Abstract
Intercepting a moving object requires prediction of its future location. This complex task has been solved by dragonflies, who intercept their prey in midair with a 95% success rate. In this study, we show that a group of 16 neurons, called target-selective descending neurons (TSDNs), code a population vector that reflects the direction of the target with high accuracy and reliability across 360°. The TSDN spatial (receptive field) and temporal (latency) properties matched the area of the retina where the prey is focused and the reaction time, respectively, during predatory flights. The directional tuning curves and morphological traits (3D tracings) for each TSDN type were consistent among animals, but spike rates were not. Our results emphasize that a successful neural circuit for target tracking and interception can be achieved with few neurons and that in dragonflies this information is relayed from the brain to the wing motor centers in population vector form.
Keywords: vision, invertebrate, predatory behavior, electrophysiology, confocal microscopy
Dragonflies continuously track their prey (1) during short predatory flights (∼200–500 ms) (1–3) by keeping the image of the moving prey on the specialized dorsal area of their eyes (1). If the prey image drifts on the retina, compensatory motor signals sent to the wings adjust the dragonfly body position to bring the prey image back. This process allows a dragonfly to visually track the prey by locking on to it, a process also named fixation (4).
The ability to fixate on a moving target is a common feature among most predatory animals. Once a pursuer’s eyes are fixated on the prey, it can aim toward the current or the future prey location. The first choice results in classical pursuit, whereas the second one yields interception. Dragonflies are thought to intercept their prey by keeping a constant bearing to their target. This strategy is also used by other species, e.g., a human catching a ball (5, 6), but the nervous system of dragonflies presents a favorable substrate for studying the neural basis of this behavior. This ancient prey-targeting system is tractable and tuned for extreme performance, as evidenced by the accuracy (around 95%) (1, 2) and speed at which it functions.
We aimed to understand the information sent to the wings when a target moves across the dragonfly visual field. In particular, we have tested whether the population vector algorithm can successfully decode the directional component of the descending information. The population vector is the weighted sum activity of an ensemble of neurons with directional tuning. It was first shown to predict the direction of an upcoming arm movement in monkeys (7, 8). Since then, the population vector algorithm has described successfully the directional responses to mechanical stimulation in several invertebrate species (9–12). The dragonfly predatory flight provides a challenge for a reliable vector code. Not only are these animals highly maneuverable, with independent control of the fore and hind wings (13) and the ability to hover and even fly backward (14), but prey interception also demands a highly accurate representation of the prey’s direction vector (15). Nonetheless, computer modeling by Vaidyanathan et al. (15) suggested that this algorithm could drive an airborne target interception system successfully.
In the dragonfly brain, a group of neurons called small target movement detectors (STMDs) respond when a small target moves in a broad range of directions (16). Although there is no evidence showing that STMDs code a population vector, they are believed to be upstream of the target-selective descending neurons (TSDNs) (16, 17). TSDNs are thought to command the dragonfly trajectory during fast aerial attacks (5) for the following reasons: (i) each TSDN type, identifiable by its profile in the brain, behaves as a distinct feature detector, with a particular preference for target location, direction, and size (18, 19); (ii) TSDNs receive the information about the position and direction of a small target and relay it to all three (prothoracic, mesothoracic, and metathoracic) ganglia (18, 19); (iii) the axon diameters of the TSDNs are among the largest found in the ventral nerve cord (VNC) (18), affording a high conduction speed necessary for the behavior; and (iv) electrical stimulation of individual TSDNs alters the position/angle of the wings (20). Therefore, it is important to examine whether TSDNs use a population vector to accurately code prey direction and control wing movement.
In the CNS, there are only eight pairs of TSDNs with receptive fields facing dorsofrontally. We hypothesized that this small number of TSDNs is sufficient to transmit a finely tuned population vector that codes the direction of prey movement. To test this, we carried out intracellular recordings and confocal imaging of the TSDNs in the dragonfly Libellula luctuosa. Our results show that the population vector of the TSDNs codes with extreme accuracy the prey direction in all of the 360° tested.
Results
Electrophysiology.
We recorded from a total of 51 TSDNs from 38 different animals as follows: MDT1, 7 recordings; MDT2, 4 recordings; MDT3, 13 recordings; MDT4, 6 recordings; MDT5, 2 recordings; DIT1, 7 recordings; DIT2, 5 recordings; DIT3, 7 recordings. TSDNs respond only to small moving targets within the dorsofrontal visual field (Fig. 1 A–C) and not at all to wide-field movement such as grating patterns (21). The identity of each recorded TSDN was ascertained from its target-direction preference and receptive field area. To sample TSDN responses in an unbiased manner and with high precision, we projected upon a white screen a sequence of 3,497 trajectories, each composed of a moving dot that appeared in a random location and moved in a random direction, but always with a straight trajectory (Fig. S1A). To allow long recordings and prevent TSDN inhibition, the target size was small, subtending 1.48° (diameter) on the dragonfly’s eye, its speed fast (134°/s) and the trajectory time short (100 ms).
Fig. 1.
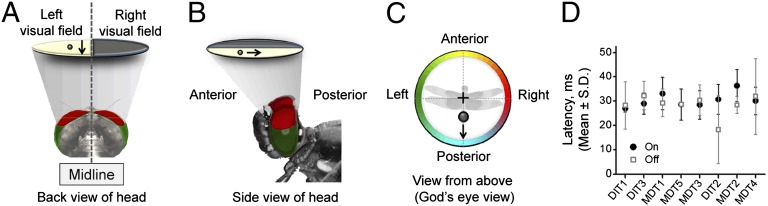
Small moving targets rapidly activate TSDNs. View of a L. luctuosa head from behind (A) and side (B) where the gray cone symbolizes the visual field of the dorsal area. The eyes are false colored in red (dorsal eye) and green (rest of the eye). A target (flying insect) moving above the dragonfly from front to back crossing the left visual field (yellow area) in the direction indicated by the arrow, excites the TSDN MDT1. (C) Polar plot with the “God’s eye view” reference used to display target directions, i.e., the target movement shown in A and B is here shown as a vector pointing down to the blue zone. (D) Summary of the latencies. The latencies obtained for each TSDN type were not significantly different from each other (one-way ANOVA).
The latency between stimulus onset (first target movement) and the earliest TSDN response (Fig. S1B) was not significantly different among TSDN types (Fig. 1D). Thus, the timing of information arrival was similar among all TSDNs at the temperature tested (23–25 °C). This was also the case for the off latency (timing of last response – timing target movement cessation). After pooling, the TSDN onset latency (29.94 ± 5.75 ms, mean ± 1SD) and offset latency (28.82 ± 9.11 ms) were not significantly different (paired Student t test, P = 0.848, n = 51) (Fig. 1D). These results indicate that, within the 100 ms time window of each target trajectory, the latency between stimulus onset and cell response did not vary significantly.
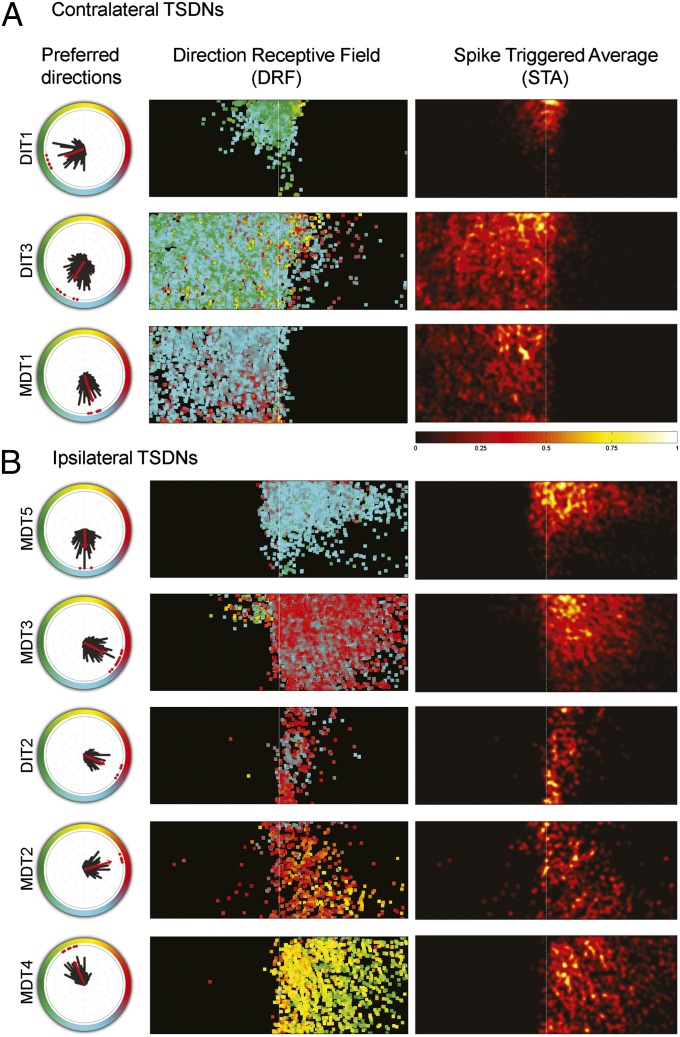
The responses from 51 cells were grouped into eight TSDN types, whose electrophysiological properties (Fig. 2) and brain traces (Fig. S2) show homology to those described in the dragonfly Anax junius (18, 19). For example, DIT1, DIT3, and MDT1 all have receptive fields contralateral to their descending axons, whereas the rest (MDT5, MDT3, DIT2, MDT2, and MDT4) have receptive fields ipsilateral to their descending axons (Fig. 2 and Fig. S2).
Fig. 2.
Each TSDN type shows a unique direction tuning curve and receptive field consistent across animals. (A) Contralateral and (B) ipsilateral TSDN receptive fields whose axons were in the right connective of the ventral nerve cord (VNC). The polar plots show the directional preference of each recorded TSDN (red dots) and their mean direction tuning distribution (black bars). The red arrow indicates mean preferred direction. The color-coded direction receptive field (DRF) maps show the mean direction preference at each pixel, which was calculated by averaging the direction peak, at each pixel, for all recordings of each TSDN type. In addition, spike-triggered average (STA) maps are shown for each TSDN type. Note that, because number of spikes was normalized before and after computing the average, the same scale applies to all STA maps.
Although all TSDNs were excited by a wide range of target directions and the total number of spikes for a TSDN type varied among animals (Fig. S3), the peak direction preferences were TSDN-type specific and consistent (Fig. 2 A and B, polar plots). The tuning width of the directional tuning curves varied between 32 and 60° (Fig. S4A). The widest direction-tuning curve is that of DIT3, which may result from its sensitivity to an expanding stimulus (Fig. S4B). A similar range of tuning widths has been reported in the cells coding for direction in the monkey motor cortex (22). The basis for such cell-to-cell tuning-width differences remains to be clarified in both cases.
In addition to the consistency in the location and size of the receptive field, the zones of increased sensitivity (where a higher number of spikes were recorded in response to a moving target) also remained similar among replicas of each TSDN type (Fig. S5). To assess whether the preferred target direction is a function of a specific area within the receptive field, for each screen pixel the mean target direction was calculated from all of the targets that triggered a spike. For most cells, the preferred direction was homogeneous across the receptive field. However, DIT3 and MDT3 also displayed responses that were not directionally sensitive [Fig. 2 A and B, direction receptive field (DRF) maps, area beyond the midline with highly variable coloration]. In the case of MDT3, such responses were tightly localized, and thus correlated with the location of the target stimulus. Hence, these responses are not likely to reflect intrinsic cellular noise. Instead, it is probable that these two cells receive additional, nondirectional, inputs from both eyes. This is important because only DIT3 responded significantly to an expanding target, a stimulus type that included moving edges in all directions (Fig. S4B). Moreover, preliminary data from the Libellulid dragonfly Libellula pulchella indicates that, although MDT3 does not respond to a linearly expanding stimulus, it does so when an accelerated expansion, quasi-looming stimulus is presented in the middle of the visual field (MDT3 responses per accelerated expansion stimulus, 4 spikes; n = 3). We note here that receptive field laterality was inadvertently switched for two TSDNs, MDT1 and MDT3, in the previous description of TSDN receptive fields (19). The corrected receptive field maps for Aeshnids agree with those reported here for Libellulids.
In all TSDNs, the “hot spots” of increased sensitivity are located near the visual midline. Although the size of our screen did not cover the receptive fields in the frontal-caudal axis entirely, the hot spots seemed to be placed dorsofrontal. This trend becomes more apparent when all of the spike-triggered average (STA) maps for each TSDN type are added and mirrored. This operation yields a sensitivity map for the space sampled by both eyes (Fig. 3A; see Fig. S6 for dragonfly head angles during the recordings). Our results show that dragonflies boast a matched detection system, with increased sensitivity to moving targets in the area of the retina where the prey image is focused during aerial hunts (1). In addition, plotting the directional tuning curve for each TSDN demonstrates that the directions pointing toward the back of the animal are sampled more intensely (Fig. 3B).
Fig. 3.
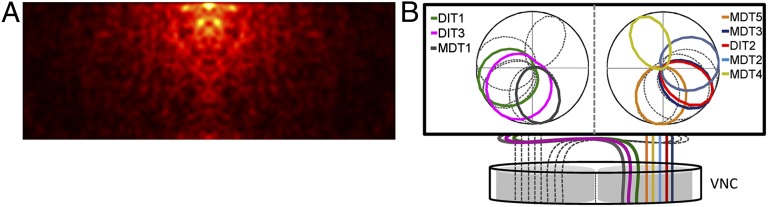
TSDN sensitivity is matched to the predatory behavior. (A) The visual receptive fields of the TSDNs combine to create an area of increased sensitivity to target movement near midline. For more details on eye coordinates, see Fig. S6. (B) DIT1, DIT3, and MDT1 transmit motion information in the visual field contralateral to their axons in the VNC. MDT5, MDT3, DIT2, MDT2, and MDT4 transmit motion information in the visual field ipsilateral to their axons in the VNC. Together, all eight axons travel through the ventral connective and thoracic ganglia.
Morphology.
The overlapping direction tuning curves and receptive fields of the different TSDNs suggest that the population vector from all TSDN responses could code a change in the prey’s bearing. However, to calculate a plausible population vector for each wing, knowledge of the TSDN arborization in wing motor centers is necessary. Moreover, to make morphological comparisons among the different TSDNs, the level of anatomical consistency of each TSDN type across different animals within the same species must be assessed first.
Earlier reports were studies of the TSDNs of the large “hawking” dragonflies of the family Aeshnidae, but the species used in this study belongs to a family of smaller “perching” dragonflies, the Libellulidae. We have confirmed, by electrophysiology (Fig. 2) and brain morphology (Fig. S2), homology between TSDNs from Aeshnids and Libellulids, but with one striking difference. In the prothoracic ganglia of L. luctuosa, MDT2, MDT3, and MDT4 travel through the dorsal intermediate tract (DIT) (Fig. S7).
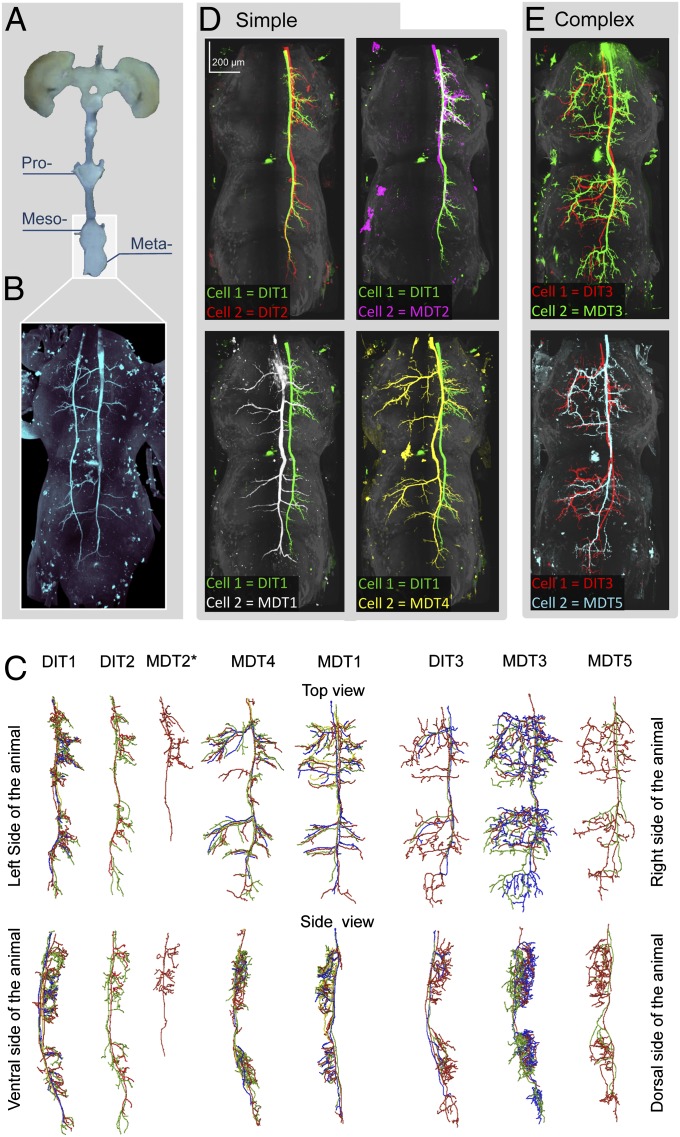
In all but one preparation, only one TSDN was injected in each animal. Three-dimensionally imaged volumes of the complex formed by the mesothoracic and metathoracic ganglia and the first abdominal ganglion were registered and warped for comparison (Fig. 4 A and B). The traces for each TSDN of the same type are remarkably consistent across all injected samples (Fig. 4C). All of the TSDNs targeted the dorsal part of mesothoracic and the metathoracic ganglia (Fig. 4C) dedicated to motor/efferent fibers in insects. Thus, if the TSDN branching pattern does not filter the electrical activity in a ganglion-specific manner, the information provided by each TSDN should be available nearly simultaneously to both the fore and hind wings of the targeted side. Once the TSDN morphology was confirmed across several animals, we chose the cell with the most extensive fill for comparisons among TSDN types. According to the complexity of their branching, we divided TSDNs into two categories: simple or complex.
Fig. 4.
In the wing motor centers, all TSDNs share morphological features. (A) The mesothoracic, metathoracic, and first abdominal ganglia were imaged and warped to allow morphological comparison. (B) Unilateral DIT2 on the left and bilateral MDT1 on the right, injected in the same animal, target the same locations. (C) Traces within each TSDN type (grouped according to the electrophysiological results) are consistent, so the most complete fill from each TSDN type was used for comparison. TSDNs were categorized into “simple” or “complex” cells. A pairwise comparison (dorsal views) shows that unilateral simple cells, DIT1 (green), DIT2 (red), and MDT2 (magenta), are indistinguishable from each other (D, Upper), but the bilateral simple cells MDT1 (white) and MDT4 (yellow) display specific branching patterns (D, Lower). However, all simple TSDNs target the same location. In contrast, pairwise comparisons between the complex cells, DIT3 (red), MDT3 (green), and MDT5 (cyan) (E, all panels) are less informative because their additional intricate branching exhibits higher variability, particularly in the medial region of the ganglia (traces in C).
Unilateral simple TSDNs, DIT1, DIT2, and MDT2, consistently target four ipsilateral locations in the lateral region of the mesothoracic and the metathoracic ganglia (Fig. 4C). DIT1 morphology is closely matched by DIT2 and MDT2 (Fig. 4D), although our best MDT2 fill is incomplete. MDT1 and MDT4 show bilateral asymmetry in their outputs (Fig. 4C), but a pairwise comparison with unilateral DIT1 reveals that the branches of MDT1 and MDT4 target the four stereotypical lateral locations in both sides of the ganglia (Fig. 4D). Indeed, when we injected the DIT2 cell on the left connective and the MDT1 on the right connective of the same animal, the contralateral branches of MDT1 extended to the same location as those of the unilateral DIT2 (Fig. 4B).
We have categorized DIT3, MDT5, and MDT3 as complex cells due to the additional number of short and highly variable branches found in the medial region of the ganglia (Fig. 4C). However, within the side of the connective in which they descend from the brain, the morphology of these complex cells also targets the four lateral locations mentioned previously (Fig. 4E).
TSDN Population Vector Coding.
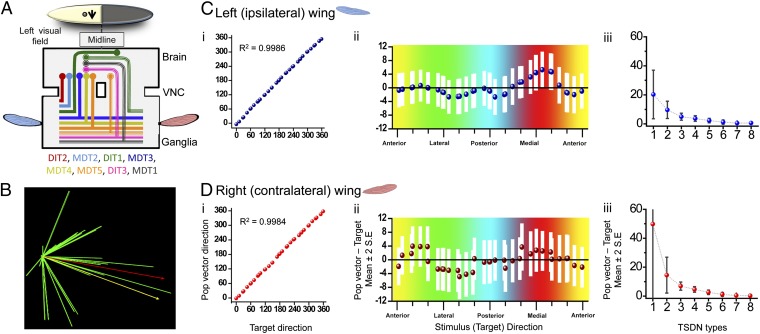
Because all of the TSDNs terminals arborize in similar areas, it is plausible that their outputs code a population vector that is converted into wing motor movements. However, a plausible population vector calculation must take into account the receptive fields and unilateral/bilateral TSDN branching morphology. Simmons (23) reported that the major motor neurons involved in flight control have extensive branching in the ipsilateral side, but rarely cross the midline. Therefore, we chose a parsimonious approach in which the branching side of the TSDNs indicated the input to the corresponding wing. To understand which TSDNs would form part of a population vector, we focused on the TSDN activity elicited by targets moving on the left side of the dragonfly’s visual field. We looked at the TSDNs with left receptive fields (Fig. 2 plus the corresponding mirror plots that would account for the TSDNs present in the opposite connective) and noted whether their outputs feed the ipsilateral (left) and/or contralateral (right) wing (Fig. 4 plus the corresponding mirror traces that would account for the TSDNs present in the opposite connective). Hence, we obtained the population of TSDNs contributing to the vectors for the wings ipsilateral (left) and contralateral (right) to the target movement (Fig. 5A). We then binned the target directions (and corresponding cell activity) every 18° and calculated the population vector (Fig. 5B) (Materials and Methods and SI Text). The results show that both population vectors, for the left and the right wing, are strongly correlated with target movement in all 360 directions tested (Fig. 5 C, i and D, i). Moreover, the accuracy is such that the population vectors and target movements are not significantly different in any of the tested directions (Fig. 5 C, ii and D, ii). Thus, the impressive ability of the system to code the target motion direction accurately is unaffected by the fact that the TSDN tuning curves do not sample all possible directions equally.
Fig. 5.
The TSDN population vector codes direction of the prey with high accuracy. (A) A target moving in the left side of the visual field activates the subset of TSDNs shown in the diagram. The population of TSDNs providing inputs to the left (nine cells) and right (seven cells) wings differ. (B) Graphical representation of a dragonfly TSDN population vector. Contributing TSDN vectors (green), stimulus direction (yellow), and population vector (red) are shown. (C and D) Population vectors for the left (C) and right (D) wings were calculated for all presented trajectories. (C, i and D, i) The population vectors, binned in 18°, are strongly correlated with the target direction. Left wing circular R = 0.9986 (P < 0.001) and right wing R = 0.9984 (P < 0.001). (C, ii and D, ii) The direction of the target and the direction of the population vector are not significantly different in any direction because the difference between these two parameters is close to zero (dotted values) and because zero is within the 95% confidence interval (white bars). Colors refer to the direction of the presented target. The data concern targets moving in the left side of the visual field, so targets that traveled toward the medial part of the animal correspond to the red section. (C, iii and D, iii) On average, removing two types of TSDN from the population (six cell types used) does not impact the accuracy of the population vector significantly. For the left wing, three TSDN types are, on average, sufficient to provide a population vector whose bias is within 10° of the presented target direction.
In the absence of a moving target, the basal activity of TSDNs is zero. Therefore, a target moving in the antipreferred direction does not reduce the spiking rate. For such a system to code target direction equally well in all 360°, a minimum of four cells with cosine directional tuning are needed (24). We investigated whether all eight TSDN types are necessary for an accurate code of the target direction. We removed various numbers of TSDN types, from one (i.e., seven TSDN types left) to seven (i.e., one TSDN type left), from the population vector calculation and assessed the resulting performance. We found that, on average, if the population vector is calculated with six or more TSDN types, the vector is not significantly different from the target direction (t test, mean difference left wing, 1.23 ± 3.72° SD, P = 0.089; right wing, 1.19469 ± 3.49° SD, P = 0.07624; Fig. 5C, iii and D, iii), and thus its accuracy is not affected. The two exceptions arise when DIT2+MDT3 are removed from the ipsilateral wing calculation and MDT3+MDT4 are removed from the contralateral wing calculation; these changes result in large coding errors (population vector direction – target direction = 20.06 and 19.28°, respectively). When only two TSDN types were used in the calculations, the constant bias in the population vector outcome, also called constant error, was not far from the target direction (mean population vector direction − target direction, left wing, 9.67°, and right wing, 14.50°). However, the variable error, which can be regarded as a plus-or-minus confidence interval on the constant error, was large (mean error, left wing, 29.52°, and right wing, 40.12°), making the code unreliable. On average, at least three types of TSDNs are needed to keep the coding error with a constant bias below the 10° of the presented target direction. To describe the accuracy of this system in more detail, the activity of all TSDNs will have to be recorded with extracellular probes in the same animal. This is because although the preferred direction for a type of TSDN is highly constant between animals, the magnitude (the spike rate) of the cell response is not (Fig. S3). The properties acquired in this study would allow appropriate TSDN type identification on extracellular traces. This information is necessary to investigate simultaneous responses from several TSDNs within one animal.
Discussion
We have shown that TSDN responses are suited to act as matched filters for the retinal slip of the prey during the predatory behavior. TSDNs are most sensitive to targets moving in the frontal-dorsal region of the eye, where dragonflies fixate the prey image during interception flights (1). It would be informative to study if the other insects known to visually track and fixate on targets, such as mantis (25) and hover flies (26), also have a group of neurons most sensitive to target motion away from the retinal fovea. Moreover, although a descending looming neuron has been recorded extracellularly in a mantis species (27), TSDNs in this taxa are yet to be reported.
Our results show that the TSDN properties make these cells excellent information channels for a prey-direction population vector that steers wing movements during predatory flights. These properties include the following: (i) overlapping receptive fields, (ii) wide and overlapping direction tuning curves, (iii) latencies of similar duration, and (iv) shared branching organization and location at the wing motor centers.
We have confirmed homology between the TSDNs in representatives of two dragonfly families with markedly different foraging behavior. The subject of this study was L. luctuosa (family Libellulidae, most of whose members forage from a perch). TSDNs were originally characterized in dragonflies of family Aeshnidae, which detect and pursue prey during continuous flight. We showed that, in L. luctuosa, three of the TSDNs travel in a different tract of the thoracic ganglia. We have also found that MDT3 travels through the DIT in another libellulid species (Libellula lydia; Fig. S8). Given that such tracts are laid down early in development and are highly conserved among insect groups (28), it is possible that this variation in pathway reflects a change in the neural “wiring” of the wing control circuitry. For example, a homologous neuron involved in swimming pattern generation has been found to carry out different functions in two closely related sea slugs (nudibranchs) that exhibit similar swimming modalities (29). To address such evolutionary questions about the dichotomy between the TSDNs in hawker and percher dragonflies, the mesothoracic and metathoracic branching patterns of the Aeshnid TSDNs also need to be investigated.
It is interesting that all TSDNs target the mesothoracic and metathoracic ganglia with four identifiable locations in the most lateral region of each ganglion. These results raise two questions. First, as the dragonfly fore and hind wings are controlled independently (30), what additional information alters the TSDN motor inputs to yield a specific movement for the rear and the fore wings? Further studies on the central pattern generators underlying the dragonfly flight motor are necessary to answer this question. Second, because the projection from leg sensory neurons into the thoracic ganglia of locusts follows a 3D somatotopic map (31), do the four locations of TSDN branching reveal a pattern of signal integration with other localized sensory inputs? Each dragonfly wing has four sensory structures called crevice organs. Crevice organs resemble the campaniform sensilla of insects and are believed to act as mechanoreceptors for the degree of wing twist (32). In the locust, campaniform sensilla measure lift and twisting amplitude of the hind wings (33). Because TSDN activation alters the attack angle of the wing (20), it seems likely that information from the TSDNs and input from the crevice organs is combined in some manner, but a follow-up investigation on the information flow from the TSDNs to the wing motor neurons is needed.
Previous reports have shown that the underlying principles used by dragonflies to calculate motion (17) and orientation (16) are similar to those found in vertebrate systems. Here, we have shown that population vector code could successfully direct target interception in these ancient animals. The population vector algorithm has been coined the “geometrical universal actuator” because sensory information coded in vector form can be used directly for appropriate motor actuation, without a need for a time-consuming transformation (12). Our data further support this view. Even though the visual information flows through the brain, the TSDN population vector carries the information about prey direction, and the appropriate compensatory forces are implemented in the wing motor centers. Thus, the population vector could allow this steering system to act as an “autopilot” (see ref. 34 for a discussion on the feasibility of automated steering in insects), providing the fast reaction speed displayed by dragonflies during predatory behavior [∼30 ms from stimulus onset to compensatory wing strokes (1)].
The dragonfly system shows a significant level of robustness; even when only six TSDN types are used for the calculation, the population vector is still not significantly different from the target direction. Unlike the population vector coding in the leech (9), TSDNs do not sample the visual space and target directions homogenously. Bias in direction sampling has also been reported in the cells of the monkey motor cortex, which code population vectors for arm movements (35). Moreover, in insects, the visual performance of motion detection is matched to the species-specific flight movements (36), a topic recently reviewed by Taylor and Krapp (37). Regardless of such bias, for the 2D plane tested in this study, TSDN population vector coding performs equally well across all directions. It is possible that the bias in the preferred direction of the different TSDNs becomes significant once the animal is presented with a more realistic target showing freedom of movement in all three dimensions (i.e., including contraction or expansion of the prey image).
Although we have focused on prey pursuit, dragonflies also pursue larger targets, such as conspecifics in male/male territorial chases. In such nonpredatory flight trajectories, collision may not be desirable, and an interception strategy may not be beneficial. It remains to be elucidated whether the TSDNs are used in an exclusively predatory context, or whether they play a part in flights with different strategies.
In summary, the fast and highly precise prey interception flight system of dragonflies is particularly amenable to investigation at many levels: electrophysiological, morphological, and behavioral. The ultimate goal is to elucidate the neural underpinnings from molecular basis to behavior, something already achieved for the swimming movement of the lamprey (38). The electrophysiological and morphological evidence presented here support the notion that TSDNs provide population vector information about target movement that can be used to direct wing movement during a predatory flight. This system relies on a low number of cells performing with high accuracy. The functional significance of this result is that, even in an insect that displays a behavior as complex as interception, the integration of signals and calculation of motor output seem to occur at the thoracic ganglia, and not in the brain. A process as seemingly complicated as interception appears automated to yield the high speed and performance necessary for insect predation. This study is of particular importance because dragonflies are the only insect group that exclusively sport direct wing muscles, allowing independent control of both sets of wings. With the view to developing bio-inspired robots, recent studies have investigated dragonfly vision (39) and wing dynamics (40). Thus, understanding the visually driven neural control of the dragonfly wings may prove invaluable for future technological applications. The 3D atlas presented here forms a basis for further understanding of this circuit.
Materials and Methods
The ventral nerve cord of adult L. luctuosa was impaled with a sharp electrode while the animal viewed a visual display (360 Hz). TSDNs were recognized initially by robust responses to a vertical or horizontal small moving target of fixed size, increased response for certain target directions, and lack of responses to a wide-field stimulus. A stimulus composed of 3,497 consecutive small moving targets was presented, and the resultant cell activity was used to calculate its latency, directional tuning (see refs. 41 and 42) and receptive field. Looming sensitivity was tested with a linearly expanding stimulus. Using negative current (−2 nA), we injected 32 of the TSDNs with Lucifer yellow. The preparations were processed and imaged with a confocal microscope (Zeiss 710; see ref. 43). To visualize the cell morphology, we used Vaa3D (www.vaa3d.org) (44, 45) and BrainAligner (46). The population vector was calculated with a bootstrap method, with tuning curves calculated by binning target directions every 18° and averaging cell activity for those bins. The population vector calculation was based on weight 12 in appendix 1 of ref. 47. See Fig. S9 and SI Materials and Methods for more information.
Supplementary Material
Acknowledgments
We thank Anthony Leonardo and the Howard Hughes Medical Institute for providing resources necessary for this study; Elliot Imler and Paul Herold for animal husbandry; Hang Xiao, Yinan Wan, and Yan Zhuang for helping with our analysis of neuron morphology; Trevor Wardill for suggestions for the manuscript; and Ian Stevenson, who provided help with application of the MATLAB analytical toolbox. We also thank the Air Force Office of Scientific Research for support (Grant FA9550-10-0472 to R.M.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 389.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210489109/-/DCSupplemental.
References
- 1.Olberg RM, Seaman RC, Coats MI, Henry AF. Eye movements and target fixation during dragonfly prey-interception flights. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2007;193(7):685–693. doi: 10.1007/s00359-007-0223-0. [DOI] [PubMed] [Google Scholar]
- 2.Combes SA, Rundle DE, Iwasaki JM, Crall JD. Linking biomechanics and ecology through predator-prey interactions: Flight performance of dragonflies and their prey. J Exp Biol. 2012;215(Pt 6):903–913. doi: 10.1242/jeb.059394. [DOI] [PubMed] [Google Scholar]
- 3.Olberg RM, Worthington AH, Venator KR. Prey pursuit and interception in dragonflies. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2000;186(2):155–162. doi: 10.1007/s003590050015. [DOI] [PubMed] [Google Scholar]
- 4.Land MF. Visual tracking and pursuit—Humans and arthropods compared. J Insect Physiol. 1992;38(12):939–951. [Google Scholar]
- 5.Olberg RM. Visual control of prey-capture flight in dragonflies. Curr Opin Neurobiol. 2012;22(2):267–271. doi: 10.1016/j.conb.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 6.McBeath MK, Shaffer DM, Kaiser MK. How baseball outfielders determine where to run to catch fly balls. Science. 1995;268(5210):569–573. doi: 10.1126/science.7725104. [DOI] [PubMed] [Google Scholar]
- 7.Georgopoulos AP, Schwartz AB, Kettner RE. Neuronal population coding of movement direction. Science. 1986;233(4771):1416–1419. doi: 10.1126/science.3749885. [DOI] [PubMed] [Google Scholar]
- 8.Georgopoulos AP, Caminiti R, Kalaska JF, Massey JT. Spatial coding of movement: A hypothesis concerning the coding of movement direction by motorcortical populations. Exp Brain Res. 1983;(Suppl 7):327–336. [Google Scholar]
- 9.Lewis JE, Kristan WB., Jr A neuronal network for computing population vectors in the leech. Nature. 1998;391(6662):76–79. doi: 10.1038/34172. [DOI] [PubMed] [Google Scholar]
- 10.Levi R, Camhi JM. Population vector coding by the giant interneurons of the cockroach. J Neurosci. 2000;20(10):3822–3829. doi: 10.1523/JNEUROSCI.20-10-03822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yono O, Shimozawa T. Synchronous firing by specific pairs of cercal giant interneurons in crickets encodes wind direction. Biosystems. 2008;93(3):218–225. doi: 10.1016/j.biosystems.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 12.van Hemmen JL, Schwartz AB. Population vector code: A geometric universal as actuator. Biol Cybern. 2008;98(6):509–518. doi: 10.1007/s00422-008-0215-3. [DOI] [PubMed] [Google Scholar]
- 13.Wakeling J, Ellington C. Dragonfly flight. II. Velocities, accelerations and kinematics of flapping flight. J Exp Biol. 1997;200(Pt 3):557–582. doi: 10.1242/jeb.200.3.557. [DOI] [PubMed] [Google Scholar]
- 14.Usherwood JR, Lehmann FO. Phasing of dragonfly wings can improve aerodynamic efficiency by removing swirl. J R Soc Interface. 2008;5(28):1303–1307. doi: 10.1098/rsif.2008.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaidyanathan R, et al. A reflexive control architecture based on a neural model of the cockroach escape response. Journal of Systems and Control Engineering. 2012;226(5):699–718. [Google Scholar]
- 16.O'Carroll D. Feature-detecting neurons in dragonflies. Nature. 1993;362(6420):541–543. [Google Scholar]
- 17.Nordström K. Neural specializations for small target detection in insects. Curr Opin Neurobiol. 2012;22(2):272–278. doi: 10.1016/j.conb.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Olberg RM. Identified target-selective visual interneurons descending from the dragonfly brain. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1986;159(6):827–840. [Google Scholar]
- 19.Frye MA, Olberg RM. Visual receptive-field properties of feature detecting neurons in the dragonfly. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1995;177(5):569–576. [Google Scholar]
- 20.Olberg RM. 1978. Visual and multimodal interneurons in dragonflies. PhD dissertation (Univ of Washington, Seattle, WA)
- 21.Olberg RM. Object- and self-movement detectors in the ventral nerve cord of the dragonfly. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1981;141(3):327–334. [Google Scholar]
- 22.Amirikian B, Georgopoulos AP. Directional tuning profiles of motor cortical cells. Neurosci Res. 2000;36(1):73–79. doi: 10.1016/s0168-0102(99)00112-1. and correction (2000) 37(1):83. [DOI] [PubMed] [Google Scholar]
- 23.Simmons P. The neuronal control of dragonfly flight. I. Anatomy. J Exp Biol. 1977;71(DEC):123–140. doi: 10.1242/jeb.71.1.123. [DOI] [PubMed] [Google Scholar]
- 24.van Hateren JH. Directional tuning curves, elementary movement detectors, and the estimation of the direction of visual movement. Vision Res. 1990;30(4):603–614. doi: 10.1016/0042-6989(90)90071-r. [DOI] [PubMed] [Google Scholar]
- 25.Rossel S. Foveal fixation and tracking in the praying mantis. J Comp Physiol. 1980;139(4):307–331. [Google Scholar]
- 26.Collett TS, Land MF. Visual control of flight behaviour in the hoverfly, Syritta pipiens L. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1975;99(1):1–66. [Google Scholar]
- 27.Yamawaki Y, Toh Y. Responses of descending neurons to looming stimuli in the praying mantis Tenodera aridifolia. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2009;195(3):253–264. doi: 10.1007/s00359-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 28.Truman JW, Ball EE. Patterns of embryonic neurogenesis in a primitive wingless insect, the silverfish, Ctenolepisma longicaudata: Comparison with those seen in flying insects. Dev Genes Evol. 1998;208(7):357–368. doi: 10.1007/s004270050192. [DOI] [PubMed] [Google Scholar]
- 29.Sakurai A, Newcomb JM, Lillvis JL, Katz PS. Different roles for homologous interneurons in species exhibiting similar rhythmic behaviors. Curr Biol. 2011;21(12):1036–1043. doi: 10.1016/j.cub.2011.04.040. [DOI] [PubMed] [Google Scholar]
- 30.Dileo C, Deng X. Design of and experiments on a dragonfly-inspired robot. Adv Robot. 2009;23(7–8):1003–1021. [Google Scholar]
- 31.Burrows M, Newland PL. Correlation between the receptive fields of locust interneurons, their dendritic morphology, and the central projections of mechanosensory neurons. J Comp Neurol. 1993;329(3):412–426. doi: 10.1002/cne.903290311. [DOI] [PubMed] [Google Scholar]
- 32.Simmons PJ. Crevice organs: Sensory structures on the wings of dragonflies (Insecta, Odonata) Zoomorphology. 1978;89(3):251–255. [Google Scholar]
- 33.Elson RC. Interneuronal processing of inputs from the campaniform sensilla of the locust hindwing. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1987;161(5):761–776. [Google Scholar]
- 34.Wolf R, Voss A, Hein S, Heisenberg M, Sullivan GD. Can a fly ride a bicycle? Philos Trans R Soc Lond B Biol Sci. 1992;337(1281):261–269. [Google Scholar]
- 35.Naselaris T, Merchant H, Amirikian B, Georgopoulos AP. Large-scale organization of preferred directions in the motor cortex. I. Motor cortical hyperacuity for forward reaching. J Neurophysiol. 2006;96(6):3231–3236. doi: 10.1152/jn.00487.2006. [DOI] [PubMed] [Google Scholar]
- 36.O’Carroll DC, Bidwell NJ, Laughlin SB, Warrant EJ. Insect motion detectors matched to visual ecology. Nature. 1996;382(6586):63–66. doi: 10.1038/382063a0. [DOI] [PubMed] [Google Scholar]
- 37.Taylor GK, Krapp HG. 2008. Sensory systems and flight stability: What do insects measure and why? Advances in Insect Physiology, eds Casas J, Simpson SJ (Academic Press, London), Vol 34, pp 231–316.
- 38.Grillner S. The motor infrastructure: From ion channels to neuronal networks. Nat Rev Neurosci. 2003;4(7):573–586. doi: 10.1038/nrn1137. [DOI] [PubMed] [Google Scholar]
- 39.Chahl J, et al. Bioinspired engineering of exploration systems: A horizon sensor/attitude reference system based on the dragonfly ocelli for Mars exploration applications. J Robot Syst. 2003;20(1):35–42. [Google Scholar]
- 40.Vargas A, Mittal R, Dong H. A computational study of the aerodynamic performance of a dragonfly wing section in gliding flight. Bioinspir Biomim. 2008;3(2):026004. doi: 10.1088/1748-3182/3/2/026004. [DOI] [PubMed] [Google Scholar]
- 41.Berens P. CircStat: A MATLAB toolbox for circular statistics. J Stat Softw. 2009;31(10):1–21. [Google Scholar]
- 42.Cronin B, Stevenson IH, Sur M, Körding KP. Hierarchical Bayesian modeling and Markov chain Monte Carlo sampling for tuning-curve analysis. J Neurophysiol. 2010;103(1):591–602. doi: 10.1152/jn.00379.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez-Bellido PT, Wardill TJ. 2012. Labeling and confocal imaging of neurons in thick invertebrate tissue samples. Cold Spring Harb Protoc 2012(9):969–983.
- 44.Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat Biotechnol. 2010;28(4):348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, Peng H. 2011. Automated high speed stitching of large 3D microscopic images. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium (IEEE, Chicago, IL), pp 238–241.
- 46.Peng H, et al. BrainAligner: 3D registration atlases of Drosophila brains. Nat Methods. 2011;8(6):493–500. doi: 10.1038/nmeth.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Georgopoulos AP, Kettner RE, Schwartz AB. Primate motor cortex and free arm movements to visual targets in three-dimensional space. II. Coding of the direction of movement by a neuronal population. J Neurosci. 1988;8(8):2928–2937. doi: 10.1523/JNEUROSCI.08-08-02928.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.