the diagnosis of Becker muscular dystrophy with rod domain deletions. Neuropathol Appl Neurobiol. 2004;30(5):540–545. doi: 10.1111/j.1365-2990.2004.00561.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.