Abstract
The plasma glycoprotein von Willebrand factor (VWF) exhibits fivefold antigen level variation across the normal human population determined by both genetic and environmental factors. Low levels of VWF are associated with bleeding and elevated levels with increased risk for thrombosis, myocardial infarction, and stroke. To identify additional genetic determinants of VWF antigen levels and to minimize the impact of age and illness-related environmental factors, we performed genome-wide association analysis in two young and healthy cohorts (n = 1,152 and n = 2,310) and identified signals at ABO (P < 7.9E-139) and VWF (P < 5.5E-16), consistent with previous reports. Additionally, linkage analysis based on sibling structure within the cohorts, identified significant signals at chromosome 2q12–2p13 (LOD score 5.3) and at the ABO locus on chromosome 9q34 (LOD score 2.9) that explained 19.2% and 24.5% of the variance in VWF levels, respectively. Given its strong effect, the linkage region on chromosome 2 could harbor a potentially important determinant of bleeding and thrombosis risk. The absence of a chromosome 2 association signal in this or previous association studies suggests a causative gene harboring many genetic variants that are individually rare, but in aggregate common. These results raise the possibility that similar loci could explain a significant portion of the “missing heritability” for other complex genetic traits.
Keywords: genome-wide association study, linkage study, venous thromboembolic disease, von Willebrand disease, quantitative trait loci
Von Willebrand factor (VWF) is a multimeric plasma glycoprotein that plays a central role in hemostasis by acting as a molecular bridge tethering platelets to injured endothelium and as a carrier molecule for coagulation factor VIII (1). Quantitative or qualitative deficiencies in VWF lead to von Willebrand Disease (VWD), the most common inherited bleeding disorder, with an estimated prevalence of 0.002–0.01% worldwide (1, 2). Type I VWD is characterized by mild to moderate bleeding and low circulating VWF levels. This form of VWD is generally associated with haploinsufficiency for VWF and is characterized by incomplete penetrance. In contrast, elevated levels of plasma VWF are an independent risk factor for venous thromboembolic disease (3), myocardial infarction (4), stroke (5, 6), and also complicate anticoagulant management (7).
Plasma VWF levels vary by approximately fivefold in healthy populations and are influenced by both environmental and inherited factors. Increased levels of VWF occur with advancing age (8), may rise acutely because of inflammation or infection, and may serve as a surrogate marker for endothelial dysfunction and atherosclerosis (9–11). Estimates for the heritability of plasma VWF levels in the general population from previous family-based studies range from 32–75%. A 1985 study in Norwegian twins reported the heritability of VWF at 66%, with 30% of this effect attributable to ABO blood type (12). More recent studies estimated the heritability of VWF levels to be as high as 75% in United Kingdom twins (13) and as low as 53% in elderly Danish twins (14). In contrast, analysis in 21 Spanish families calculated VWF level heritability at only 32% (15), identifying with significant linkage only observed at the ABO locus (LOD = 3.46).
The connection between VWF antigen levels and ABO blood group has been well studied, with VWF levels reduced by ∼30% in type O individuals compared with most other individuals with non-O blood types (16). Approximately 20% of the mass of circulating VWF is composed of carbohydrate side chains, with N-linked ABO blood-group glycans representing ∼13% of the total glycan structures (17). The ABO enzyme is a glycosyltransferase that attaches N-acetyl galactosamine (A allele) or simply galactose (B allele) to the H antigen (oligosaccharide that ends in a fucose linked to galactose) on target proteins. The common O allele results from a single nucleotide deletion (G261del) in exon 6 of ABO, which creates an early stop codon and a nonfunctioning transferase (18). Although the precise molecular mechanisms remain unclear, individuals with type O blood group (35–45% of the population) exhibit a shorter VWF half-life, suggesting that the major effect of ABO on VWF level is through alteration of clearance rates (16). The A2 allele encodes a hypomorphic version of A1 with 30- to 50-fold less enzyme activity (19). The relationship of ABO variants and VWF levels fits an autosomal recessive pattern, with similarly elevated VWF levels observed in A1O, BO, and AB individuals.
A genome-wide association study (GWAS) meta-analysis of 17,596 individual adults (average age 58 y) from several large cardiovascular disease cohorts confirmed the influence of common variants at the ABO locus on plasma VWF level and demonstrated additional smaller association signals at VWF and six other loci (20). However, the total variance in plasma VWF explained by all of the significant loci in this study was estimated at only 12.8%, notably less than the heritability estimates of 32–75% previously reported in literature.
To identify additional quantitative trait loci (QTL) for VWF antigen variation and to minimize the impact of age and illness-related environmental factors, we studied two independent healthy young cohorts, the Genes and Blood-Clotting Study (GABC, n = 1,152) and the Trinity Student Study (TSS, n = 2,310) (21–23). In addition to confirming the association signals at ABO and VWF, linkage analysis identified a previously unidentified QTL on chromosome 2 (Chr2) that was undetected in our GWAS or previous studies. This locus explains 19.2% of additional plasma VWF variance and suggests that the use of cohorts with family structure can identify missing genetic determinants for other complex traits with similar allelic architecture.
Results
Study Cohorts.
Table 1 displays demographic information for the two study cohorts. The median age of participants was 21 y (Q1:19, Q3:23) for the GABC cohort and 22 y (Q1:21, Q3:24) for the TSS. Although the TSS cohort was 100% ethnically Irish, the GABC cohort was of mixed ancestry, consistent with the University of Michigan student population from which participants were recruited (24). Genotyping data identified 81.5% (940 of 1,152) of the GABC cohort as European ancestry, in close agreement with self-report. Of the 502 GABC families (1,152 individuals) who passed genotyping quality control, 13 were singletons, 366 were pairs, and 94, 22, 5, and 2 were comprised of 3, 4, 5, and 6 members, respectively. In the TSS cohort, 2,310 participants were successfully genotyped, including 71 sibling pairs and one sibling trio, for a total of 145 full siblings.
Table 1.
Characteristics of study participants
| Cohort | GABC | TSS |
| Subject counts, n | 1,152 | 2,310 |
| Age, y (Q1, Q3) | 21 (19, 23) | 22 (21, 24) |
| Female (%) | 721 (63) | 1,310 (59) |
| Weight, lb (Q1, Q3) | 145 (125, 165) | 148 (133, 167) |
| Body mass index, kg/m2 (Q1, Q3) | 22.5 (20.7, 25.0) | 22.6 (21.0, 24.4) |
| Current smoker, n/total cohort (%) | 55/1,152 (4.8) | 737/2,299 (32.1) |
| Sibship size, (n sibships) | 2 (366); 3 (94); 4 (22);5 (5); 6 (2) | 2 (71); 3 (1) |
| VWF, IU/dL (Q1, Q3) | 108.1 (78.3, 129.7) | 108.4 (79.3, 128.2) |
Median values for age, weight, body mass index, and VWF levels are reported with quartiles in parentheses.
VWF Antigen Levels.
SI Appendix, Fig. S1 displays the distribution of untransformed VWF antigen levels. The median VWF levels were 108.1 IU/dL and 108.4 IU/dL for the GABC and TSS cohorts, respectively (Table 1). The 5th and 95th percentiles of the distribution spanned a 3.4-fold difference, from 54.3 IU/dL to 187.1 IU/dL (GABC), and 3.2-fold difference, from 59.1 IU/dL to 187.2 IU/dL (TSS). There was no significant difference in the distributions of VWF levels between GABC and TSS (Kolmogorov–Smirnov test, P = 0.16). Although 23% were regular tobacco smokers (4.8% GABC, 32% TSS) based on self-report and confirmed with serum cotinine levels (TSS only), there was no significant difference in the distribution of VWF levels between smokers and nonsmokers (Kolmogorov–Smirnov test, P = 0.40). Narrow sense heritability (h2) of the VWF antigen level in the combined GABC and TSS dataset was first estimated by using the known sibship relationships, yielding an intraclass correlation-based upper-bound estimate of 64.5%. This finding is consistent with the estimates derived from the genome-wide genotyping data for all individuals: 66.3% using MERLIN, and 64.9% using GCTA (Materials and Methods) (25, 26).
Association Studies, GABC Cohort.
In the GABC dataset, 38 SNPs were significantly associated with VWF antigen level (P < 5.0E-8) after adjustment for age, sex, and population structure (SI Appendix, Fig. S2A). All 38 SNPs reside in a 300-kb region on Chr 9q34, with 31 of 38 located within the ABO gene (SI Appendix, Fig. S2B), consistent with the association signal for ABO reported in a previous meta-analysis (20). The G allele of the top SNP, rs687289, was associated with decreased VWF antigen level and tags the O allele of ABO (β = −0.36 ± 0.022 IU/dL per allele in an additive model, P = 1.7E-52) (SI Appendix, Table S1), consistent with the known association between type O blood group and the shorter half-life of VWF. The lowest P value outside of the 9q34-associated region was 1.5E-7, not reaching genome-wide significance. The Q-Q plot (SI Appendix, Fig. S2C) of observed versus expected –log10(P value) demonstrates a large deviation from expectation as a result of the significant signals at the ABO locus (in red) and a slight signal inflation, possibly because of family structure. As the initial analysis treated all samples as unrelated, we applied multiple approaches to examine the impact of sample relatedness. These approaches include GWAF (genome-wide association analyses with family data) and EMMAX (Materials and Methods), the former using a linear mixed effect model and the latter using a variance components-based model. Log-scatter pair-wise comparisons (SI Appendix, Fig. S2 D and E) and Q-Q plot comparisons (SI Appendix, Fig. S2 F and G) showed that results obtained by considering relatedness were highly similar to those not considering relatedness. We therefore proceeded to use the original results (assuming unrelated samples) in the meta-analysis below.
Association Studies, TSS Cohort.
The TSS cohort revealed two significantly associated regions (SI Appendix, Fig. S3A), with 31 SNPs overlapping the ABO locus (SI Appendix, Fig. S3B) and 10 SNPs overlapping the VWF gene (SI Appendix, Fig. S3C). The Q-Q plot (SI Appendix, Fig. S3D) of observed versus expected –log10(P value) exhibits a large deviation from expectation as a result of the significant signals at the ABO and VWF loci. The association statistics for the lead SNPs are shown in SI Appendix, Table S2. The top ABO SNP in TSS, rs687289, was the same as the top ABO SNP in GABC, and demonstrated a similar effect size for the G allele (β = −0.33 ± 0.016, P = 3.7E-89). In the VWF region, the highest associated SNP was rs1063856 (β = −0.12 ± 0.015, P = 8.5E-14). This SNP did not reach genome-wide significance in GABC (β = 0.095 ± 0.024, P = 1.1E-4) likely because of the latter’s 2.4-fold smaller sample size compared with TSS.
Meta-Analysis of GABC and TSS.
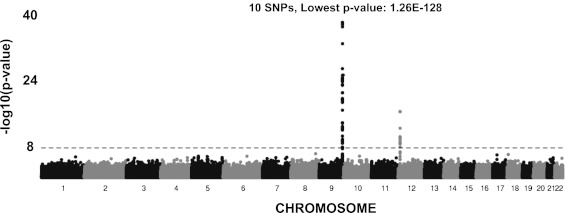
Seventy-three SNPs were significantly associated (P < 5.0E-8) with VWF level in the meta-analysis of GABC and TSS (Fig. 1 and Table 2). These SNPs collectively explained ∼18.7% of the VWF level variation in an analysis using GCTA (a tool for genome-wide complex trait analysis). Of the 73 SNPs, 58 were in the associated region surrounding the ABO gene (including ADAMTS13) (SI Appendix, Fig. S4A), with the remaining 15 overlapping the VWF locus (SI Appendix, Fig. S4B). The strongest signal outside of the ABO and VWF loci had a P value of 1.3E-6, not reaching genome-wide significance.
Fig. 1.
GABC and TSS association meta-analysis (∼724 K SNPs) using age-, sex-, and principal component-adjusted VWF values. Genome-wide −log10(P value) plot. The horizontal line marks the 5E-08 threshold of genome-wide significance.
Table 2.
Top meta-analysis SNPs
| Meta-analysis |
GABC |
TSS |
|||||||||
| Chr | SNP | Position* | Allele† | P value‡ | Closest gene | Freq | β (SE) | P value§ | Freq | β (SE) | P value§ |
| 9 | rs687289 | 135126927 | T | 1.3E-128 | ABO | 0.643 | 0.36 (0.022) | 1.6E-52 | 0.751 | 0.33 (0.016) | 3.7E-89 |
| 9 | rs11244079 | 135174347 | A | 4.4E-36 | LOC653163 | 0.949 | 0.33 (0.055) | 4.6E-09 | 0.937 | 0.35 (0.030) | 3.9E-31 |
| 9 | rs4962153 | 135313575 | A | 1.9E-31 | ADAMTS13 | 0.839 | 0.208 (0.032) | 1.6E-10 | 0.894 | 0.25 (0.024) | 1.2E-24 |
| 9 | rs11244035 | 135071140 | T | 1.9E-25 | OBP2B | 0.885 | 0.27 (0.039) | 3.2E-12 | 0.915 | 0.23 (0.027) | 3.4E-17 |
| 9 | rs45618736 | 137580983 | A | 4.6E-24 | OBP2A | 0.890 | 0.26 (0.040) | 1.3E-10 | 0.918 | 0.24 (0.029) | 5.4E-17 |
| 9 | rs28602591 | 135236487 | T | 3.3E-20 | C9orf96 | 0.886 | 0.19 (0.038) | 5.7E-07 | 0.928 | 0.24 (0.029) | 4.8E-16 |
| 12 | rs1063856 | 6023795 | A | 4.9E-16 | VWF | 0.360 | −0.095 (0.024) | 1.1E-04 | 0.395 | −0.12 (0.015) | 8.5E-14 |
| 9 | rs4507838 | 135092020 | A | 1.54E-13 | LOC286310 | 0.875 | 0.081 (0.036) | 0.025 | 0.867 | 0.17 (0.022) | 4.8E-14 |
| 9 | rs554710 | 135171669 | T | 4.7E-12 | SURF6 | 0.281 | 0.14 (0.027) | 1.5E-07 | 0.286 | 0.088 (0.017) | 1.8E-07 |
| 9 | rs11655 | 135197664 | A | 1.9E-09 | SURF5 | 0.885 | −0.16 (0.037) | 3.2E-05 | 0.885 | −0.11 (0.024) | 1.8E-06 |
Genome-wide significant (P value < 5.0E-8) SNPs in GABC+TSS meta-analysis and individual cohorts, sorted by P value, with respect to their relationship to the nearest gene.
*Positions are in National Center for Biotechnology Information Build 36 coordinates.
†Tested allele.
‡Meta-analysis P value.
§Cohort specific P value.
Thirty-nine of the 73 significant SNPs in the meta-analysis were not significant in the GABC cohort, whereas 12 of the 73 were not significant in TSS. However, the allelic effect sizes and directions of the top SNPs in each cohort showed excellent agreement (SI Appendix, Fig. S5). R2 values were 0.95 and 0.90 for the β-values of the top 38 GABC and top 49 TSS SNPs, respectively, suggesting that the apparent difference in significance levels are mainly due to sample size differences.
Conditional Analysis.
To screen for potential secondary signals masked by the strong primary signal at ABO, the top SNP, rs687289, was introduced as a covariate in a conditional association analysis in the joint set of GABC and TSS cohorts (SI Appendix, Fig. S6 A and D). A new signal emerged at chromosome 9 in the ABO locus, with rs8176704 as the lead SNP (P = 4.1E-34), which was not significant in the original test (P = 0.18) (SI Appendix, Fig. S6 B and C). The A allele of rs8176704 tags the A2 allele of ABO and is associated with a decrease in VWF levels (β = −0.34), consistent with the hypomorphic effect of the A2 allele. The significant SNPs in the ADAMTS13 locus in the meta-analysis were no longer significant in the conditional analysis, suggesting that the ADAMTS13 signal may be because of its linkage disequilibrium with ABO. No other new significant genome-wide signals were detected in the conditional analysis.
ABO Haplotype Effects.
Four major ABO SNP haplotypes, tagged by three ABO SNPs: rs8176704, rs8176749, and rs687289 (27), determine the majority of the ABO blood group serotypes (A1, A2, B, and O). We investigated their association with VWF antigen levels. Using the haplotypes inferred from genotype data we deduced each subject’s most likely ABO serotypes. The overall ABO blood group allele frequencies were similar to those reported in the Atherosclerosis Risk in Communities Study (ARIC) (28) and Framingham Heart Study (FHS) (29) cohort (Table 3), and were in concordance with the expected frequencies in populations with European ancestry (30). As expected, the A1 and B alleles were positively and significantly associated with the VWF antigen levels (β = 0.36 and P = 3.5E-98; β = 0.36 and P = 7.2E-50, respectively) and O and A2 alleles were negatively associated, but only the O allele was significant (β = −0.34 and P = 2.3E-138; β = −0.038 and P = 0.18, respectively).
Table 3.
ABO haplotype association
| Allele/haplotype frequencies |
GABC+TSS haplotype association |
|||||
| ABO-allele | Haplotype* | ARIC | FHS | GABC + TSS | β | P value |
| O | GGC | 0.650 | 0.673 | 0.720 | −0.34 | 2.3E-138 |
| A1 | AGC | 0.209 | 0.196 | 0.155 | 0.36 | 3.5E-98 |
| B | AGT | 0.0730 | 0.0770 | 0.0722 | 0.36 | 7.2E-50 |
| A2 | AAC | 0.0680 | 0.0540 | 0.0531 | −0.038 | 1.8E-01 |
ABO-alleles, tagging haplotypes, and inferred frequencies in the FHS, ARIC, and GABC+TSS cohorts, and association results in GABC+TSS.
*rs687289, rs8176704, rs8176749.
Focused Analysis at the VWF Locus.
The second significant region in the TSS cohort and in the meta-analysis was the VWF locus. The lead SNP in the meta-analysis was rs1063856 (P = 5.5E-16), a coding nonsynonymous SNP at the VWF gene. It is in strong linkage disequilibrium (LD) with the second highly associated SNP, rs1063857 (r2 = 1.00, P = 6.5E-16), a coding synonymous SNP. Although this region was not significant in the GABC cohort, a power analysis using the Genetic Power Calculator (31) showed that given the effect size of β = 0.012, as seen in TSS, and given the sample size of GABC, the power to detect this VWF signal was only 38.4% in GABC.
Similar to the haplotype analysis at ABO, we extended the association analysis at VWF to haplotypes using the combined set of GABC and TSS. From the genotype data for the 71 available VWF SNPs we constructed a haplotype map of the VWF gene using Haploview and identified 12 LD blocks (SI Appendix, Fig. S7). Of the 15 most highly associated SNPs in the meta-analysis, 11 reside in block 6, two reside in block 7 (the top two SNPs), and the remaining two reside in block 8. These three blocks span exons 14–22, which encode the D′, D2, and D3 domains of VWF and contain the VWF propeptide and factor VIII binding domains. Haplotype association tests of these blocks yielded P values of 8.8E-3, 6.3E-7, and 7.9E-3 for the most frequent haplotypes in block 6, block 7, and block 8, respectively.
Linkage Analysis.
To scan for linkage signals for plasma VWF levels we analyzed the genotype data of siblings from the GABC and TSS cohorts, for a total of 561 sibships and 1,284 individuals. Linkage LOD scores from an initial scan using the unpruned dataset of ∼760 K SNP were likely inflated (SI Appendix, Fig. S8A), because of strong LD between many SNPs and missing parental genotypes (32). This explanation was supported by the reduced significance levels observed in a revised scan using an LD pruned dataset (1 SNP/Mb, r2 < 0.001, 2.7 K SNPs) (SI Appendix, Fig. S8B).
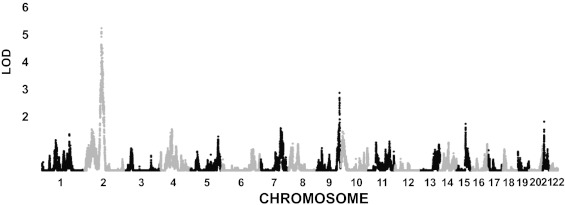
To incorporate data-derived LD patterns, we applied the LD-modeling algorithm in MERLIN (25) to define independent LD clusters and conducted linkage analysis using inferred haplotype frequencies within clusters (Materials and Methods). First, markers were organized into clusters using an LD criterion of r2 = 0.001. This process produced ∼37 K nearly independent clusters; and the linkage analysis on each yielded its LOD score and P value. The per-cluster LOD scores (Fig. 2) revealed a strong linkage signal (peak LOD ∼5.3) crossing the centromere of Chr2 (2q12-2p13), as well as a strong signal on Chr9 (9q34, peak LOD ∼2.9). The latter region contains the ABO gene.
Fig. 2.
Linkage analysis results for GABC+TSS using the age-, sex-, and principal component-adjusted VWF values. Genome-wide LOD Scores for 37 K “clusters,” defined in MERLIN to model the LD among SNPs (Materials and Methods).
We evaluated the genome-wide significance of the linkage results with a locus-counting approach proposed by Wiltshire et al. (33) comparing the observed LOD scores of the top independent regions of linkage (IRLs) with the null distributions of LOD scores for their corresponding equal-ranked IRLs over 1,000 simulated datasets with randomized phenotypes. The IRLs were defined as the 40-cM interval around the location of the maximum LOD score. This analysis showed that the three highest IRLs in our study had higher LOD scores than the 95th percentile of their respective equal-ranked LOD score null distributions (SI Appendix, Fig. S9A). The highest signal, at ∼82.4 Mb on Chr2, had a LOD score of 5.27 (original MERLIN P value = 4.2E-7; simulation-based empirical P value = 0.0015) (Fig. 2, and SI Appendix, Fig. S9A). The second strongest signal, at ∼135.3 Mb on chromosome 9, had a LOD score of 2.87 (P = 1.4E-4; empirical P = 0.048) (Fig. 2 and SI Appendix, Fig. S9A). The third strongest signal was at ∼104.46 Mb on Chr2, with a LOD score of 2.687 (P = 2.2E-4; empirical P = 0.020) (Fig. 2 and SI Appendix, Fig. S9A).
The analysis above used a fixed width (± 20 cM) to define IRL boundaries. An alternative approach is to apply a LOD score cutoff, such as one unit below the maxima. This LOD-based approach yielded an overlapping interval for the first and third significant IRLs, suggesting that the two intervals may be part of the same linkage region. The combined linkage region spans 34 Mb on 2q12-2p13 (positions 74.98 Mb–108.95 Mb), crossing over the centromere (SI Appendix, Fig. S9B). Using the same LOD score cutoff, the second top significant IRL spans 790 Kb on 9q34 (positions 135.03 Mb–135.82 Mb) and includes the ABO locus (SI Appendix, Fig. S9C).
By using GCTA we estimated that the LOD score-based linkage intervals on Chr2 and 9 explained 19.2% and 24.5% of the variation in the VWF levels, respectively. Furthermore, by expanding from the core region on 2q12–2p13, we found that the variance explained changed minimally beyond the 34-Mb core interval, increasing from 19.2–19.7% when expanded to 48 cM, suggesting that the 34-Mb interval contains the majority of the linkage signal.
Focused Analysis in the Chr2 Linkage Region.
For the Chr2 linkage region (74.98 Mb–108.95 Mb), the phased genotype data defined 886 haplotype blocks, containing 4,074 distinct haplotypes with frequency >1% in our study (SI Appendix, SI Methods). We performed haplotype-based association tests for VWF levels in each block. None of the tests reached region-wide significance (P < 5.6E-5) (SI Appendix, Fig. S10A), and few blocks produced substantially smaller haplotype-based P values than single-SNP P values in the same block (SI Appendix, Fig. S10B). Thus, these results did not provide clear evidence of association to common haplotypes that could explain the linkage signal in Chr2.
Next, we screened for sets of at least three SNPs with different LD structures between high-VWF and low-VWF individuals in the GABC cohort and identified six SNP sets in the linkage interval (SI Appendix, SI Methods). We performed association tests using the long-range haplotypes formed by these SNP sets and discovered two sets with P values smaller than 0.001 (SI Appendix, Table S3). Simulation results showed that these two sets reached an empirical significance level of 0.05. The first set (empirical P = 0.006) was anchored by the index SNP rs6547231 (association P = 5.8E-4) and spanned a 987-kb interval from 78.963 Mb to 79.951 Mb. The second set (empirical P = 0.002), anchored by the index SNP rs7566719 (association P = 3.3E-4), covered an ∼973-kb interval from 76.240 Mb to 77.214 Mb.
Discussion
Our association analysis of VWF variation in healthy young subjects identified two major signals at the ABO and VWF loci, confirming results from a previously published meta-analysis. Taking advantage of the sibling structure of our cohorts, we also identified a unique QTL on Chr2, which was undetected in previous studies (15, 20, 34, 35).
With a total sample size of 3,250, our GWAS was underpowered to detect the several smaller-effect loci identified in the larger CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) consortium meta-analysis (n = 17,596). However, the total variation explained by the genetic loci identified in our GWAS analysis (18.7%) was notably larger than that explained by the similar analysis in the CHARGE study (12.8%). The young age and healthy status of our subjects may have increased the contribution of the heritable components of VWF levels by limiting the impact of known environmental effects on VWF levels, including age and common illnesses, such as diabetes and heart disease. For example, contrary to studies in older cohorts (36, 37), we found no associations between smoking and VWF levels in this study, suggesting that the previously observed association may be a proxy for vascular disease related to chronic smoking. Consistent with this notion, the estimated heritability of VWF variation of ∼65% from our study is at the upper end of the range previously reported (13–15).
The strongest association signals in our analysis were at the ABO locus. ABO haplotype analysis confirmed that SNPs tagging the O and A2 alleles were associated with lower VWF levels, but those tagging the B and A1 alleles were associated with higher VWF levels. Thus, the association signals at ABO are consistent with known alteration of VWF levels through specific ABO glycosylation patterns. Although we were able to detect the association of the A2 allele (minor allele frequency 0.05) after a conditional analysis, we were unable to detect several other less common alleles at ABO (30), likely because of their lower allele frequencies. The significantly associated SNPs at the VWF locus tag three of seven haplotype blocks, consistent with a similar analysis of the ARIC cohort (38, 39). Although the causative SNPs are not yet at this point, they likely act via alterations in VWF biosynthesis, secretion, or clearance from plasma (40).
Previous studies have reported significant single-gene associations between plasma VWF levels and variants at the FUT2 locus (41, 42) lipoprotein receptor-related protein (LRP1) (43), angiotensin-converting enzyme (ACE) (44), and arginine vasopressin 2 receptor (AVPR2) (45). None of these loci were detected in our analysis or in the CHARGE GWAS (20), although the low minor allele frequency for the LRP1 variant (<2%) and X-chromosomal location of AVPR2 limited the power for their detection.
In addition to GWAS, the sibling structure of our GABC and TSS cohorts enabled linkage analysis, which identified a highly significant unique QTL on Chr2. The effect size of this locus on VWF variation (19.2% variance explained) was comparable to the effect of the ABO locus (24.5%). Variants in this linkage interval were not detected in our GWAS analysis of the same subjects, nor were they detected in the CHARGE study (20). A previously reported linkage analysis in 21 families (the GAIT study) only identified significant linkage at the ABO locus (LOD = 3.46). Although a peak with LOD = 1.65 was reported on Chr2 (2q33.2), it did not overlap with the Chr2 peak in our study (15).
The linkage peak on Chr2 spans ∼34 Mb and contains over 100 annotated genes, with plausible candidates for altering plasma VWF including several glycosyltransferases, sialyltransferases, and SNARE complex proteins potentially involved in protein secretion. Conventional haplotype-based analyses revealed no region-wide significant association in the Chr2 linkage region. However, analysis of differences in LD structure between high- and low-VWF individuals (SI Appendix, SI Methods) identified two ∼1-Mb intervals that might harbor rare causal variants contributing to the linkage signal. The first region contained seven RefSeq genes (REG3G, REG1B, REG1A, REG1P, REG3A, CTNNA2, MIR4264) and the second region one RefSeq gene, LRRTM4. Although none of the genes play an obvious role in VWF biology, these regions represent targets for future study.
We estimate that variants in the Chr2 linkage interval alter steady-state VWF level with a magnitude similar to ABO (20–30%). Therefore, detection as a Mendelian bleeding disorder similar to VWD (VWF levels reduced by >50%) would be unlikely. However, this locus, together with ABO blood group, could represent an important modifier of VWD severity and penetrance, as well as the thrombosis risk associated with elevated VWF. Identification of the underlying gene and its mechanism of action could also lead to improved treatment for VWD and venous thromboembolic disease.
Mendelian genes for complex traits, such as type 2 diabetes, are often undetected by GWAS (46, 47). However, mutations at these loci are generally very rare and contribute only a small part of the overall population variance for that trait. The large contribution of the Chr2 locus to plasma VWF variation, comparable to the effect of ABO but undetectable even by a well-powered GWAS, is surprising and not readily explained by known mechanisms of VWF homeostasis. We hypothesize that this locus may contain a critical VWF regulatory gene harboring rare causal variants in a situation analogous to allelic heterogeneity, thus having reduced power of detection in association tests. However, these variants might have sufficiently large effect size in each individual family and accrued linkage signals across families, thus contributing to the observed Chr2 linkage results. A similar pattern of a strong linkage signal without corresponding evidence for association was recently reported for a QTL for cystic fibrosis disease severity (48). Additionally, in silico experiments predict the presence rare variants detected by linkage that contribute to variation in complex genetic traits (49). Taken together, these findings suggest that similar loci could explain a significant portion of the “missing heritability” for other complex genetic traits.
Materials and Methods
Genes and Blood-Clotting Study.
A cohort of healthy siblings was recruited from the University of Michigan, Ann Arbor, between June 26, 2006 and January 30, 2009. Participants were between the ages of 14 and 35 y, and had at least one eligible healthy sibling. Subjects who indicated that they were pregnant, had a known bleeding or blood-clotting disorder, or any illness requiring regular medical care were excluded. All participants provided informed consent by a process that was previously described (22). Subjects completed an online phenotyping survey and donated a blood sample for DNA extraction and plasma biochemical phenotyping. Details of the sample collection, genotyping, and data-cleaning process for the GABC cohort are described in SI Appendix.
Trinity Student Study.
A cohort of 2,524 healthy, ethnically Irish individuals, attending the University of Dublin, Trinity College, with ages between 18 and 28 y, was recruited over one academic year in 2003–2004 (21, 23). Ethical approval was obtained from the Dublin Federated Hospitals Research Ethics Committee, which is affiliated with the Trinity College, and reviewed by the Office of Human Subjects Research at the National Institutes of Health. Written informed consent was obtained from participants before recruitment. Details of the sample collection, genotyping, and data-cleaning process are described in SI Appendix.
VWF Antigen Level.
VWF antigen levels were measured in both the GABC and TSS cohorts using a custom AlphaLISA (Perkin-Elmer) assay. For further details on this assay, refer to SI Appendix.
Phenotype Data Processing.
Consistent with previous studies (12, 20), the raw VWF antigen level distribution in our cohorts was not normal. Therefore, the antigen-level distribution was normalized by log transformation. The associations of smoking with raw VWF level distribution were analyzed with Kolmogorov–Smirnov tests. The effects of age, sex, and population structure on the VWF antigen levels were determined by separate single-factor linear regression analysis. SI Appendix provides further details of phenotype data processing.
Heritability of VWF Antigen Levels.
For the combined set of GABC and TSS, the sib-pair intraclass correlation coefficient (ICC) was calculated using the log-transformed, age-, sex-, and PC3-adjusted values in GABC, and age- and sex-adjusted values in TSS. As there were sibships with more than two members, we randomly sampled two sibs from each family, calculated ICC, and repeated the process 500 times to obtain the average ICC. The upper-bound of the narrow sense heritability (h2) was estimated by multiplying the average ICC by 2. In addition to the pedigree-based estimate described above, independent estimates were also calculated using the genotype and phenotype data in MERLIN-REGRESS and GCTA using all individuals (25, 26).
Computational Analyses.
A detailed description of the computational methods used to assess population substructure, association analysis, meta-analysis, linkage analysis, variance-explained calculations, and haplotype-based association analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank the participants in the Genes and Blood-Clotting Study and the Trinity Student Study; and the high-throughput screening laboratory in the University of Michigan’s Center for Chemical Genomics for their assistance with the performance of high-throughput von Willebrand factor assays. This study was supported by the Intramural Research Programs of the National Institutes of Health, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the National Human Genome Research Institute; and by National Institute of Health Grants 5K12HD028820-15 (to K.C.D.) and 2RO1HL039693 (to D.G.). D.G. is a Howard Hughes Investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the database of Genotypes and Phenotypes, www.ncbi.nlm.nih.gov/gap (dbGaP study accession phs000304.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219885110/-/DCSupplemental.
References
- 1.Sadler JE. von Willebrand factor: Two sides of a coin. J Thromb Haemost. 2005;3(8):1702–1709. doi: 10.1111/j.1538-7836.2005.01369.x. [DOI] [PubMed] [Google Scholar]
- 2.Nichols WL, et al. von Willebrand disease (VWD): Evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA) Haemophilia. 2008;14(2):171–232. doi: 10.1111/j.1365-2516.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 3.Koster T, Blann AD, Briët E, Vandenbroucke JP, Rosendaal FR. Role of clotting factor VIII in effect of von Willebrand factor on occurrence of deep-vein thrombosis. Lancet. 1995;345(8943):152–155. doi: 10.1016/s0140-6736(95)90166-3. [DOI] [PubMed] [Google Scholar]
- 4.Morange PE, et al. PRIME Study Group Endothelial cell markers and the risk of coronary heart disease: The Prospective Epidemiological Study of Myocardial Infarction (PRIME) study. Circulation. 2004;109(11):1343–1348. doi: 10.1161/01.CIR.0000120705.55512.EC. [DOI] [PubMed] [Google Scholar]
- 5.Wieberdink RG, et al. High von Willebrand factor levels increase the risk of stroke: The Rotterdam study. Stroke. 2010;41(10):2151–2156. doi: 10.1161/STROKEAHA.110.586289. [DOI] [PubMed] [Google Scholar]
- 6.van Schie MC, et al. Active von Willebrand factor and the risk of stroke. Atherosclerosis. 2010;208(2):322–323. doi: 10.1016/j.atherosclerosis.2009.07.047. [DOI] [PubMed] [Google Scholar]
- 7.Roldán V, et al. Plasma von Willebrand factor levels are an independent risk factor for adverse events including mortality and major bleeding in anticoagulated atrial fibrillation patients. J Am Coll Cardiol. 2011;57(25):2496–2504. doi: 10.1016/j.jacc.2010.12.033. [DOI] [PubMed] [Google Scholar]
- 8.de Lange M, et al. Genetic influences on fibrinogen, tissue plasminogen activator-antigen and von Willebrand factor in males and females. Thromb Haemost. 2006;95(3):414–419. doi: 10.1160/TH05-09-0596. [DOI] [PubMed] [Google Scholar]
- 9.Jilma B, et al. High dose dexamethasone increases circulating P-selectin and von Willebrand factor levels in healthy men. Thromb Haemost. 2005;94(4):797–801. doi: 10.1160/TH04-10-0652. [DOI] [PubMed] [Google Scholar]
- 10.Lip GY, Foster W, Blann AD. Plasma von Willebrand factor levels and surrogates of atherosclerosis. J Thromb Haemost. 2005;3(4):659–661. doi: 10.1111/j.1538-7836.2005.01284.x. [DOI] [PubMed] [Google Scholar]
- 11.Goligorsky MS, Patschan D, Kuo MC. Weibel-Palade bodies—Sentinels of acute stress. Nat Rev Nephrol. 2009;5(7):423–426. doi: 10.1038/nrneph.2009.87. [DOI] [PubMed] [Google Scholar]
- 12.Orstavik KH, et al. Factor VIII and factor IX in a twin population. Evidence for a major effect of ABO locus on factor VIII level. Am J Hum Genet. 1985;37(1):89–101. [PMC free article] [PubMed] [Google Scholar]
- 13.de Lange M, Snieder H, Ariëns RA, Spector TD, Grant PJ. The genetics of haemostasis: A twin study. Lancet. 2001;357(9250):101–105. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- 14.Bladbjerg EM, et al. Genetic influence on thrombotic risk markers in the elderly–A Danish twin study. J Thromb Haemost. 2006;4(3):599–607. doi: 10.1111/j.1538-7836.2005.01778.x. [DOI] [PubMed] [Google Scholar]
- 15.Souto JC, et al. Genome-wide linkage analysis of von Willebrand factor plasma levels: Results from the GAIT project. Thromb Haemost. 2003;89(3):468–474. [PubMed] [Google Scholar]
- 16.Gallinaro L, et al. A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood. 2008;111(7):3540–3545. doi: 10.1182/blood-2007-11-122945. [DOI] [PubMed] [Google Scholar]
- 17.Canis K, et al. The plasma von Willebrand factor O-glycome comprises a surprising variety of structures including ABH antigens and disialosyl motifs. J Thromb Haemost. 2010;8(1):137–145. doi: 10.1111/j.1538-7836.2009.03665.x. [DOI] [PubMed] [Google Scholar]
- 18.Nossent AY, Vos HL, Rosendaal FR, Bertina RM, Eikenboom JC. Aquaporin 2 gene variations, risk of venous thrombosis and plasma levels of von Willebrand factor and factor VIII. Haematologica. 2008;93(6):959–960. doi: 10.3324/haematol.12296. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto F, McNeill PD, Hakomori S. Human histo-blood group A2 transferase coded by A2 allele, one of the A subtypes, is characterized by a single base deletion in the coding sequence, which results in an additional domain at the carboxyl terminal. Biochem Biophys Res Commun. 1992;187(1):366–374. doi: 10.1016/s0006-291x(05)81502-5. [DOI] [PubMed] [Google Scholar]
- 20.Smith NL, et al. Wellcome Trust Case Control Consortium Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121(12):1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone N, et al. Bioinformatic and genetic association analysis of microRNA target sites in one-carbon metabolism genes. PLoS ONE. 2011;6(7):e21851. doi: 10.1371/journal.pone.0021851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desch K, et al. Analysis of informed consent document utilization in a minimal-risk genetic study. Ann Intern Med. 2011;155(5):316–322. doi: 10.1059/0003-4819-155-5-201109060-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills JL, et al. Do high blood folate concentrations exacerbate metabolic abnormalities in people with low vitamin B-12 status? Am J Clin Nutr. 2011;94(2):495–500. doi: 10.3945/ajcn.111.014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.University of Michigan, Office of the Registrar Ethnicity of University of Michigan students. 2010. www.umich.edu/~regoff/enrollment/ethnicity.php. Accessed 11-14-2012.
- 25.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—Rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbalic M, et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet. 2010;19(9):1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folsom AR, Wu KK, Rosamond WD, Sharrett AR, Chambless LE. Prospective study of hemostatic factors and incidence of coronary heart disease: The Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 1997;96(4):1102–1108. doi: 10.1161/01.cir.96.4.1102. [DOI] [PubMed] [Google Scholar]
- 29.Feng D, et al. Factor VII gene polymorphism, factor VII levels, and prevalent cardiovascular disease: The Framingham Heart Study. Arterioscler Thromb Vasc Biol. 2000;20(2):593–600. doi: 10.1161/01.atv.20.2.593. [DOI] [PubMed] [Google Scholar]
- 30.Chester MA, Olsson ML. The ABO blood group gene: A locus of considerable genetic diversity. Transfus Med Rev. 2001;15(3):177–200. doi: 10.1053/tmrv.2001.24591. [DOI] [PubMed] [Google Scholar]
- 31.Ahlen MT, et al. The development of severe neonatal alloimmune thrombocytopenia due to anti-HPA-1a antibodies is correlated to maternal ABO genotypes. Clin Dev Immunol. 2012;2012:156867. doi: 10.1155/2012/156867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyles AL, et al. Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered. 2005;59(4):220–227. doi: 10.1159/000087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiltshire S, Cardon LR, McCarthy MI. Evaluating the results of genomewide linkage scans of complex traits by locus counting. Am J Hum Genet. 2002;71(5):1175–1182. doi: 10.1086/342976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antoni G, et al. Combined analysis of three genome-wide association studies on vWF and FVIII plasma levels. BMC Med Genet. 2011;12:102. doi: 10.1186/1471-2350-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zabaneh D, et al. Genetic variants associated with Von Willebrand factor levels in healthy men and women identified using the HumanCVD BeadChip. Ann Hum Genet. 2011;75(4):456–467. doi: 10.1111/j.1469-1809.2011.00654.x. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, et al. Relationship between biomarkers of cigarette smoke exposure and biomarkers of inflammation, oxidative stress, and platelet activation in adult cigarette smokers. Cancer Epidemiol Biomarkers Prev. 2011;20(8):1760–1769. doi: 10.1158/1055-9965.EPI-10-0987. [DOI] [PubMed] [Google Scholar]
- 37.Jefferis BJ, et al. Secondhand smoke (SHS) exposure is associated with circulating markers of inflammation and endothelial function in adult men and women. Atherosclerosis. 2010;208(2):550–556. doi: 10.1016/j.atherosclerosis.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campos M, et al. Genetic determinants of plasma von Willebrand factor antigen levels: A target gene SNP and haplotype analysis of ARIC cohort. Blood. 2011;117(19):5224–5230. doi: 10.1182/blood-2010-08-300152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campos M, et al. Influence of single nucleotide polymorphisms in factor VIII and von Willebrand factor genes on plasma factor VIII activity: The ARIC Study. Blood. 2012;119(8):1929–1934. doi: 10.1182/blood-2011-10-383661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadler JE. von Willebrand factor assembly and secretion. J Thromb Haemost. 2009;7(Suppl 1):24–27. doi: 10.1111/j.1538-7836.2009.03375.x. [DOI] [PubMed] [Google Scholar]
- 41.Orstavik KH, Kornstad L, Reisner H, Berg K. Possible effect of secretor locus on plasma concentration of factor VIII and von Willebrand factor. Blood. 1989;73(4):990–993. [PubMed] [Google Scholar]
- 42.O’Donnell J, Boulton FE, Manning RA, Laffan MA. Genotype at the secretor blood group locus is a determinant of plasma von Willebrand factor level. Br J Haematol. 2002;116(2):350–356. doi: 10.1046/j.1365-2141.2002.03270.x. [DOI] [PubMed] [Google Scholar]
- 43.Morange PE, et al. Biological and genetic factors influencing plasma factor VIII levels in a healthy family population: Results from the Stanislas cohort. Br J Haematol. 2005;128(1):91–99. doi: 10.1111/j.1365-2141.2004.05275.x. [DOI] [PubMed] [Google Scholar]
- 44.Kario K, et al. Endothelial cell damage and angiotensin-converting enzyme insertion/deletion genotype in elderly hypertensive patients. J Am Coll Cardiol. 1998;32(2):444–450. [PubMed] [Google Scholar]
- 45.Nossent AY, et al. Functional variation in the arginine vasopressin 2 receptor as a modifier of human plasma von Willebrand factor levels. J Thromb Haemost. 2010;8(7):1547–1554. doi: 10.1111/j.1538-7836.2010.03884.x. [DOI] [PubMed] [Google Scholar]
- 46.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 47.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright FA, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinrichs AL, Suarez BK. Incorporating linkage information into a common disease/rare variant framework. Genet Epidemiol. 2011;35(Suppl 1):S74–S79. doi: 10.1002/gepi.20654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.