Abstract
Plants produce a wide range of allelochemicals to defend against herbivore attack, and generalist herbivores have evolved mechanisms to avoid, sequester, or detoxify a broad spectrum of natural defense compounds. Successful arthropod pests have also developed resistance to diverse classes of pesticides and this adaptation is of critical importance to agriculture. To test whether mechanisms to overcome plant defenses predispose the development of pesticide resistance, we examined adaptation of the generalist two-spotted spider mite, Tetranychus urticae, to host plant transfer and pesticides. T. urticae is an extreme polyphagous pest with more than 1,100 documented hosts and has an extraordinary ability to develop pesticide resistance. When mites from a pesticide-susceptible strain propagated on bean were adapted to a challenging host (tomato), transcriptional responses increased over time with ∼7.5% of genes differentially expressed after five generations. Whereas many genes with altered expression belonged to known detoxification families (like P450 monooxygenases), new gene families not previously associated with detoxification in other herbivores showed a striking response, including ring-splitting dioxygenase genes acquired by horizontal gene transfer. Strikingly, transcriptional profiles of tomato-adapted mites resembled those of multipesticide-resistant strains, and adaptation to tomato decreased the susceptibility to unrelated pesticide classes. Our findings suggest key roles for both an expanded environmental response gene repertoire and transcriptional regulation in the life history of generalist herbivores. They also support a model whereby selection for the ability to mount a broad response to the diverse defense chemistry of plants predisposes the evolution of pesticide resistance in generalists.
Keywords: genetic variation, lipocalin, transcriptome, major facilitator superfamily, xenosensors
Plants produce a wide variety of allelochemicals, among which are a plethora of defense compounds. These can affect herbivore fitness in subtle ways by changing behavior or in less subtle ways by causing acute toxicity. The effectiveness of plant defenses is remarkable as herbivory has evolved successfully in only about one-third of all animals (1). Nevertheless, herbivores are among the most diverse terrestrial faunas (2). The ability to metabolize and detoxify plant chemicals is considered one of the major responses that arthropod herbivores have evolved during their coevolution with plants. Thus, the vast majority of insect herbivores are associated with no more than one or a few plant species (3), potentially reflecting the need for specialized mechanisms to cope with plant chemicals. Herbivorous specialists encounter high levels of predictable toxicants and have often evolved efficient and specialized detoxification systems (4). A well-known example is the role of CYP6B enzymes in Papilio species that feed on plants containing toxic furanocoumarins (5). These enzymes, belonging to the large P450 family, can convert these compounds to nontoxic metabolites and are thought to be a key innovation allowing the “escape and radiate” diversification of Papilionidae (6). Plants too can escape and radiate by producing new chemicals that are toxic to herbivores that have not yet evolved an effective detoxification response. An extension of this reasoning is that compounds that have evolved earlier and that are taxonomically widespread should be less toxic than newer compounds and that specialist herbivores should be less affected than generalists by the toxic compounds of their host plant (4). This is the “jack of all trades, master of none” argument comparing the generalist/specialist ability to cope with plant secondary chemistry (7). However, the way generalist (polyphagous) herbivores cope with the tremendous variety of chemicals in their toxic diet is not well documented in molecular terms. The original assumption was that generalists have a greater capacity to detoxify plant chemicals than specialists (8). This has been refined to state that generalists have detoxification enzymes, in particular P450 enzymes, with broader substrate specificity (9).
Recently introduced chemical pesticides can be considered as a metaphor for newly evolved or encountered plant chemicals, and a parallel has often been drawn between the evolution of resistance to insecticides and the response to host plant chemicals. This view was presented by Gordon in 1961, who thought that resistance genes are alleles of common genes, “the normal function of which is metabolism of biochemicals present in the [diet]” (ref. 10, p. 30). The “preadaptation hypothesis” for insecticide resistance has been supported by surveys of the literature (11, 12) although the comparisons drawn between herbivores and natural enemies or between chewing and sucking herbivores may be confounded by taxonomy, thus calling for other forms of experimental and observational evidence (12). It is now well accepted that herbivore exposure to different plant allelochemicals can affect the toxicity of pesticides (13–25). Moreover, metabolic resistance to pesticides is known to commonly rely on the increased expression of one or more genes encoding detoxification enzymes and formal evidence that many of these detoxification enzymes can metabolize both plant chemicals and pesticides is accumulating (26, 27). However, it has also been argued that the pattern of selection by plant allelochemicals and by pesticides differs (27, 28), so whether the polyphagous nature of many crop pests results in a preadaptation potential to cope with pesticides remains conjectural.
To elucidate the relationship between host plant adaptation and pesticide resistance in a systematic way, the two-spotted spider mite, Tetranychus urticae, is an excellent choice. T. urticae is among the most polyphagous herbivores known: It can feed on over 1,100 different plants in more than 140 different plant families that produce a broad spectrum of chemical defenses (29). Spider mites have been shown to rapidly adapt to new or less favorable hosts without a correlated fitness cost compared with the ancestral host (30, 31). Moreover, long-term adaptation on a single host does not markedly reduce genetic variation or the capability to subsequently adapt to a different host (32, 33). Also, experimental evolution has shown that although induced plant responses to T. urticae herbivory decrease the fitness of unadapted mites, induced plant response resulted in higher fitness of adapted mites, suggesting that spider mites can overcome both constitutive and induced plant defenses (30). In parallel with an exceptionally broad host range, T. urticae has demonstrated an unprecedented ability to develop resistance to pesticides; regardless of the chemical class, the first cases of resistance are usually reported within a few years after the introduction of a new acaricide. Selection for resistance in T. urticae is accelerated by its high fecundity and very short life cycle (34) and potentially also by its haplodiploid sex-determination system (unmated females produce haploid males) (35, 36).
To date, studies of resistance in T. urticae have focused largely on target site mutations and on classical detoxifying enzyme systems, such as P450 monooxygenases (P450s), carboxyl/cholinesterases (CCEs) and glutathione-S-transferases (GSTs) (27, 34). However, these studies have not been satisfactory for understanding the scope of acaricide resistance in T. urticae. Under field conditions, multiresistant strains that are resistant to all commercially available acaricides are often encountered, and strikingly these strains also resist compounds with new modes of action that have never been used in the field (37). Here, we have taken advantage of the high-quality genome sequence of T. urticae (29, 38) to construct an expression microarray that we then used to collect genome-wide expression data over a time course ranging from hours to generations after transfer of mites to a new, challenging host. We then related changes in gene expression after host plant change to constitutive patterns of gene expression in two strains that are highly resistant to a spectrum of pesticides. In doing so, we defined a set of genes and gene families that are of potential adaptive relevance to both situations. Remarkably, our studies suggest that the polyphagous spider mite exploits a large and shared repertoire of “classical” detoxification genes as well as potential new players as a defense against plant chemicals and pesticides.
Results
Host Plant Shift Effects on Gene Expression.
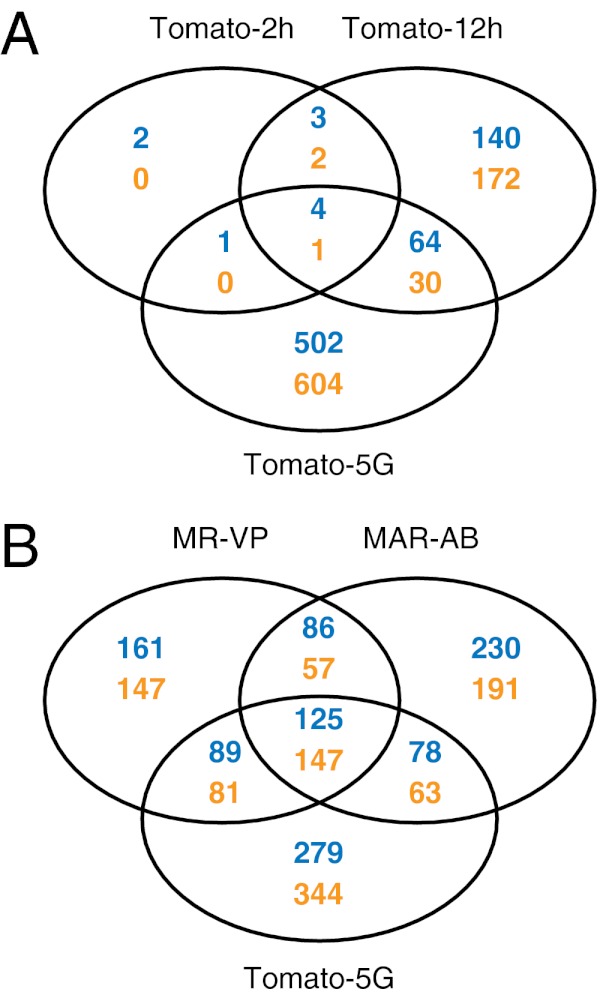
To examine genome-wide patterns of gene expression in T. urticae, we constructed an expression microarray (using the Agilent eArray platform; Materials and Methods) with long oligo probes against all predicted genes of the London reference strain. We then used this array to examine expression changes associated with host plant change, as well as expression patterns in acaricide-resistant strains. For the host plant change experiment, we transferred 1- to 3-d-old females (London strain) from their common host, Phaseolus vulgaris (kidney beans), to a more challenging and less accepted host, Solanum lycopersicum (tomato). We used young females because at this stage they actively disperse with the wind to escape kin competition and overexploitation (39, 40) and hence are expected to encounter potentially less favorable plants, on which they must immediately feed to produce eggs for colony establishment (colonies can then persist for many generations). We followed transcriptional changes over the short term to understand the initial responses, as well as after five generations on the new host. Briefly, female mites grown on beans were transferred to tomato, and transcriptional responses of mites were assessed at 2 h (Tomato-2h), at 12 h (Tomato-12h), and after propagation for five consecutive generations (Tomato-5G). As assessed by the number of differentially expressed genes [log2(fold change (FC)) ≥ 1, Benjamini–Hochberg false discovery rate (FDR) < 0.05], the transcriptional response increased with time. Thirteen and 416 genes were differentially expressed after 2 and 12 h, respectively, whereas 1,206 or about 7.5% of all predicted genes with probes on the array were differentially expressed after five generations (Fig. 1A). There was little overlap between genes associated with the early responses (Tomato-2h and Tomato-12h) and those with changed expression after five generations (Tomato-5G) (i.e., only 8.3% of Tomato-5G was shared with Tomato-2h and Tomato-12h) (Fig. 1A).
Fig. 1.
Venn diagrams depicting overlap among differentially expressed genes [log2(FC) ≥ 1, FDR < 0.05] from pairwise comparisons of mites shifted from bean to tomato and of resistant mites. Blue, up-regulated genes; orange, down-regulated genes. (A) Comparisons for shift to tomato for 2 h, 12 h, and five generations. (B) Comparisons for strains MAR-AB, MR-VP, and Tomato-5G.
Acaricide-Resistance Effects on Gene Expression.
To relate patterns of response between host adaptation and evolved pesticide resistance, we assessed gene expression patterns between two highly resistant field-collected strains (MR-VP and MAR-AB) and the reference susceptible London strain (37). These field strains, one collected on beans and the second collected on roses (Materials and Methods), are resistant when grown on bean, the host we used for assessing transcriptome variation among the three strains. Comparing them with the London strain by array hybridization, we observed differences in transcript levels [log2(FC) ≥ 1, FDR < 0.05] for 893 and 977 genes for MR-VP and MAR-AB, respectively (Fig. 1B). Our earlier work has shown that mite strains can be genetically diverse (29, 41), potentially confounding comparison of gene expression across strains (polymorphisms can affect array hybridization). However, our long oligo array is expected to be relatively robust to SNP and small indel changes (42); more importantly, we validated with quantitative PCR (qPCR) (Fig. S1) a subset of genes predicted from the array to be differentially expressed between the London strain and a second susceptible strain (LS-VL) (43), with a different genetic background.
Relationships Among Transcriptome Profiles.
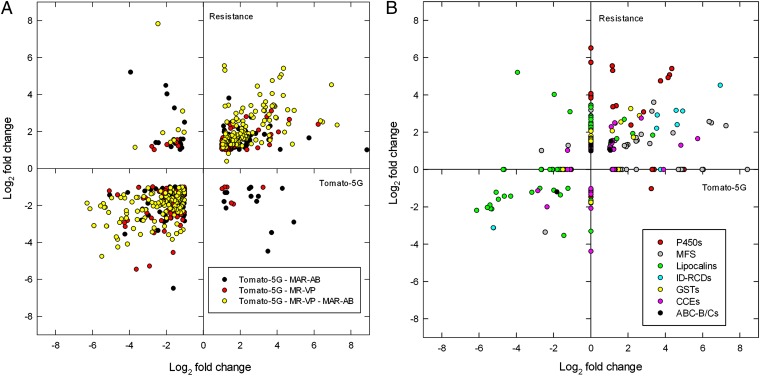
Although the resistant strains are genetically unrelated, there was an overlap of 415 differentially expressed genes (46.5% and 42.5% of the total number of differentially expressed genes in MR-VP and MAR-AB, respectively). Further, we found that 49.5% and 42.3% of differentially expressed genes in the resistant MR-VP and MAR-AB strains were also differentially expressed after the London strain was transferred to tomato for five generations. A scatter plot of the fold changes for the intersection of differentially expressed genes between the host transfer and resistance datasets revealed a high correlation for gene expression levels (Spearman’s correlation: ρ = 0.740, P < 0.001, Fig. 2A). Further, hierarchical clustering (Pearson’s centered distance metric, complete linkage rule) across all of the expression data revealed that expression patterns for the two resistant strains and for mites feeding on tomato for five generations clustered together and not with early responses for mites transferred to tomato for 2 h or 12 h. The overlap between resistance and host plant change was even more striking when genes were grouped in gene (sub)families by OrthoMCL clustering (Table 1). Shared responses were largely mediated by a few gene families, and in some cases a large proportion of all family members were included. As revealed by PFAM-domains searches of OrthoMCL clusters [48% of genes in shared OrthoMCL clusters (Table 1) have an assigned PFAM domain with E-value ≤ e−5], some responsive families belong to those that have been commonly implicated in detoxification or transport of xenobiotics [e.g., CCEs, P450s, GSTs, and ABC transporters (ABC-B/Cs)]. Among these, P450 genes stood out as being markedly differentially expressed among resistant mites and after host transfer of the susceptible London strain (Fig. 2B).
Fig. 2.
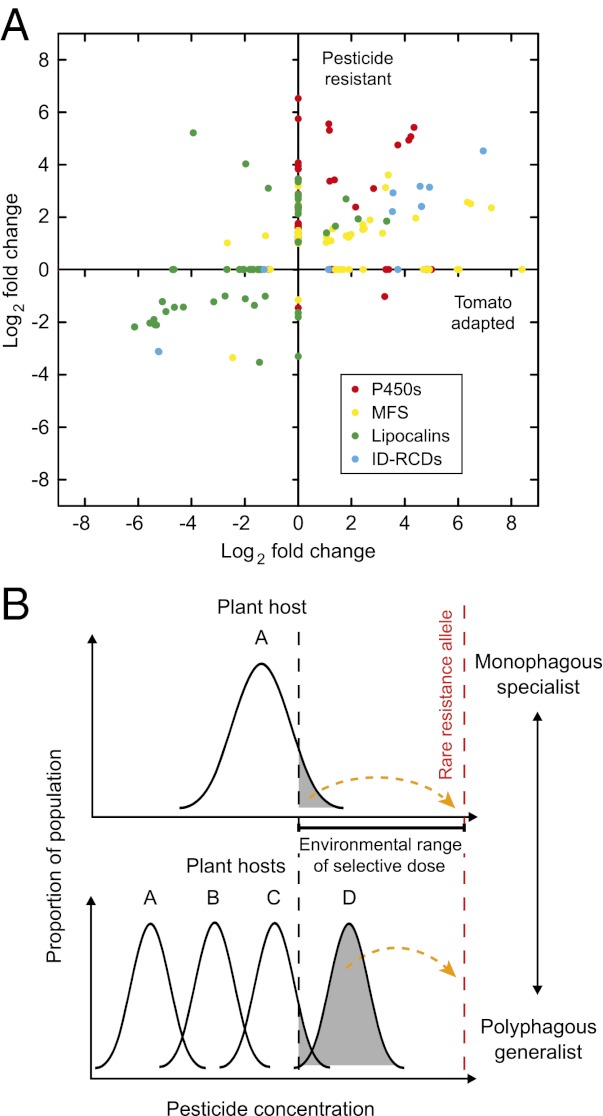
Global changes in gene expression of two multiresistant T. urticae strains (MR-VP and MAR-AB) relative to the London susceptible strain, compared with gene expression changes upon host plant change (Tomato-5G). (A) Commonly differentially expressed genes [log2(FC) ≥ 1, FDR < 0.05] in two multiresistant strains (MR-VP and/or MAR-AB: “Resistance”) and after host plant change for five generations (Tomato-5G): black, differentially expressed genes in Tomato-5G and MAR-AB; red, differentially expressed genes in Tomato-5G and MR-VP; yellow, differentially expressed genes in Tomato-5G, MR-VP, and MAR-AB (the Log2 of the average of fold changes of commonly differentially expressed genes of MR-VP and MAR-AB is plotted). (B) Fold changes of differentially expressed genes [log2(FC) ≥ 1, FDR < 0.05], known to be implicated in detoxification and transport, in two multiresistant strains (MR-VP and/or MAR-AB: Resistance) and after host plant change for five generations (Tomato-5G): red, P450 monooxygenases (P450s); black, ATP-binding cassette transporters, classes B and C (ABC-B/Cs); green, lipocalins; pink, carboxyl-cholinesterases (CCEs); yellow, glutathione S-transferases (GSTs); light blue, intradiol ring-cleavage dioxygenases (ID-RCDs); gray, MFS transporters (OrthoMCL clusters 10032, 10082, and 10236).
Table 1.
Extent of gene expression changes within gene clusters
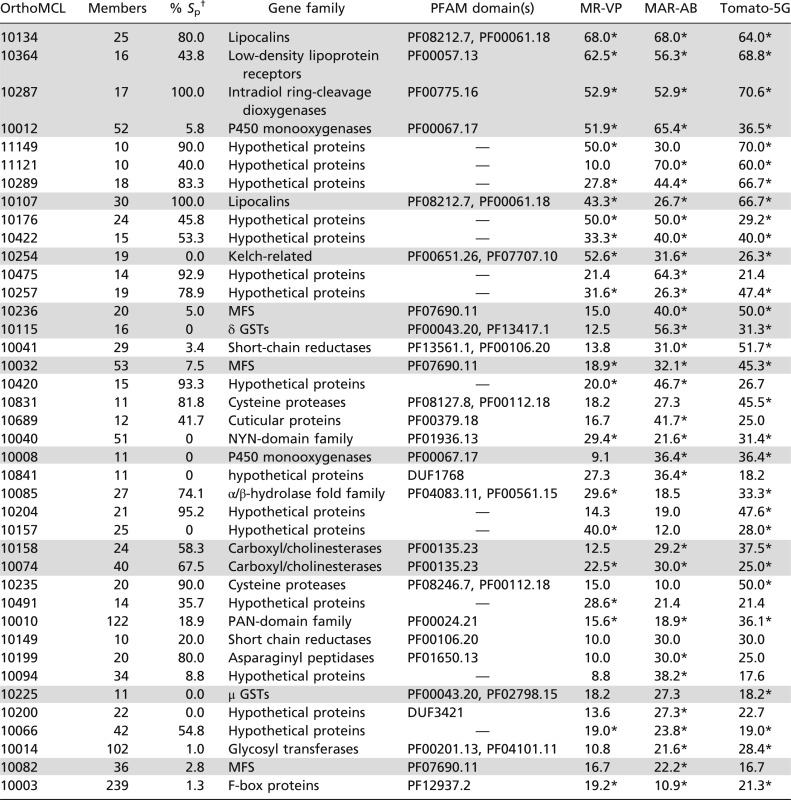 |
Gene clusters are as defined by OrthoMCL clustering. Percentages of genes differentially expressed within each OrthoMCL cluster in MR-VP, MAR-AB, and Tomato-5G are shown. Only shared OrthoMCL clusters (≥10 genes) where at least 20% of members are differentially expressed in MR-VP, MAR-AB, or Tomato-5G are shown. Gene family names have been linked to OrthoMCL clusters when possible. Many clusters consist of proteins with no PFAM hits (hypothetical proteins). OrthoMCL clusters are sorted on the basis of the average of the percentage of differentially expressed genes within each OrthoMCL cluster across MR-VP, MAR-AB, and Tomato-5G. For each OrthoMCL cluster the percentage of genes predicted with a signal peptide was calculated using SignalP 3.0 (115). Clusters mentioned in this study are shaded in gray.
*OrthoMCL clusters with an EASE (modified Fisher’s exact P value) score <0.05 (107).
†Percentage of gene members predicted with a signal peptide using SignalP 3.0 (115).
Intriguingly, some of the most strongly affected gene families in both experiments have signatures that have, until now, not been commonly associated with response to xenobiotics in arthropods. To shed insights into the T. urticae polyphagous life history, we therefore examined the composition and the nature of transcriptional responses for such families of moderate size (10 members or more, see below). We note, however, that genes of unknown function had some of the most striking expression changes. For many such genes, encoded products are predicted to be secreted (Table 1). An example is OrthoMCL cluster 10257 for which tetur11g05420, tetur11g05450, and tetur46g00020 were up-regulated by ∼700-fold upon host transfer to tomato for five generations.
Intradiol Ring-Cleavage Dioxygenases.
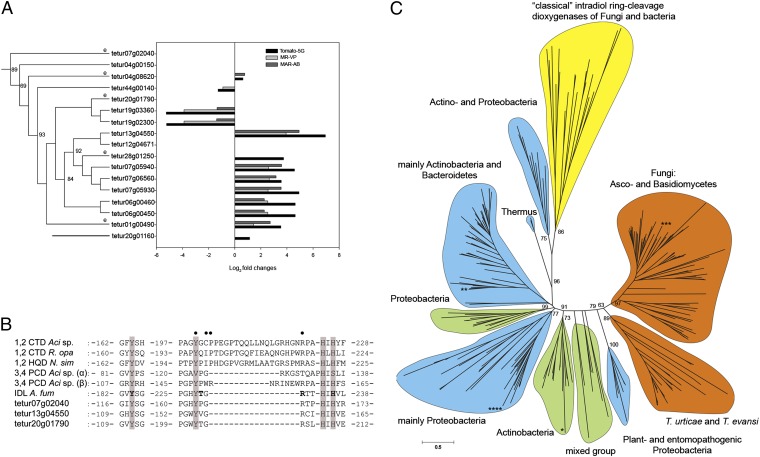
A set of 17 genes encoding secreted proteins identified as intradiol ring-cleavage dioxygenases (ID-RCDs) was among the most striking differentially expressed in our analysis (Table 1). These genes, belonging to the “intradiol dioxygenase-like” subgroup (cd03457) according to the Conserved Domain Database (44), were recently identified as a case of lateral gene transfer in the genome of T. urticae and have not been reported in other metazoan genomes to date (29). More than half of the genes in this family were differentially expressed upon host plant change and in multiresistant strains, and their expression patterns were highly correlated (Figs. 2B and 3A). ID-RCDs catalyze the oxygenolytic fission of catecholic substances, allowing bacteria and fungi to degrade aromatic rings, a crucial step in the global carbon cycle. Although bacteria usually harbor only 1–4 ID-RCD genes, this family has proliferated in T. urticae to 16 complete ID-RCDs and a pseudogene (tetur07g06560). Spider mite ID-RCDs share the conserved 2 His-2 Tyr nonheme iron (III) active site with previously described and functionally characterized ID-RCDs (such as catechol, hydroxyquinol, and protocatechuate ID-RCDs) (29, 45–48) (Fig. 3B). They are distributed over 11 genomic scaffolds and all but one (tetur07g02040) are intronless. Clusters of duplicated T. urticae ID-RCD genes were found on several scaffolds (Fig. 3A). We detected by PCR five orthologous genes in the closely related species, Tetranychus evansi, an oligophagous specialist of Solanaceae (Fig. 3 A and C). We also found ID-RCD sequences (E-value ≤ 2e−36) in the RNAseq data from the citrus red spider mite, Panonychus citri [European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) accession no. ERP000885]. However, we found no trace of their presence in the genomes of other, non–plant-feeding Acari, such as Metaseiulus occidentalis(a predatory mite), Varroa destructor (an ectoparasite of bees), or Ixodes scapularis (a blood-feeding tick). This suggests that a horizontal transfer occurred after the split of these lineages from the Tetranychidae. Phylogenetic analysis revealed that spider mite ID-RCDs cluster with a group of fungal ID-RCDs that share a common ancestor with plant and entomopathogenic bacterial ID-RCDs [such as Xenorhabdus sp. and Photorhabdus sp. (49)] (Fig. 3C). None of these fungal ID-RCDs belong to previously characterized “classical” fungal and bacterial ID-RCDs known to metabolize well-characterized substrates such as catechol and protocatechuate. In contrast to the characterized cytoplasmic enzymes, this large clade of predicted secreted forms of ID-RCDs (Fig. 3C and Table S1) has not yet been recognized and thoroughly characterized, although we found these proteins in proteomic data on fungal secretomes (50, 51).
Fig. 3.
Intradiol ring-cleavage dioxygenases (ID-RCDs) in T. urticae. (A) Phylogenetic relationship of T. urticae ID-RCDs linked to expression levels (log2(FC)) in acaricide-multiresistant strains (MR-VP and MAR-AB) and after host plant shift to tomato (S. lycopersicum) for five generations. Genes with detected orthologs in T. evansi are depicted with an “e”. (B) Alignment of conserved residues in “classical” and secreted ID-RCDs. CTD, catechol ID-RCD; PCD, protocatechuate ID-RCD; HQD, hydroxyquinol ID-RCD; IDL, intradiol dioxygenase-like (cd03457); Aci, Acinetobacter sp.; R. opa, Rhodococcus opacus; N. sim, Nocardia simplex; A. fum, A. fumigatus. Tetur07g02040, tetur13g04550, and tetur20g01790 are ID-RCD representatives of T. urticae. The two His-2 Tyr nonheme iron (III) binding sites are indicated by shading. Residues defined by crystallographic (46–48) studies to have an influence on substrate interaction in classical ID-RCDs (CTD, PCD, and HQD) are indicated by solid circles. The predicted binding residues of epicatechin in the protein sequence of A. fumigatus are indicated in boldface type (69). (C) Maximun-likelihood unrooted tree of 17 ID-RCDs of T. urticae (and five T. evansi orthologs) with 232 bacterial and fungal sequences. Color codes indicate the percentage of secretion within the clade: yellow, not secreted; blue, <55% secreted; green, 55–85% secreted; and orange, >85% secreted. All members of the T. urticae clade are secreted and form a sister clade to fungal secreted dioxygenases, sharing a most recent common ancestor with plant and entomopathogenic Proteobacteria. The classical biochemically characterized ID-RCDs (CTD, PCD, and HQD) are not secreted and cluster together as an outgroup. The phylogenetic positions of the ID-RCD protein sequences of Naegleria gruberi (Protozoa), Shistosoma mansoni (Metazoa), P. infestans (oomycete), and Haloferax volcanii (Archaea) are indicated by *, **, ***, and ****, respectively.
Lipocalins.
We also found that lipocalins, small proteins capable of binding to hydrophobic molecules, were strongly differentially expressed (68% of genes of OrthoMCL cluster 10134) between London and resistant strains and dynamically over time to host plant change (Table 1 and Fig. 4A). As revealed by hierarchical clustering, lipocalin expression patterns of resistant strains and mites feeding on tomato for five generations group together. Some of these lipocalin genes are strongly and progressively induced in mites feeding for 2 and 12 h on tomato, but are completely down-regulated after five generations, whereas stable induced expression is maintained for other lipocalin genes (Fig. S2A).
Fig. 4.
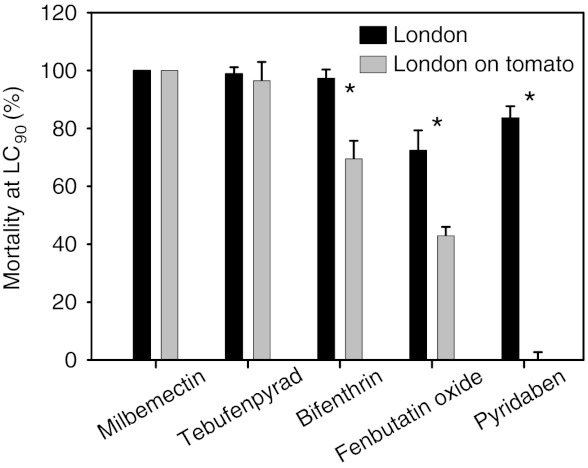
Effect of host plant on acaricide toxicity. Percentage mortality of spider mites from the London strain to pesticide treatment. Shown are LC90 values for exposure to various acaricides (pyridaben, milbemectin, tebufenpyrad, fenbutatin oxide, and bifenthrin) before (London) and after adaptation on tomato (London on tomato). An asterisk indicates significant differences determined by a t test for paired samples.
Of the 58 complete lipocalins we annotated in the T. urticae genome, more than the half are concentrated on only three scaffolds (20 on scaffold 6, 8 on scaffold 1, and 5 on scaffold 31) (Table S2). The number of T. urticae lipocalins far exceeds those reported in insects (Drosophila melanogaster, 4; Apis mellifera, 4; Rhodnius prolixus, 22) (52) and in humans (10) (53), but is in the same range as in ticks (54). Thirty-six (62.0%) T. urticae lipocalins were predicted to have an antiparallel β-barrel, whereas 15 (25.9%) had only a small deviation from the canonical lipocalin secondary structure (55) (Table S2). T. urticae lipocalins do not have a GPI-anchor signal omega site and are, with the exception of tetur31g00780, predicted to have a signal peptide (Table S2). Two main types of lipocalin gene organization were apparent. Thirty-seven genes have five exons and an intron phase pattern of 0-2-1-1, corresponding to the arthropod lipocalin gene consensus pattern, whereas 18 lipocalin genes have only four exons and a 0-1-1 intron phase pattern, a gene structure also reported for a moth lipocalin (56) (Table S2 and Fig. S2C). Most T. urticae lipocalins cluster together with a previously described clade (57, 58) comprising vertebrate apolipoprotein D (ApoD) and crustacyanins with a high bootstrap support (83%) (Fig. S2B and Tables S2 and S3). Within this large T. urticae lipocalin clade, most lipocalins cluster according to their intron phase pattern (Fig. S2 B and C). However, 5 T. urticae lipocalins (tetur01g01500, tetur01g01510, tetur01g01520, tetur01g16584, and tetur02g09610) grouped together with Karl, a lipocalin expressed in the blood cells of D. melanogaster, and 2 (tetur174g00050 and tetur07g03970) were closely related to insect biliproteins (Fig. S2B).
Major Facilitator Superfamily.
Among the genes differentially expressed in MR-VP, MAR-AB, and Tomato-5G (Table 1), three OrthoMCL clusters (10082, 10032, and 10236) with a total of 109 genes shared the PFAM-domain PF07690.11 that characterizes the major facilitator superfamily (MFS), also known as the uniporter-symporter-antiporter family. Members of OrthoMCL clusters 10236 and 10082 were highly similar (E-value ≤ e−17 and ≤ e−5, respectively) to the anion/cation symporter (ACS) family and the Na+-dependent glucose transporter family, respectively (Table S4). On the other hand, most members of cluster 10032 showed similarity (E-value ≥ e−10, Table S4) to bacterial tetracycline:H+ antiporters and their mammalian homologs, the heme-carrier proteins/thymic-folate cotransporters (59). All differentially expressed members of OrthoMCL cluster 10032 were up-regulated in both MR-VP and MAR-AB, and 87.5% of the differentially expressed members were also up-regulated on tomato for five generations. Tetur11g05410 was more than 300-fold up-regulated by transfer to tomato. Moreover, 16 members of cluster 10032 were already up-regulated in mites transferred from bean to tomato for 12 h.
Transcription Factors.
Transcription factors belonging to the nuclear receptor family, the bHLH-PAS family, and the bZIP family are known to be involved in the response to stress and xenobiotics in vertebrates and in insects (60, 61). The T. urticae genome harbors at least 700 transcription factors (29), and in a hierarchical clustering analysis (Pearson’s centered distance metric, complete linkage rule) with transcription factor expression data, Tomato-5G clustered with MR-VP and MAR-AB and formed a sister clade to Tomato-2h and Tomato-12h. Seventeen, 20, and 27 transcription factors were differentially expressed (log2(FC) ≥ 1, FDR < 0.05) in MR-VP, MAR-AB, and Tomato-5G, respectively, although only 4 (tetur03g03150, tetur07g01800, tetur24g02450, and tetur36g00260) were shared between MR-VP, MAR-AB, and Tomato-5G (Table S5). Tetur36g00260 belongs to the class of nuclear receptors and is one of the eight paralogs of the vertebrate xenosensors PXR and CAR found in spider mites (29).
In Drosophila, the xenosensor cap “n” collar isoform-C (CncC) is down-regulated by Kelch-like ECH-associated protein (Keap1) (60). Intriguingly, we found a large proportion of genes in the OrthoMCL cluster 10254, with PFAM domains typical for Kelch-like proteins (Table 1), to be differentially expressed in Tomato-5G, MR-VP, and MAR-AB, with almost all (83–100%) of these genes down-regulated. Four genes were down-regulated under all three conditions, in particular tetur24g00770 (down-regulated 34.8, 7.5, and 2.9 times in Tomato-5G, MR-VP, and MAR-AB, respectively). We also searched for homologs of the Drosophila xenosensor CncC and identified two paralogs of this bZIP transcription factor in the T. urticae genome [tetur07g06850 and 07g04600, E-value < e−31 using CncC of D. melanogaster (GenBank accession no. AAN13930) as a query]. However, neither one changed expression levels in our microarray experiments.
Effects of Host Plant Shift Effects on Acaricide Toxicity.
Because we found that expression profiles were highly correlated between mites adapted to tomato and pesticide-resistant strains, we asked whether acaricide toxicity was affected by host plant shift. We chose five acaricides with four different modes of action and compared their toxicity on susceptible (London) mites kept on beans vs. mites adapted to tomato. The adaptation to tomato was accompanied by a significant decrease in toxicity for three of the acaricides (Fig. 4). Among these, the two mitochondrial electron transport inhibitors, pyridaben and tebufenpyrad, had a different response. The toxicity of pyridaben was greatly diminished, whereas that of tebufenpyrad was not affected.
Discussion
Transcriptional Response to Host Plant Shift.
Classical studies of mite host transfer have shown that fitness on new hosts can increase rapidly over a small number of generations (32, 62). Here, we show that the response of mites to a new host is accompanied by similarly rapid changes in transcriptional profiles on a timescale of hours to a relatively small number of generations. Genes that responded in the short term (12 h or less) included those in classical detoxification families, such as P450s and CCEs. This finding is consistent with that observed for T. urticae unfed larvae that were placed for 12 h on bean, tomato, or Arabidopsis thaliana and that was ascertained with another method (RNA-seq) (29). However, compared with the short-term response, expression changes were far more dramatic at five generations. About threefold more genes were detected as differentially expressed, with ∼7.5% of all genes changed in expression. The host adaptation experiments used the London reference strain that, although partially inbred, nonetheless segregates for several haplotypes throughout much of the genome (41). Whether the pronounced differences in gene expression observed at five generations reflect selection on standing genetic variation, or alternatively physiological or epigenetic changes, remains to be determined. However, genetic variation between spider mites for characters affecting host use has been reported previously (32, 63, 64), and selection is observed in spider mite populations after a few generations (32, 62).
Host Plant Shift and Pesticide Resistance.
Remarkably, the transcriptome of pesticide-susceptible mites grown for five generations on tomato was closer to that of two acaricide-resistant strains than to that of the initial response to the host plant shift. Of note, coordinated transcriptional changes were apparent for known major environmental response genes including P450s, GSTs, and CCEs (65). This suggests that in response to different chemical challenges, the spider mite is “rounding up the usual suspects.” The mechanism involved is still uncertain, but the pattern of expression for several transcription factor genes and their regulators is also similar between Tomato-5G and the resistant strains, possibly underlying the coordinated response.
In mammals, the pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are major xenosensor genes (61). The Drosophila paralog of these nuclear receptor genes, DHR96, is also involved in the response to xenobiotics (66). One of the eight paralogs of DHR96, tetur36g00260, was more than twofold up-regulated in Tomato-5G, MAR-AB, and MR-VP (Table S5). Further, RNAi silencing of the Drosophila Keap1 gene, a negative regulator of the Drosophila transcription factor CncC, up-regulated detoxification gene expression (60). In our experiments, the down-regulation of OrthoMCL cluster 10254 transcripts (Kelch-like proteins) might similarly result in the up-regulation of T. urticae detoxification genes. However, there are multiple Kelch-like proteins and two CncC orthologs in T. urticae, and experimental studies will be required to confirm the roles of upstream transcriptional regulators in establishing the observed transcriptional profiles.
Extending the Arsenal of Environmental Response Genes.
Despite the involvement of known detoxification gene families, a major finding was that, after host plant shift or between acaricide-susceptible and -resistant strains, many genes that encode proteins without homology to proteins of known functions changed expression. In part, this may reflect the phylogenetic distance between spider mites and insects for which most functional-genetic studies in arthropods have been performed [the divergence between mites and insects is more than 450 My (67)]. However, it may also reflect the recruitment of diverse genes to contribute to T. urticae’s polyphagous lifestyle.
This interpretation is supported by our finding of strong transcriptional responses to changes in chemical exposure for some gene families either absent from insects or previously not recognized to play a major role in xenobiotic response. For instance, intradiol ring-cleavage dioxygenases, which have not been reported in other metazoa, may contribute prominently to the spider mite detoxification arsenal. The expansion of this family in the T. urticae genome (29) and the transcriptional regulation of many family members in response to host transfer or in pesticide-resistant strains are indicative of a selective advantage after the initial horizontal transfer. Currently, the substrate specificity of the spider mite ID-RCDs is not known. Spider mite ID-RCDs are most closely related to fungal enzymes, and an ID-RCD of Aspergillus fumigatus can modify an array of procyanidins, which are polymers of (+)-catechine and/or (−)-epicatechin (68). This hints that some ID-RCDs can metabolize more complex structures than simple catecholic substances. The A. fumigatus enzyme shows common features with the spider mite enzymes: It is predicted to be secreted, has a similar distance between active site residues, and contrasts with previously biochemically characterized enzymes in bacteria and other fungi (Fig. 3B). One of the T. urticae enzymes (tetur20g01790) has identical residues at the predicted binding site for epicatechin in the A. fumigatus protein sequence (Tyr184, Thr229, Arg231, and His236, A. fumigatus numbering) (69) (Fig. 3B). Interestingly, a secreted ID-RCD from the oomycete Phytophthora infestans (GenBank accession no. XP_002905783) clustered within the group of fungal secreted ID-RCDs (Fig. 3C). This is in agreement with Richards et al. (70, 71) who showed that the closely related species Phytophthora ramorum acquired its extracellular ID-RCDs from filamentous ascomycetes through horizontal gene transfer.
Other gene families are ubiquitous in their phylogenetic distribution, but their role in environmental response is striking in the spider mite. Lipocalins are small extracellular proteins and are characterized by (i) their ability to bind to hydrophobic molecules, (ii) a conserved secondary structure (an antiparallel β-barrel, with a repeated +1 topology, with an internal ligand-binding site), (iii) low sequence conservation (<20%), and (iv) a conserved exon–intron structure (55, 56). Members of the lipocalin family are found in a wide range of species, with roles in metabolism, coloration, perception, reproduction, developmental processes, and modulation of immune and inflammatory responses (72), resulting in a very diverse nomenclature for each specific lipocalin (e.g., apolipoprotein D, crustacyanins, biliproteins, and salivacalins). As lipocalins typically bind hydrophobic molecules, they may bind pesticides/allelochemicals in mites, resulting in sequestration of these toxic, generally hydrophobic compounds. Moreover, the feeding strategy of mites may favor sequestration in dispensable phagocytes, as suggested by Mullin and Croft (73). Lipocalins may also protect against oxidative stress, as loss of the Drosophila homolog of ApoD, Glial Lazarillo, increased sensitivity to oxidative stress, whereas overexpression increased hyperoxia tolerance (74). Similar findings were also reported for ApoD in mice and in Arabidopsis (75, 76). In plants, the oxidative burst is one of the earliest observable aspects of the plant’s defense strategy against herbivores (77, 78). Whether mite lipocalins up-regulated specifically in the early response to transfer to tomato (2 and 12 h) are involved in resistance to the oxidative response warrants investigation.
In addition to enzymes and small secreted proteins, membrane-binding proteins and transporters featured prominently in our analysis, including the MFS family, one of the largest families of membrane transporters along with ABC transporters. Our experiments did not reveal an increased expression of many ABC transporter genes, although this gene family is highly expanded in the spider mite (29). Multiple MFS family members were differentially regulated in both Tomato-5G and resistant strains. MFS transporters are single-polypeptide carriers that work in symport/antiport (79, 80), and studies in bacteria and fungi have identified roles in the transport of toxic substances (81–83). For example, overexpression of the mfsM2 gene in a sensitive strain of Botrytis cinerea, a fungal plant pathogen, led to drug-resistance levels similar to those of a fungicide-resistant B. cinerea strain (82). If spider mite MFS proteins function as efflux transporters, their up-regulation might result in a higher efflux of acaricides/toxic allelochemicals or their metabolites out of spider mite cells.
A third group of noncatalytic binding proteins/transporter proteins with a high percentage of genes changing expression in Tomato-5G, MR-VP, and MAR-AB was low-density lipoprotein receptor proteins (LDLRs) (OrthoMCL cluster 10364, Table 1). In humans, these endocytic receptors bind hydrophobic molecules (84) and participate in a wide range of physiological processes. Some members of this family have also multiligand-binding properties (85, 86). In our experiments, all these genes were down-regulated. If hydrophobic pesticides and allelochemicals interact with these LDLRs, a down-regulation could result, through lower receptor-mediated endocytosis, in a lower uptake into spider mite cells.
The role of binding proteins/transporters in the response of insects to chemically challenging environments has until now been generally overlooked and would merit closer attention. Earlier work using dedicated microarrays (87, 88) could not, by definition, uncover the importance of these new players. However, microarray studies with restricted random sets of ESTs have already pointed out that transporters were differentially regulated in lepidopteran larvae that were fed with plants that differed in their chemical defense profile (89). Our study with a pangenomic array extends this early observation and emphasizes the importance of (largely) unbiased approaches for studies to understand the basis of generalist herbivore life histories. Obtaining formal evidence that some members of these gene families actually contribute to xenobiotic tolerance when up- or down-regulated should be a priority for future research.
Host Plant Change and Acaricide Tolerance.
We show here that adaptation to tomato not only changes the transcript levels of many detoxification enzymes, but also results in a decreased acute toxicity for three of five acaricides tested. The transcriptional remodeling we observed after adaptation to tomato may thus be the proximal cause for the well-known effect of host plants on the efficacy of acaricides in T. urticae (17, 24, 90, 91). Host plants are also known to affect the toxicity of insecticides to insects (13–16, 18–23, 25). Feeding on alternative hosts changes the activity of mite detoxification enzymes measured with some model substrates (24, 73), and performance of mites on tomato was negatively affected by a P450 inhibitor (92). These findings indicate that the transcriptomic changes we observed may be essential for performance. The importance of active (expressed) herbivore detoxification enzymes to survival on chemically challenging hosts is also well documented in insects (e.g., refs. 15, 93).
Polyphagy and Pesticide Resistance.
We found a common pattern of gene expression between mites that adapted to a new host and those constitutively resistant to diverse pesticides. Moreover, the unexpected gene families that figure so prominently in our analysis, as well as the usual suspects, form a coherent whole from transcription factors to effector genes in detoxification, binding, and transport. This indicates an orchestrated response rather than a random deregulation caused by the toxic effects of the plant or acaricide challenge. Many gene families we found to have strong transcriptional responses are large, reflecting a (presumed) long evolutionary history of gene duplications, sometimes in a lineage-specific manner (29, 94). In the response to plants, such patterns are expected given the long evolutionary timescale over which plant–herbivore interactions are fine tuned. In contrast, the number of detoxification genes with altered transcription in the resistant strains, and with fold changes mimicking those of host plant transfer, is not expected from classical theory. It has long been argued that field-evolved resistance would select single genes with major effects rather than many with limited effect (95, 96). In some cases of target site resistance and metabolic resistance, the monogenic inheritance has been established experimentally (41, 97). Additionally, there is a substantial difference between the moderate tolerance level of the tomato-adapted mites and the high resistance level of the acaricide-resistant mites. If all common genes in the transcriptomic signature contributed directly to resistance, then Tomato-5G should be much more resistant. This difference between the high resistance level of the MR-VP and MAR-AB strains and the more moderate tolerance level of the Tomato-5G mites may be due to the presence of target site mutations in the resistant strains. Indeed the presence of a mutation in the glutamate-gated chloride channel of the MAR-AB strain has already been documented (98).
We propose an explanation for these apparent paradoxes that may be relevant to the rapid development of resistance in polyphagous pests (Fig. S3). Whereas a single resistance gene with major effect may eventually be selected from rare alleles, initial survival in an environment with a heterogenous dose or distribution of the pesticide may depend on multiple alleles that confer moderate resistance. Such multiple alleles may be present in modules and include genes controlling detoxification/binding/transport processes, thus affecting all aspects of the toxicokinetic balance. Intraspecific genetic variation in host preference is a common aspect of polyphagy (99, 100) and has been repeatedly shown in the spider mite (32, 101). To the mite, encountering a plant treated with an acaricide may be akin to encountering a new host plant. There would be rapid selection for a genotype carrying a set of genes whose expression would best buffer against the chemical signature of the new hostile environment. That genetic variation in environmental response can come in groups of connected genes has been recently documented. Phenotypic variation in the transcriptome profile of 40 inbred Drosophila strains was shown to consist of groups of interconnected genes. This formed hundreds of “modules” of ecologically relevant correlated genetic variation (102). In a polyphagous pest such as the spider mite, such modules of coregulated genes may provide an explanation for the common transcriptional patterns of MR-VP, MAR-AB, and Tomato-5G. In this sense, when polyphagy is seen as genetic polymorphism in the response to different chemical environments, it may represent a preadaptation to xenobiotic resistance as suggested previously (10, 12). The selection of the rare resistance allele to the acaricide would be facilitated by the initial, higher survival rate of a subset of the population harboring it. A plant-specific transcriptional response has been observed before in polyphagous Lepidoptera and in the spider mite (29, 89, 103). Specialist herbivores, on the other hand, are characterized by a xenobiotic response that is more constitutive and more targeted to the favorite host plants (65), and the transcriptional response to change in plant chemistry is much more restricted in a specialist than in a generalist (89). It remains to be shown that intraspecific genetic variation in the spider mite includes the differential regulation of specific subsets of genes involved in fitness on different host plants, such as detoxification, binding, and transport as we predict here. Such experimental verification is as important as it is difficult to obtain. However, the rapid development of resistance in polyphagous herbivores and its relative absence in specialists such as natural enemies and predators is well known (11). Our results provide an unprecedented insight into the transcriptional correlation that may link polyphagy with development of pesticide resistance. They also highlight the need to study not just detoxification enzymes, but also binding proteins and transporters as major contributors to survival in a toxic environment. The spider mite is a polyphagous jack of all trades, but has evolved to be a master of some as well.
Materials and Methods
Plant Rearing.
Tomato seeds (S. lycopersicum cv. “Moneymaker”) and kidney bean (P. vulgaris cv. “Prelude”) were potted in black earth (Structural Professional, pH 5.0–6.5, 20% organic matter; Snebbout NV) and allowed to grow in a growth chamber at 26 + 0.5 °C, 60% relative humidity (RH) and a 16:8-h light:dark (L:D) photoperiod. Tomato plants were used for experiments when they had at least four completely developed leaves (about 35 d old) whereas bean plants were used for either experiments or spider mite rearing when they had two completely developed leaves (about 14 d old).
Mite Strains.
The London reference strain originates from a wild-collected T. urticae population from the Vineland region (Ontario, Canada) and DNA from an inbred line of this strain was used for T. urticae genome sequencing (29). This strain is susceptible to commercially available acaricides (37). The LS-VL laboratory reference strain, originally collected in 2000 near Ghent (Belgium), has been previously described as highly susceptible to acaricides (43). The MR-VP resistant strain was originally collected from different cultivars of bean plants in a greenhouse at the national botanical garden (Brussels) in September 2005, where spider mite control was reported to be extremely problematic. The strain was controlled by regular foliar applications of commercial formulations of the following acaricides: tebufenpyrad, pyridaben, clofentezine, hexythiazox, bifenthrin, fenbutatin oxide, abamectine, and oxamyl. Resistance to mitochondrial electron transport inhibitor (METI) acaricides is well characterized in MR-VP (104). The Marathonas (MAR-AB) strain was isolated from a heavily sprayed rose greenhouse near Athens (Greece) in 2009. The strain is highly resistant to abamectin, bifenthrin, clofentezine, hexythiazox, fenbutatin oxide, and pyridaben (98).
All T. urticae strains were mass reared on potted kidney bean plants in a climatically controlled room at 26 °C (±0.5 °C), 60% RH, and a 16:8-h L:D photoperiod. The strains were offered fresh bean plants weekly.
Host Change Experiment.
For each time point (2 h, 12 h, 80 d) a tomato plant was infested with about 150 female mites from the London strain (cultured on bean plants). Two hours (Tomato-2h), 12 h (Tomato-12h), and 80 d (about five generations, Tomato-5G) after infestation 100 mites were recollected for total RNA extraction. All experiments were performed at 26 + 0.5 °C, 60% RH, and a 16:8-h L:D photoperiod and four biological replicates were performed for each time point. The Tomato-2h and Tomato-12h experiments were performed during the 16-h light photoperiod.
Toxicity Tests.
Toxicity bioassays on the London strain with and without adaptation to tomato (eight generations) were performed in a similar way to that described by Van Leeuwen et al. (43). First, adult female mites were transferred to square kidney bean leaf discs (P. vulgaris cv. “Prelude”), placed on wet cotton in a Petri dish. Subsequently, 800 μL of the lethal concentration killing 90% of the population (LC90) of acaricides with different modes of action [pyridaben (Sanmite 200 g/kg WP), tebufenpyrad (Pyranica 200 g/L EC), mitochondrial complex I electron transport inhibitors; milbemectin (Milbeknock 9.3 g/L EC), chloride channel activator; fenbutatin oxide (Torque 550 g/L SC), inhibitor of mitochondrial ATP synthase and bifenthrin (Talstar 80 g/L SC), sodium channel modulator] was sprayed on the mites. Mites sprayed with double-distilled water were used as a control. Finally, bioassays were placed in a climatically controlled room at 26 ± 0.5 °C, 60% RH, and a 16:8-h L:D photoperiod. Four replicates were performed for each strain and for each acaricide. Mortality was assessed 24 h after acaricide application and corrected for control mortality using Abott’s equation (105). Mites were considered dead when they were drowned or when they did not move after prodding with a fine hair brush.
Microarray Experiments and qPCR.
A custom Sureprint genome-wide G3 Gene Expression 8 × 60K microarray was designed using the Agilent eArray platform (Agilent Technologies) based on the T. urticae genome annotation (version from April 20, 2010). RNA was isolated with the RNeasy Mini kit (Qiagen) followed by DNase treatment (Turbo DNase; Ambion). RNA was labeled using the Agilent Low-Input Quick Amp Labeling kit (dual color) (version 6.5; Agilent Technologies). Arrays were scanned by an Agilent Microarray High-Resolution Scanner with default settings for 8 × 60K G3 microarrays. For detailed description of the design, target preparation, hybridization, and analysis see SI Materials and Methods. A subset of differentially expressed genes as identified by microarray experiments was validated by qPCR analysis (details in SI Materials and Methods and Table S6).
Clustering Analysis.
The GeneSpring GX11.0 software (Agilent Technologies) was used to perform a hierarchical clustering analysis of microarray expression data, using the Pearson centered distance metric and complete linkage rule. For clustering of all T. urticae predicted protein sequences, the OrthoMCL (106) software version 2.0 was used (details in SI Materials and Methods). A modified Fisher’s exact P value (EASE score) was calculated (107) to measure the gene enrichment in OrthoMCL clusters of our microarray expression data.
Gene Family Analysis.
Major gene families identified by microarray experiments were analyzed in detail using tBLASTn, BLASTp (108), and PFAM-domain searches (109). Alignments were constructed using Clustal X (110) and MUSCLE (111). For phylogenetic analysis, model selection was performed with Prottest 1.4 (112) and maximum-likelihood analysis was performed with Treefinder (113). Phylogenetic trees were visualized and edited using MEGA5 (114). Signal peptides were predicted with SignalP 3.0 (115) and Jpred 3 (116) was used to predict secondary structures for lipocalins. A detailed description of procedures is described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Lies Vantomme for helping with microarray hybridizations and Luc Tirry (L.T.) and Vojislava Grbić for valuable remarks on the manuscript. T.V.L. is a postdoctoral fellow of the Fund for Scientific Research Flanders (FWO). This work was supported by FWO Grants 3G061011 (to T.V.L.) and 3G009312 (to T.V.L. and L.T.), Ghent University Special Research Fund Grant 01J13711 (to T.V.L. and L.T.), the Government of Canada through Genome Canada and the Ontario Genomics Institute Grant OGI-046 (to M.G.), and a University of Utah Funding Incentive Seed Grant (to R.M.C.). N.W. was supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT, Grant IWT/SB/101451). Part of this research was supported by the “THALIS” project 377301, Operational Programme “Education and Life-Long Learning,” which is cofunded by the European Social Fund and National Resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE39869). Tetranychus evansi sequences reported in this paper have been deposited in the GenBank database (accession nos. JQ736355–JQ736359).
See Author Summary on page 393 (volume 110, number 2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213214110/-/DCSupplemental.
References
- 1.Strong DR, Lawton JH, Southwood TRE. Insects on Plants: Community Patterns and Mechanisms. Oxford: Blackwell Scientific; 1984. [Google Scholar]
- 2.Futuyma DJ, Agrawal AA. Macroevolution and the biological diversity of plants and herbivores. Proc Natl Acad Sci USA. 2009;106(43):18054–18061. doi: 10.1073/pnas.0904106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernays EA, Chapman RF. Host-Plant Selection by Phytophagous Insects. New York: Chapman & Hall; 1994. [Google Scholar]
- 4.Cornell HV, Hawkins BA. Herbivore responses to plant secondary compounds: A test of phytochemical coevolution theory. Am Nat. 2003;161(4):507–522. doi: 10.1086/368346. [DOI] [PubMed] [Google Scholar]
- 5.Berenbaum MR, Favret C, Schuler MA. On defining key innovations in an adaptive radiation - cytochrome P450s and Papilionidae. Am Nat. 1996;148:S139–S155. [Google Scholar]
- 6.Thompson JN. The Coevolutionary Process. Chicago: Univ of Chicago Press; 1994. [Google Scholar]
- 7.Ali JG, Agrawal AA. Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci. 2012;17(5):293–302. doi: 10.1016/j.tplants.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Krieger RI, Feeny PP, Wilkinson CF. Detoxication enzymes in the guts of caterpillars: An evolutionary answer to plant defenses? Science. 1971;172(3983):579–581. doi: 10.1126/science.172.3983.579. [DOI] [PubMed] [Google Scholar]
- 9.Berenbaum MR, Cohen MB, Schuler MA. Cytochrome-P450 monooxygenase genes in oligophagous Lepidoptera. ACS Symp Ser. 1992;505:114–124. [Google Scholar]
- 10.Gordon HT. Nutritional factors in insect resistance to chemicals. Annu Rev Entomol. 1961;6:27–54. [Google Scholar]
- 11.Croft BA, Strickler K. In: Pest Resistance to Pesticides. Georghiou GP, Saito T, editors. New York: Plenum; 1983. pp. 669–702. [Google Scholar]
- 12.Rosenheim JA, Johnson MW, Mau RFL, Welter SC, Tabashnik BE. Biochemical preadaptations, founder events, and the evolution of resistance in arthropods. J Econ Entomol. 1996;89:263–273. [Google Scholar]
- 13.Ahmad S. Enzymatic adaptations of herbivorous insects and mites to phytochemicals. J Chem Ecol. 1986;12:533–560. doi: 10.1007/BF01020571. [DOI] [PubMed] [Google Scholar]
- 14.Berry RE, Yu SJ, Terriere LC. Influence of host plants on insecticide metabolism and management of variegated cutworm (Lepidoptera, Noctuidae) J Econ Entomol. 1980;73:771–774. [Google Scholar]
- 15.Brattsten LB, Wilkinson CF, Eisner T. Herbivore-plant interactions: Mixed-function oxidases and secondary plant substances. Science. 1977;196(4296):1349–1352. doi: 10.1126/science.196.4296.1349. [DOI] [PubMed] [Google Scholar]
- 16.Castle SJ, Prabhaker N, Henneberry TJ, Toscano NC. Host plant influence on susceptibility of Bemisia tabaci (Hemiptera: Aleyrodidae) to insecticides. Bull Entomol Res. 2009;99(3):263–273. doi: 10.1017/S0007485308006329. [DOI] [PubMed] [Google Scholar]
- 17.Gould F, Carroll CR, Futuyma DJ. Cross-resistance to pesticides and plant defenses: A study of the two-spotted spider-mite. Entomol Exp Appl. 1982;31:175–180. [Google Scholar]
- 18.Kennedy GG. 2-tridecanone, tomatoes and Heliothis zea - potential incompatibility of plant antibiosis with insecticidal control. Entomol Exp Appl. 1984;35:305–311. [Google Scholar]
- 19.Li X, Baudry J, Berenbaum MR, Schuler MA. Structural and functional divergence of insect CYP6B proteins: From specialist to generalist cytochrome P450. Proc Natl Acad Sci USA. 2004;101(9):2939–2944. doi: 10.1073/pnas.0308691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Zangerl AR, Schuler MA, Berenbaum MR. Cross-resistance to alpha-cypermethrin after xanthotoxin ingestion in Helicoverpa zea (Lepidoptera: Noctuidae) J Econ Entomol. 2000;93(1):18–25. doi: 10.1603/0022-0493-93.1.18. [DOI] [PubMed] [Google Scholar]
- 21.Lindroth RL. Chemical ecology of the luna moth effects of host plant on detoxification enzyme-activity. J Chem Ecol. 1989;15:2019–2029. doi: 10.1007/BF01207434. [DOI] [PubMed] [Google Scholar]
- 22.Sasabe M, Wen ZM, Berenbaum MR, Schuler MA. Molecular analysis of CYP321A1, a novel cytochrome P450 involved in metabolism of plant allelochemicals (furanocoumarins) and insecticides (cypermethrin) in Helicoverpa zea. Gene. 2004;338(2):163–175. doi: 10.1016/j.gene.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 23.Zeng RS, Wen Z, Niu G, Schuler MA, Berenbaum MR. Allelochemical induction of cytochrome P450 monooxygenases and amelioration of xenobiotic toxicity in Helicoverpa zea. J Chem Ecol. 2007;33(3):449–461. doi: 10.1007/s10886-006-9238-1. [DOI] [PubMed] [Google Scholar]
- 24.Yang XM, Margolies DC, Zhu KY, Buschman LL. Host plant-induced changes in detoxification enzymes and susceptibility to pesticides in the twospotted spider mite (Acari: Tetranychidae) J Econ Entomol. 2001;94(2):381–387. doi: 10.1603/0022-0493-94.2.381. [DOI] [PubMed] [Google Scholar]
- 25.Yu SJ, Berry RE, Terriere LC. Host plant stimulation of detoxifying enzymes in a phytophagous insect. Pestic Biochem Physiol. 1979;12:280–284. [Google Scholar]
- 26.Feyereisen R. In: Insect Molecular Biology and Biochemistry. Gilbert LI, editor. London: Academic Press, Elsevier; 2012. pp. 236–316. [Google Scholar]
- 27.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 28.Després L, David JP, Gallet C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol Evol. 2007;22(6):298–307. doi: 10.1016/j.tree.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Grbić M, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479(7374):487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agrawal AA. Host-range evolution: Adaptation and trade-offs in fitness of mites on alternative hosts. Ecology. 2000;81:500–508. [Google Scholar]
- 31.Magalhães S, Blanchet E, Egas M, Olivieri I. Are adaptation costs necessary to build up a local adaptation pattern? BMC Evol Biol. 2009;9:182. doi: 10.1186/1471-2148-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fry JD. Evolutionary adaptation to host plants in a laboratory population of the phytophagous mite Tetranychus urticae Koch. Oecologia. 1989;81:559–565. doi: 10.1007/BF00378969. [DOI] [PubMed] [Google Scholar]
- 33.Magalhães S, Fayard J, Janssen A, Carbonell D, Olivieri I. Adaptation in a spider mite population after long-term evolution on a single host plant. J Evol Biol. 2007;20(5):2016–2027. doi: 10.1111/j.1420-9101.2007.01365.x. [DOI] [PubMed] [Google Scholar]
- 34.Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem Mol Biol. 2010;40(8):563–572. doi: 10.1016/j.ibmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Carrière Y. Haplodiploidy, sex, and the evolution of pesticide resistance. J Econ Entomol. 2003;96(6):1626–1640. [PubMed] [Google Scholar]
- 36.Denholm I, Cahill M, Dennehy TJ, Horowitz AR. Challenges with managing insecticide resistance in agricultural pests, exemplified by the whitefly Bemisia tabaci. Philos Trans R Soc B Biol Sci. 1998;353(1376):1757–1767. [Google Scholar]
- 37.Khajehali J, Van Nieuwenhuyse P, Demaeght P, Tirry L, Van Leeuwen T. Acaricide resistance and resistance mechanisms in Tetranychus urticae populations from rose greenhouses in the Netherlands. Pest Manag Sci. 2011;67(11):1424–1433. doi: 10.1002/ps.2191. [DOI] [PubMed] [Google Scholar]
- 38.Van Leeuwen T, Dermauw W, Grbic M, Tirry L, Feyereisen R. Spider mite control and resistance management: Does a genome help? Pest Manag Sci. 2012 doi: 10.1002/ps.3335. [DOI] [PubMed] [Google Scholar]
- 39.Bitume EV, et al. Heritability and artificial selection on ambulatory dispersal distance in Tetranychus urticae: Effects of density and maternal effects. PLoS ONE. 2011;6(10):e26927. doi: 10.1371/journal.pone.0026927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy GG, Smitley DR. In: Spider Mites. Their Biology, Natural Enemies and Control. Helle W, Sabelis MW, editors. Vol 1A. Amsterdam: Elsevier; 1985. pp. 233–242. [Google Scholar]
- 41.Van Leeuwen T, et al. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc Natl Acad Sci USA. 2012;109(12):4407–4412. doi: 10.1073/pnas.1200068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes TR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19(4):342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 43.Van Leeuwen T, Van Pottelberge S, Tirry L. Comparative acaricide susceptibility and detoxifying enzyme activities in field-collected resistant and susceptible strains of Tetranychus urticae. Pest Manag Sci. 2005;61(5):499–507. doi: 10.1002/ps.1001. [DOI] [PubMed] [Google Scholar]
- 44.Marchler-Bauer A, et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaillancourt FH, Bolin JT, Eltis LD. The ins and outs of ring-cleaving dioxygenases. Crit Rev Biochem Mol Biol. 2006;41(4):241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 46.Ferraroni M, et al. Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation. J Biol Chem. 2005;280(22):21144–21154. doi: 10.1074/jbc.M500666200. [DOI] [PubMed] [Google Scholar]
- 47.Matera I, et al. Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts. J Struct Biol. 2010;170(3):548–564. doi: 10.1016/j.jsb.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 48.Vetting MW, Ohlendorf DH. The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure. 2000;8(4):429–440. doi: 10.1016/s0969-2126(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 49.Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: Two roads to the same destination. Mol Microbiol. 2007;64(2):260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- 50.Paper JM, Scott-Craig JS, Adhikari ND, Cuomo CA, Walton JD. Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics. 2007;7(17):3171–3183. doi: 10.1002/pmic.200700184. [DOI] [PubMed] [Google Scholar]
- 51.Yang F, et al. Secretomics identifies Fusarium graminearum proteins involved in the interaction with barley and wheat. Mol Plant Pathol. 2012;13(5):445–453. doi: 10.1111/j.1364-3703.2011.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganfornina MD, Kayser H, Sanchez D. In: Lipocalins. Åkerström B, Borregaard N, Flower DR, Salier JP, editors. Austin, TX: Landes Bioscience; 2006. pp. 49–74. [Google Scholar]
- 53.Breustedt DA, Schönfeld DL, Skerra A. Comparative ligand-binding analysis of ten human lipocalins. Biochim Biophys Acta. 2006;1764(2):161–173. doi: 10.1016/j.bbapap.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Ribeiro JMC, et al. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. 2006;36(2):111–129. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Flower DR, North ACT, Sansom CE. The lipocalin protein family: Structural and sequence overview. Biochim Biophys Acta. 2000;1482(1–2):9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez D, et al. In: Lipocalins. Åkerström B, Borregaard N, Flower DR, Salier JP, editors. Austin, TX: Landes Bioscience; 2006. pp. 5–16. [Google Scholar]
- 57.Ganfornina MD, Gutiérrez G, Bastiani M, Sánchez D. A phylogenetic analysis of the lipocalin protein family. Mol Biol Evol. 2000;17(1):114–126. doi: 10.1093/oxfordjournals.molbev.a026224. [DOI] [PubMed] [Google Scholar]
- 58.Wade NM, Tollenaere A, Hall MR, Degnan BM. Evolution of a novel carotenoid-binding protein responsible for crustacean shell color. Mol Biol Evol. 2009;26(8):1851–1864. doi: 10.1093/molbev/msp092. [DOI] [PubMed] [Google Scholar]
- 59.Shayeghi M, et al. Identification of an intestinal heme transporter. Cell. 2005;122(5):789–801. doi: 10.1016/j.cell.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 60.Misra JR, Horner MA, Lam G, Thummel CS. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011;25(17):1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pascussi J-M, et al. The tangle of nuclear receptors that controls xenobiotic metabolism and transport: Crosstalk and consequences. Annu Rev Pharmacol Toxicol. 2008;48:1–32. doi: 10.1146/annurev.pharmtox.47.120505.105349. [DOI] [PubMed] [Google Scholar]
- 62.Yano S, Takabayashi J, Takafuji A. Trade-offs in performance on different plants may not restrict the host plant range of the phytophagous mite, Tetranychus urticae. Exp Appl Acarol. 2001;25(5):371–381. doi: 10.1023/a:1017926017081. [DOI] [PubMed] [Google Scholar]
- 63.Gotoh T, Bruin J, Sabelis MW, Menken SBJ. Host race formation in Tetranychus urticae - Genetic differentiation, host-plant preference, and mate choice in a tomato and a cucumber strain. Entomol Exp Appl. 1993;68:171–178. [Google Scholar]
- 64.Kant MR, Sabelis MW, Haring MA, Schuurink RC. Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proc R Soc B Biol Sci. 2008;275(1633):443–452. doi: 10.1098/rspb.2007.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berenbaum MR. Postgenomic chemical ecology: From genetic code to ecological interactions. J Chem Ecol. 2002;28(5):873–896. doi: 10.1023/a:1015260931034. [DOI] [PubMed] [Google Scholar]
- 66.King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4(1):37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Dunlop JA. Geological history and phylogeny of Chelicerata. Arthropod Struct Dev. 2010;39(2–3):124–142. doi: 10.1016/j.asd.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Roopesh K, et al. Biotransformation of procyanidins by a purified fungal dioxygenase: Identification and characterization of the products using mass spectrometry. Process Biochem. 2010;45:904–913. [Google Scholar]
- 69.Roopesh K, et al. Dioxygenase from Aspergillus fumigatus MC8: Molecular modelling and in silico studies on enzyme-substrate interactions. Mol Simul. 2012;38:144–151. [Google Scholar]
- 70.Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: Gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16(18):1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- 71.Richards TA, et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci USA. 2011;108(37):15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chudzinski-Tavassi AM, et al. A lipocalin sequence signature modulates cell survival. FEBS Lett. 2010;584(13):2896–2900. doi: 10.1016/j.febslet.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Mullin C, Croft B. Host-related alterations of detoxification enzymes in Tetranychus urticae (Acari: Tetranychidae) Environ Entomol. 1983;12:1278–1282. [Google Scholar]
- 74.Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr Biol. 2006;16(7):674–679. doi: 10.1016/j.cub.2006.01.057. [DOI] [PubMed] [Google Scholar]
- 75.Charron J-BF, Ouellet F, Houde M, Sarhan F. The plant Apolipoprotein D ortholog protects Arabidopsis against oxidative stress. BMC Plant Biol. 2008;8:86. doi: 10.1186/1471-2229-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ganfornina MD, et al. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell. 2008;7(4):506–515. doi: 10.1111/j.1474-9726.2008.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bi JL, Felton GW. Foliar oxidative stress and insect herbivory - Primary compounds, secondary metabolites, and reactive oxygen species as components of induced resistance. J Chem Ecol. 1995;21:1511–1530. doi: 10.1007/BF02035149. [DOI] [PubMed] [Google Scholar]
- 78.Hildebrand DF, Rodriguez JG, Brown GC, Luu KT, Volden CS. Peroxidative responses of leaves in 2 soybean genotypes injured by 2-spotted spider-mites (Acari, Tetranychidae) J Econ Entomol. 1986;79:1459–1465. [Google Scholar]
- 79.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279(11):2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 82.Kretschmer M, et al. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 2009;5(12):e1000696. doi: 10.1371/journal.ppat.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saidijam M, et al. Microbial drug efflux proteins of the major facilitator superfamily. Curr Drug Targets. 2006;7(7):793–811. doi: 10.2174/138945006777709575. [DOI] [PubMed] [Google Scholar]
- 84.Krieger M, et al. Reconstituted low density lipoprotein: A vehicle for the delivery of hydrophobic fluorescent probes to cells. J Supramol Struct. 1979;10(4):467–478. doi: 10.1002/jss.400100409. [DOI] [PubMed] [Google Scholar]
- 85.Herz J, Strickland DK. LRP: A multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108(6):779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.May P, Woldt E, Matz RL, Boucher P. The LDL receptor-related protein (LRP) family: An old family of proteins with new physiological functions. Ann Med. 2007;39(3):219–228. doi: 10.1080/07853890701214881. [DOI] [PubMed] [Google Scholar]
- 87.Daborn PJ, et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297(5590):2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- 88.David JP, et al. The Anopheles gambiae detoxification chip: A highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc Natl Acad Sci USA. 2005;102(11):4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Govind G, et al. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS ONE. 2010;5(1):e8735. doi: 10.1371/journal.pone.0008735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ibrahim MMS. Effect of the host plant on susceptibility of the two-spotted spider mite, Tetranychus urticae Koch, (Acari: Tetranychidae) to some acaricides. J Agric Sci Mansoura Univ. 2009;34:10735–10744. [Google Scholar]
- 91.Neiswander CR, Rodriguez JG, Neiswander RB. Natural and induced variations in two-spotted spider mite populations. J Econ Entomol. 1950;43:633–636. [Google Scholar]
- 92.Agrawal AA, Vala F, Sabelis MW. Induction of preference and performance after acclimation to novel hosts in a phytophagous spider mite: Adaptive plasticity? Am Nat. 2002;159(5):553–565. doi: 10.1086/339463. [DOI] [PubMed] [Google Scholar]
- 93.Snyder MJ, Glendinning JI. Causal connection between detoxification enzyme activity and consumption of a toxic plant compound. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1996;179(2):255–261. doi: 10.1007/BF00222792. [DOI] [PubMed] [Google Scholar]
- 94.Feyereisen R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim Biophys Acta. 2011;1814(1):19–28. doi: 10.1016/j.bbapap.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 95.Roush RT, McKenzie JA. Ecological genetics of insecticide and acaricide resistance. Annu Rev Entomol. 1987;32:361–380. doi: 10.1146/annurev.en.32.010187.002045. [DOI] [PubMed] [Google Scholar]
- 96.McKenzie JA, Batterham P. The genetic, molecular and phenotypic consequences of selection for insecticide resistance. Trends Ecol Evol. 1994;9(5):166–169. doi: 10.1016/0169-5347(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 97.ffrench-Constant RH, Daborn PJ, Le Goff G. The genetics and genomics of insecticide resistance. Trends Genet. 2004;20(3):163–170. doi: 10.1016/j.tig.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 98.Dermauw W, et al. The cys-loop ligand-gated ion channel gene family of Tetranychus urticae: Implications for acaricide toxicology and a novel mutation associated with abamectin resistance. Insect Biochem Mol Biol. 2012;42(7):455–465. doi: 10.1016/j.ibmb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 99.Futuyma DJ, Peterson SC. Genetic variation in the use of resources by insects. Annu Rev Entomol. 1985;30:217–238. [Google Scholar]
- 100.Via S. Ecological genetics and host adaptation in herbivorous insects: The experimental study of evolution in natural and agricultural systems. Annu Rev Entomol. 1990;35:421–446. doi: 10.1146/annurev.en.35.010190.002225. [DOI] [PubMed] [Google Scholar]
- 101.Gould F. Rapid host range evolution in a population of the phytophagous mite Tetranychus urticae Koch. Evolution. 1979;33:791–802. doi: 10.1111/j.1558-5646.1979.tb04735.x. [DOI] [PubMed] [Google Scholar]
- 102.Ayroles JF, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41(3):299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Celorio-Mancera Mdl P, Heckel DG, Vogel H. Transcriptional analysis of physiological pathways in a generalist herbivore: Responses to different host plants and plant structures by the cotton bollworm, Helicoverpa armigera. Entomol Exp Appl. 2012;144:123–133. [Google Scholar]
- 104.Van Pottelberge S, Van Leeuwen T, Nauen R, Tirry L. Resistance mechanisms to mitochondrial electron transport inhibitors in a field-collected strain of Tetranychus urticae Koch (Acari: Tetranychidae) Bull Entomol Res. 2009;99(1):23–31. doi: 10.1017/S0007485308006081. [DOI] [PubMed] [Google Scholar]
- 105.Abbott WS. A method of computing the effectiveness of an insecticide. J Econ Entomol. 1925;18:265–267. [Google Scholar]
- 106.Van Dongen S. 2000. Graph clustering by flow simulation. PhD thesis (Univ of Utrecht, Utrecht, The Netherlands)
- 107.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 108.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 109.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Larkin MA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 111.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 113.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 116.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue):W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]