Abstract
Despite the enormous proliferation of bacterial genome data, surprisingly persistent collections of bacterial proteins have resisted functional annotation. In a typical genome, roughly 30% of genes have no assigned function. Many of these proteins are conserved across a large number of bacterial genomes. To assign a putative function to these conserved proteins of unknown function, we created a physical interaction map by measuring biophysical interaction of these proteins. Binary protein-–protein interactions in the model organism Streptococcus pneumoniae (TIGR4) are measured with a microfluidic high-throughput assay technology. In some cases, informatic analysis was used to restrict the space of potential binding partners. In other cases, we performed in vitro proteome-wide interaction screens. We were able to assign putative functions to 50 conserved proteins of unknown function that we studied with this approach.
Keywords: high-throughput screening, unknown proteins
Protein interactions are a hallmark of all living organisms, and research on protein interactions has begun to contemplate complete interactome approaches (1). Interactomes are the whole set of protein interactions in a cell; they are regarded as the framework for future systems and synthetic biology engineering approaches (2). It was noticed that more than 70% of physically interacting proteins share functional annotations (3). This empirical observation has led to statistical models to annotate proteins with unknown function by observing the functions of their interaction partners (4, 5). The most simple yet effective method in this respect is the majority rule (4, 6, 7): a protein with an unknown function is grouped to a functional class with the highest statistical count found among its interaction partners. The success and accuracy of the majority method is dependent on the number and completeness of the available protein-interaction networks and existing functional information about the proteins. Protein functional assignments are often obtained by sequence similarity searches of newly discovered ORFs in sequenced genomes against experimentally characterized homologous proteins (8). The coverage of functional assignments for genomes based on sequence similarity searches varies between organisms. The actual fraction of possible assignments is controversial (9). Additionally, genome information, such as operon structure (10), gene neighborhoods (11), and conserved protein domain structures (12), can mainly be used for prokaryotes to increase the coverage of functional protein assignments. Much of the genome-wide functional annotations are based on in silico methods, which are fast and cost-effective. Physical protein interactions on a proteome scale can fill the gaps left by in silico methods. Furthermore, this process can concomitantly bring experimental evidence to the functional annotation problem.
The yeast two-hybrid (Y2H) and tandem-affinity purification methods coupled with a mass spectrometer (AP-MS) are generally used for protein–protein interaction studies on the proteome scale (13, 14); this is mainly because of their optimized workflows for higher throughput. The combinatorial use of both techniques together with orthogonal screening strategies and computational quantification has to some extent overcome the problem of higher error rates for protein-interaction data (15). For this procedure, however, the protein-interaction screening space has to be oversampled, which led to high experimental costs (16, 17). As an alternative to the prevailing techniques, we recently developed a microfluidic chip implementing a miniaturized immunoprecipitation (mIP) assay to test for binary protein–protein interactions in a parallel fashion (640 measurements per chip) (18). Microfluidics, which refers to the study and control of fluidic properties and their content in structures of micrometer dimensions, provides a powerful platform to interrogate protein interactions (19). Here, we have increased the throughput by an order of magnitude to 4,000 measurements per chip, extensively benchmarked the interaction assay, and performed proteome-wide protein–protein interaction measurements in a model organism, namely Streptococcus pneumoniae (SP strain TIGR4).
SP-TIGR4 is a Gram-positive bacterium and is annotated with 2,105 predicted ORFs. For approximately one-third of the genome (742 proteins), no functional assignments are found. From this set we chose 112 widely conserved proteins and determined in three consecutive screening strategy protein-interaction partners. In the first round we tested predicted functional interaction partners based on the genomic context of the conserved protein with unknown function (cPUF); in the second round we determined the interaction network between cPUFS; and in the last round we selected interaction partners from the first two rounds and screened them against the expressed SP proteome on chip. The screening space of binary protein interactions was gradually increased with the screening round from 103 to 104 to 105 binary-tested protein interactions per round. The newly found protein interactions allowed us to compare the functional information for the cPUFs that were derived from genome information with the functional information derived from the physical protein interaction by applying the majority rule. Furthermore, we tested the connectivity between cPUFs in SP and gained basic information about their distributions over the functional space in the SP proteome that demonstrates the biological relevance of the found interactions.
Results
Microfluidic Chip Platform.
The microfluidic principles of the protein-interaction assay have been developed previously (18); however, to reach higher throughput, changes to the chemical and microarchitecture of the chip were made (SI Materials and Methods). In general, the technology is a combination of a microfluidic chip and a DNA microarray. A large number of binary combinations of cDNAs encoding for proteins of interest were spotted on a glass slide. The resulting microarray was aligned and bound to a polydimethyl siloxane (PDMS) chip with 4,096 identical unit cells. All unit cells were operated in parallel fashion. One unit cell had a size of about 750 × 300 µm2 (Fig. S1). With the help of an automated fluid control system and ∼8,400 integrated microvalves, two biochemical steps were integrated on a chip in each unit cell (i.e., the in vitro expression of two proteins from their cDNA and a mIP assay to test for interaction between the two expressed proteins). Flush sequences with chemical composition of the fluids are listed in Table S1. The mIP assay works in analogy to a direct surface ELISA, with a pull down and detection antibody specific for sequence tags encoded in the corresponding proteins. Hereafter, we name the protein that was pulled down to the glass surface of the chip “bait” and the protein that was tested to coprecipitate “prey.” Fluorescence signals indicating the protein interactions were determined with a microarray scanner.
Benchmark of the Interaction Assay on Chip.
Before screening for binary interactions of proteins with unknown functions within the proteome of SP-TIGR4, the microfluidic protein-interaction technology was benchmarked using a previously published binary interaction set from yeast (Table S2) (15). The benchmark set combines 94 positive and randomly generated binary interactions from the model organism Saccharomyces cervisiae. The positive interaction set was derived from five comprehensive protein-interaction databases. Protein interactions from high-throughput screens or associated to larger protein complexes were excluded. A further selection criterion for the positive set was that each protein interaction needed at least four independent literature entries. The random interaction set was the same size as the positive set and was generated from ∼14 × 106 possible yeast protein pairs, where no interactions had been detected before. From 376 Gateway clones, we were able to express on chip 356:87 positive and 91 negative protein-interaction pairs. Protein interactions from the benchmark set were measured on the chip platform.
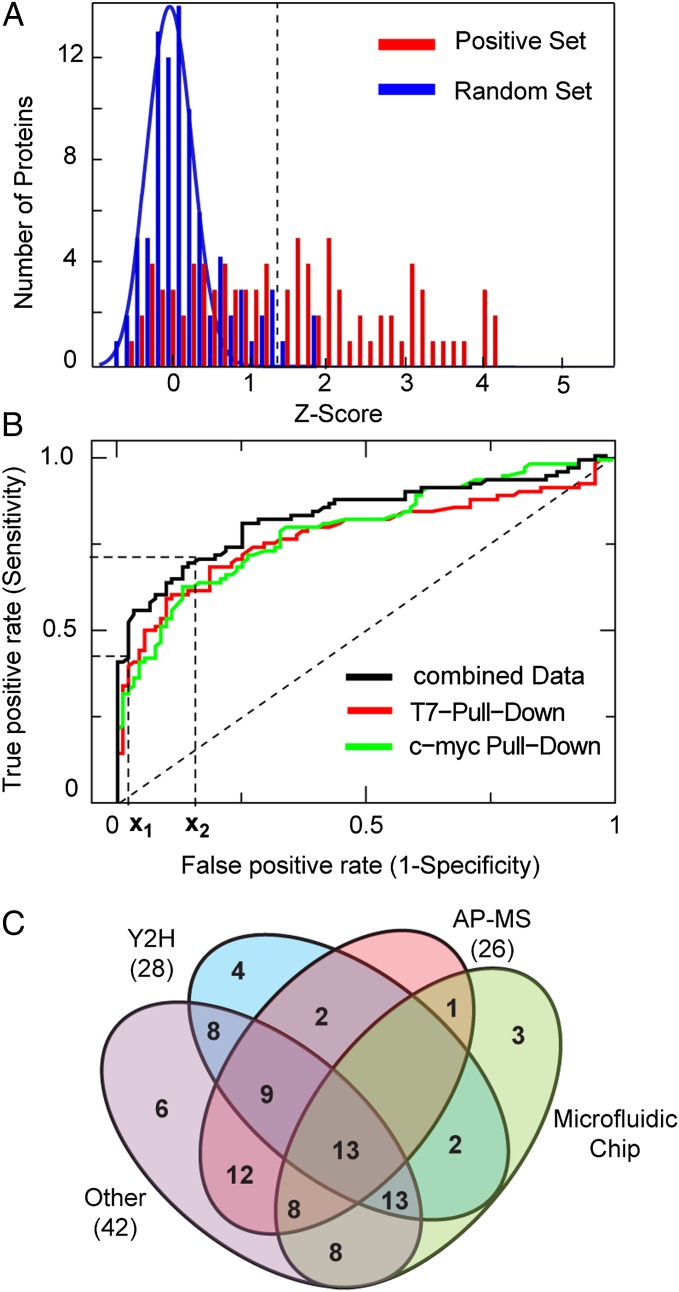
Fig. 1A shows the statistical mean z-score values for the positive and random interaction assays obtained from two independent experiments with different pull-down directions (i.e., reversed bait/prey order). The z-score indicates by how many SDs the fluorescence signal of a positive protein-interaction measurement is above the mean of the control experiments. A clear separation of the z-score between the positive and random interaction set is observed. The likelihood that these two populations exhibit the same mean z-score was tested with Wilcoxon’s signed-rank test and resulted in a P value of 3.6 × 10−14. A detailed description of the consistency of the on chip-interaction assay for the benchmark set is given in SI Materials and Methods, which includes correlations between protein-interaction repeats on chips, between independent chip experiments, and between different pull-down antibodies (Fig. S2).
Fig. 1.
Benchmark of the protein-interaction chip technology. (A) Histogram of the z-scores of the random and positive benchmark set obtained from on chip protein-interaction measurements. The blue line is a Gaussian fit to the z-scores of the random set (r = 0.91). The dotted line defines the cutoff value obtained from the x1 value in B. (B) The receiver operator curves for protein-interaction measurements with different pull-down antibodies (reverse bait/prey order) and the combined dataset. x1 and x2 denote the high specificity cutoff level and the value for the error determination, respectively. (C) Comparison of the microfluidic chip to established interaction technologies based on the benchmark set. The Venn diagram shows the number of protein interactions from the benchmark set positively identified by the Y2H, AP-MS, other techniques and the microfluidic chip technology. Numbers in brackets denote the overlap between the microfluidic chip technology with corresponding techniques.
For the identification of positive protein interaction and determination of error rates of the assay system, a receiver operator analysis was used in which the sensitivity (true positive rate) of the assay is plotted against its specificity (false-negative rate) at various thresholds. The resulting receiver operating characteristic curve (Fig. 1B) was analyzed at two cutoff points. The first cutoff point is the closest point of the receiver operating characteristic curve to the sensitivity and specificity values of [0;1]. In contrast, the second cutoff point was arbitrarily chosen with a fixed high specificity value of 0.02. The first value indicated objectively the characteristics of the on chip-protein interaction test system, whereas the second was used as a stringent threshold value to identify protein interaction with a low false-positive rate in the unknown SP-TIGR4 protein screen. The false-positive and false-negative rate for the first cutoff point was 0.22 and 0.2, respectively, and for the second cutoff point, 0.02 and 0.48, respectively. Taking the second cutoff point for identifications of the number of positive interaction, we found 46 of the 87, and three false-positive interactions within the benchmark set. Differences between immuno-pull down directions existed and are shown in detail in Table S3 with corresponding error rates for each used antibody. Nevertheless, analysis of the separate pull-down experiments revealed a reciprocal discovery rate of 0.56 for the positive interaction in the benchmark set. The obtained false-positive and false-negative error rates for the protein interaction mIP-assay on chip are about 25% and 20%, respectively. One possible explanation for the false-negative rate for the mIP-assay is the misfolding of the protein after in vitro synthesis. Both error rates found here are of the same order as determined for high-throughput protein interaction data yield from Y2H and AP-MS screens (20).
Besides internal benchmarking, the positive protein-interaction set can be used for comparison of the microfluidic chip to the established protein interaction assay technologies. The Venn diagram in Fig. 1C shows the overlap and differences for the positive identified protein interactions from the benchmark set between the microfluidic, Y2H, AP-MS, and Other technology. “Other” denotes all protein binding assay technologies different from Y2H and AP-MS. No single assay is expected to detect all protein interactions, and the actual fraction of positive detected is inherently linked to assay and the stringency at which the assay is implemented (15–17). The microfluidic chip technique exhibited the largest overlap with the assay group Other, followed by Y2H and AP-MS. The order of the overlap of the positive identified protein interactions between the microfluidic chip and Y2H or AP-MS assay technologies is comparable to the overlap between AP-MS and Y2H.
SP-TIGR4 Proteome Library Construction.
To perform proteome-wide protein-interaction screens for proteins with unknown functions on chip, we constructed two cDNA library sets of the SP-TIGR4 proteome with different pull-down tag sequences. The first cDNA library set contained a c-Myc/6xHis at the N and C terminus, respectively. The second library set only contained a T7 tag at the N terminus. From 2,105 available ORFs (J. Craig Venter Institute, JVCI), we successfully amplified about 1,500 cDNAs for each of the two library sets. In vitro expression analysis (Fig. S3) of all cDNA constructs revealed that it was possible to express 1,030 proteins with a c-Myc/6xHis tag combination and 870 proteins with a T7 tag on chip. A cell location analysis with the PSORTb 3.0 algorithm (21) revealed that the fraction of expressed to total number of proteins in the proteome with cytosolic location is larger (0.51) than the fraction of proteins with a membrane assigned location (0.29).
Selection of Proteins with Unknown Functions and Sequence Similarities.
From the 742 predicted ORFs annotated with unknown functions [for annotation sources, see JVCI, National Center for Biotechnology Information (NCBI), and RAST], we concentrated on proteins with conserved sequences because of their presumed general interest in biology (22) and to reduce the chances of measuring false predicted reading frames (23). Functionally unknown proteins with similar sequences (normalized bitscore > 0.4) (Fig. S4) found in more than 40 organisms regardless of taxonomy are termed in the following as cPUFs. Under the given constraints, we clustered 178 SP-TIGR4 proteins as conserved from the 742 SP-TIGR4 proteins with unknown function. Although all homologous proteins to the 178 cPUFs are termed as functionally unknown, about half of the cPUFs exhibit domains with similarities to a functional characterized protein family (pfam). Those sets were not excluded from the study because the proteome-wide protein-interaction screens are expected to clarify the context of these cPUFs in the cell network of SP-TIGR4. Of the 178 clustered cPUFs, 112 could be expressed on chip, which were then selected for the protein-interaction screen in the following.
Protein-Interaction Screen Among Predicted Functional Associated Partners.
After sequence similarity searches, we exploited the genomic information of the cPUFs to find functional association partners. For this scoring, algorithms were used that analyzed the proximity (300 bp), the operon structure of the cPUF gene, genes with the same co-occurrence profiles in different organisms, and genes that may have fused during evolution to form the cPUF. The prediction algorithms for functional association partners based on such genomic information in bacteria had shown some degree of success (11). It is important to note that functional association partners deduced from genome information do not necessarily have to overlap with physical protein interactions because both are in distinct relationships. Nevertheless, information about an overlap between computational and experimental techniques will lead to better appraisal knowledge of the quality of prediction algorithms.
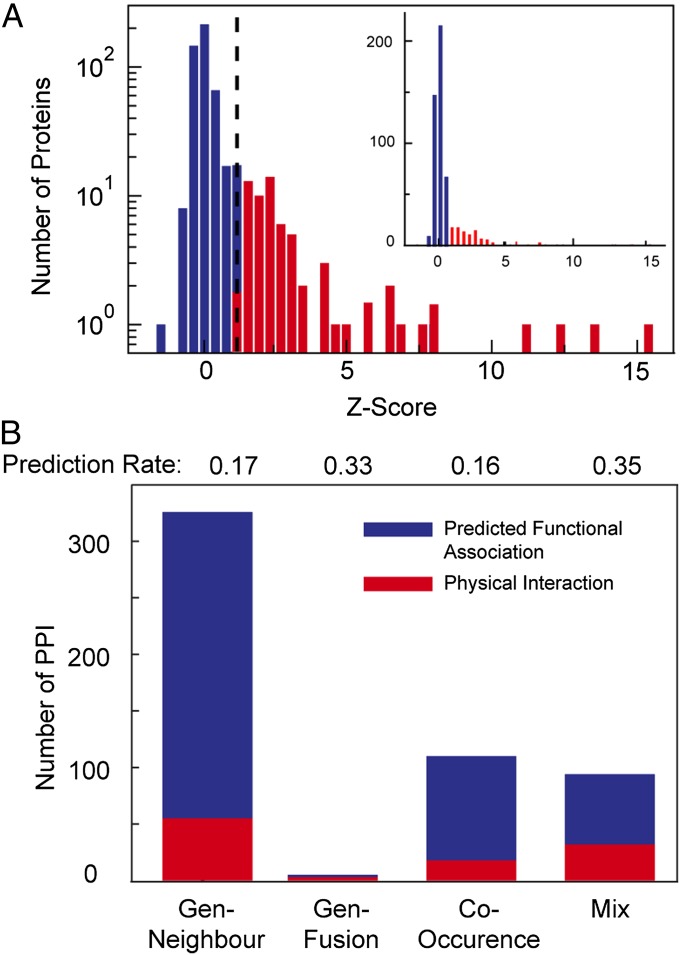
To predict functional associated partners for each cPUF we exploited the STRING database. For 100 cPUFs, we found five predicted functional association partners with a high confident score and concomitantly a positive in vitro expression result on chip. For the remaining 12 cPUFs, at least one association partner was found that was expressed on chip. Thus, we were able to measure in total 772 binary protein interactions in reciprocal order (switched bait/prey), with two repeats for each antibody direction on chip. Fig. 2 shows a corresponding histogram of the z-scores from the interaction screen of the predicted protein associations to the cPUFs. Clearly, two z-score distributions are observed within the histogram, where the optimal threshold separating the two z-score groups from each other matched the above obtained cutoff value for the benchmark set. Protein interactions above the cutoff value were assigned to the algorithm used for their prediction. We found that a fraction of around 0.17 of each predicted functional association was a physical protein interaction. The overlap of the predicted functional associated proteins with physical protein interaction increased up to 0.35, if several algorithms indicated a functional association. This result is comparable to or greater than that found in previous studies, which used in silico and experimental approaches (24), and is therefore a valuable approach to reduce the screening space if functional knowledge about a protein is unknown.
Fig. 2.
Screen of predicted protein interactions for functionally unknown proteins of S. pneumoniae. (A) Mean z-scores of the two pull-down experiments of 532 predicted interactions for the 112 conserved unknown SP proteins. The dashed lines mark the cutoff level for positive protein interactions (determined by the benchmark set). (B) The staggered bar chart shows the number of found physical interactions among the predicted interaction from different algorithms. Mix, predicted protein interactions scored by two of the other three algorithms. The blue and red bars denote negative and positive physical interactions.
Protein Interactions Among cPUFs.
For the second screening round, we concentrated on the group of cPUFs itself and addressed the question of the number and degree of interactions among the cPUFs. Possible interactions in the group of cPUFs could reveal unknown protein clusters, parts of a single pathway, or regulatory networks. To find interactions within the cPUFs a matrix-screening approach was applied in which all 112 cPUFs were screened against each other. The binary interaction tests resembled the screen from the previous round. The z-score of all screens is shown in the histogram of Fig. S5. The screen revealed 19 interactions among cPUFs, where the degree of connectivity between these interactions is 17.
To further extrapolate these findings, we compared the number of interactions for the cPUFs to the number of interactions between randomly generated protein samples from model bacteria, namely Campylobacter jejuni (25), Helicobacter pylori (26), Treponema pallidum (27), and Escherichia coli K12 (28). The corresponding number of interactions in each random protein set from one organism was extracted from corresponding large-scale Y2H screens. The mean number of interactions in random protein sets was 15 ± 3, except for C. jejuni, which was 44. The first number of interaction is close to the found number of interactions within the cPUFs set. Thus, it is reasonable to assume that the physical relationship between cPUFs resembles that sampled from a random protein set and larger unknown complexes or subnetworks within the set of cPUFs could not be detected.
The distributions of the number of interactions found for random sample sets from each of the four organisms are given in Fig. S6. Apart from the low connectivity, this comparison indicates similarities between the microfluidic and Y2H screens because both technologies work with threshold values for the identification of protein interactions (20). The close match of the mean number of interaction between random sample sets and the cPUFs sets suggest that comparable threshold values for the identification of positive protein interactions were used in the studies. A lower threshold value may then explain the higher mean interaction value for random samples observed for C. jejuni, although we used the core interaction set of this study for our comparison.
Proteome-Wide Interaction Screens.
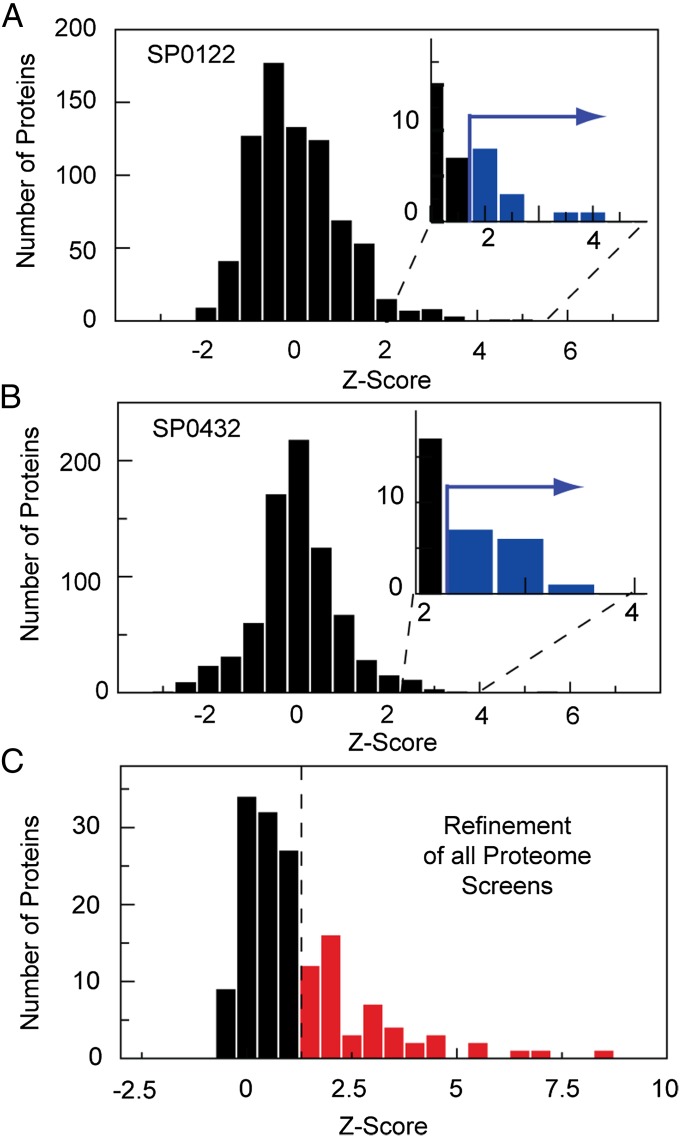
For the last screen we performed proteome-wide interaction screening of selected cPUFs. We chose 18 cPUFs for which no protein interaction was observed in the functional prediction screen. Additionally, 12 cPUFs were selected for which a protein interaction to another cPUF or a functional annotated protein could be detected in the matrix or prediction screen, respectively. Protein interactions of the latter protein set were used for benchmarking of the proteome-wide screen because all interaction partners are included in the expressed proteome on chip and, thus, should be found again. Fig. 3 A and B represents the measured z-score distributions for the two cPUFs when screened against the SP proteome on the microfluidic platform. Measurements were performed in only one pull-down direction. The z-score distributions obtained from proteome-wide screens are broader than those measured in all previous experiments. The reduced number of repeats can explain the broader z-score distribution. An additional factor contributing to a higher noise level of the mIP assay was the large spectrum of proteins with different physical properties. Nevertheless, we found all previous interactions detected for the 12 cPUFs within the matrix screen in the top 2% of the z-score distribution within the proteome-wide screen.
Fig. 3.
Proteome-wide protein–protein interaction screens. (A and B) The histogram of the z-score distributions obtained for the protein interaction measurement on chip between the Streptococcus pneumonia proteome set with cmyc/His tag against the unknown conserved proteins SP0122 and SP0432, respectively. Measurements were performed unidirectionally. (Insets) The z-scores for the top ∼1% of interactions, which were selected for refinement measurements. (C) The histogram of the mean z-score distribution of the top ∼1% of identified protein interactions from all performed proteome-wide screens repeated in bidirectional order with four repeats. The dotted line shows the cutoff value determined from the benchmark set.
To work consistently in the evaluation of positive interactions, we pooled all proteins found in the top 0.5–2% of the z-score distributions of the single proteome-wide screens of the cPUFs and subjected them to an additional refinement screen. The refinement screen resembled the previous screens in number of repeats and two pull-down directions. The z-score distribution of the refinement screen for all 30 cPUFs is shown in Fig. 3C. It was then possible to extract the positive cPUF interactions from the refinement screen with a high confidence level by using the threshold value determined in the yeast benchmark experiments. For the 12 cPUFs with 49 previous identified interactions from the matrix and prediction screen, 28 were again identified within the proteome-wide screens. The new detected protein interactions for all cPUFs are listed in Table S4.
Discussion
The three screening rounds lead to 163 new identified protein interactions for the group of cPUFs from the SP TIGR4 strain. Fig. 4A shows a network representation of the determined interactions. For 78 of 112 investigated cPUFs, at least one interaction partner was found. Before using an algorithm to assign functional annotations, we manually inspected the protein-interaction data for biological and functional consistency. One example is given here. The cPUFs SP1748, a predicted RNA binding protein, and SP1746 form an interaction cluster with SP1079, which belongs to the obg GTPase family. Obg family proteins play a role in the biogenesis of ribosomes, regulation of the cell cycle, and stress response (29). We found additional proteins interacting or located one edge next to SP1746 and SP1748, which are associated with ribosome modification and stress response, namely: SP0434, a stress-response protein; FtsJ, a protein methylating the 23S rRNA for ribosome regulation; nusB, a termination factor for transcription; the 50S ribosomal protein L21; and the transcription factor MarR with unknown activation response. Accumulation of similar functional groups was also found in other clusters. To increase the confidence of the acquired protein interactions we selected 16 protein interactions out of the positive set of the cPUFs and validated them off chip with classic ELISA in one pull-down direction only. Of the 16 protein interactions, 10 were positively identified in the ELISA experiments (Table S4).
Fig. 4.

(A) Graph of the protein interaction network determined for the cPUFs of S. pneumoniae by microfluidic chip technology. Black and red nodes denote the cPUFs and proteins with functional annotation, respectively. Proteins in the dashed box are ribosome associated; the grey node is cPUF SP1748). All genes with annotations and interaction partners are given in detail in SI Materials and Methods. (B) The functional categories found for the cPUFs based on the majority rule and functional prediction.
General functional annotation for the cPUFs was derived by applying the majority rule to the acquired protein-interaction set. For this process, we followed the method as described in ref. 4, in which interaction between two nonfunctional classified proteins are taken into consideration. Although there are numerous alternative algorithms to the majority rule for assigning protein functions based on their interactions, we use the majority rule because of its simplicity coupled with high accuracy. cPUFs were classified into the main functional categories of the Clusters of Orthologous Groups (COG) (NCBI) nomenclature. Fig. 4B summarizes all determined functional classes for the cPUFs (blue bars). The number of found members in each class is normalized to the total number of SP proteins currently assigned to the class (NCBI). The analysis leads to putative functional assignments of 48 cPUFs to COG categories. The cPUFs are almost evenly distributed over all of the general functional classes. This result supports the finding that the group of cPUFs as a whole resembles a randomly sampled collection of proteins. We further added to the bar graph in Fig. 4B functional assignments for the cPUFs calculated with the majority rule based on the functional predicted genomic association partners (red bars). The overall representation of functional groups among the cPUFs is only slightly changed; however, the annotation COG class for a particular cPUF overlaps only for 42% of the investigated proteins.
The rapidly increasing number of full genome sequences and the correspondingly increasing number of ORFs with no functional information about the encoded protein are raising the demand for experimental methods to characterize protein function. None of the identified 163 SP cPUF protein interactions discovered in this work were reported in prior interaction data repositories. That only 42% of the in silico annotation overlap with the annotations resulted from the protein-interaction approach can be an indicator for uncertainties in the various approaches. Despite uncertainties and global assignments, our collected data are unique experimental data for these proteins, and the annotations can be used as a launching point for deeper functional experiments. The comparable degree of connectivity among the cPUFs compared with random protein samples indicates that the major signaling networks and functional groups common to many organisms have been exhaustively determined. Finally, we demonstrated that the microfluidic technology for measuring protein interactions is comparable with other high throughput technologies. Because of high error rates of the high-throughput protein interaction technologies, including the microfluidic chip technique, oversampling of datasets will still be required in future. The benchmark experiments of the microfluidic chip revealed a larger overlap with classic protein-interaction assay techniques than with Y2H and AP-MS techniques. Therefore, the chip technology is working orthogonal to the prevailing techniques with the advantage of lowering the experimental costs of protein–protein interaction sampling.
Materials and Methods
Microfluidic Chip.
The microfluidic protein-interaction assay and the general chip layout are described in SI Materials and Methods and are available upon request. The production of the multilayer PDMS chip followed standard protocols as published in ref. 19 and given on the Stanford Microfluidic Foundry Wep page (http://www.stanford.edu/group/foundry). Additionally, the Stanford Microfluidics Foundry will supply fabricated chips at cost to the academic community.
cDNA Library Construction.
The construction of linear cDNA expression templates for both the yeast benchmark set (pDONR221 clones provided by Haiyuan Yu, Cornell University, Ithaca, NY) and S. pneumoniae (pDONR223 clone library obtained from the J. Craig Venter Institute) were generated by a two-step extension PCR. In the first step, the target sequence was amplified with general primers for the source vectors, and in the second step, the ORF sequences were extended with appropriate 5′ UTR and 3′ UTR sequences required for the IVTT (Promega TNT7-specific) and pull-down system (6xHis, T7, and cMyc tags) used on chip. PCR conditions for both steps resembled the vendor description (Platinum, Invitrogen and Phusion, New England Biolabs). All primer sequences are given in SI Materials and Methods.
Microarray Production.
Selected DNA expression templates with a concentration between 0.1 and 0.2 µg/µL in a 0.8% BSA, 10 mM phosphate buffer (pH 7.2) solution, were spotted onto epoxy-coated glass slides (2 inches × 3 inches) (ThermoFisher). OmniGrid Micro microarray printer with a custom-made print-head holding 2 × 5 silicon pins (Parallel Synthesis Technologies) were used for contact printing of the cDNAs on the glass slides. Microarrays between experiments varied between two set-ups. Within the first set-up we spotted one layer of cDNAs on each spot location, whereas in the second set-up a second layer was deposited on top of the first. In the latter set-up cDNAs of both bait and prey proteins were colocalized on the microarray. The print for the second layer was started after drying the first layer for 4 h. After printing, the microarrays were aligned and bounded to the PDMS chip for 5 h at 65 °C. Bound chips were used within 1 wk and not stored for longer.
Data Evaluation of the Protein-Interaction Chip.
Fluorescence images were analyzed using GenePix v.6.0 software to determine median fluorescence intensities from the pull-down area of the bait protein on the glass slide. The area corresponds to the button valve area (∅22 µm) and position within one unit cell of the PDMS chip and can clearly be visualized on the images. Protein-interaction spots with obvious nonuniform high-intensity signals caused by accumulated impurities during the chip run were flagged and excluded from further analysis. For each binary interaction measurement, two negative control experiments were included on chip (i.e., the no-bait and no-prey experiments). Controls were arranged such that they were locally close to real binary interaction measurement. Downstream analysis was performed using Matlab 7.0 (Mathworks). Local intensity variation of all spots on the images, resulting from the scanner and other fluorescence background sources, were corrected by subtracting a median local background value. The local background was measured in the surrounding of the pull-down spot (three-times the radius of the spot). For normalization of the experiments, we calculated a z-score. For this process, we used the intensity signal of the no-prey and no-bait experiments, which were normally distributed with some outliners. Normalization of the interaction data were then archived by fitting a Gaussian function to the distribution of all control spots. The mean value of the Gaussian fit was subtracted from all measurements and the resulting value was then divided by the SD of the Gaussian fit, resulting in a z-score for each interaction measurement. Outliners of the control spots with an intensity four times higher than the mean of all control spots were excluded, together with corresponding interaction spots because they indicate cross reactivity of proteins with the antibodies or precipitation of the in vitro-synthesized protein.
Supplementary Material
Acknowledgments
We thank D. Gerber and D. Tran for help in constructing the SP cDNA library. H. Yu kindly provided the yeast clones for the benchmark set. This work was funded by the National Institute of Health and Howard Hughes Medical Institute, and the Alexander von Humboldt Foundation (M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper are archived in the IntAct database.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210634110/-/DCSupplemental.
References
- 1.Kelly W, Stumpf M. Protein-protein interactions: From global to local analyses. Curr Opin Biotechnol. 2008;19(4):396–403. doi: 10.1016/j.copbio.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. Systems biology: A brief overview. Science. 2002;295(5560):1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 3.Mayer ML, Hieter P. Protein networks-built by association. Nat Biotechnol. 2000;18(12):1242–1243. doi: 10.1038/82342. [DOI] [PubMed] [Google Scholar]
- 4.Vazquez A, Flammini A, Maritan A, Vespignani A. Global protein function prediction from protein-protein interaction networks. Nat Biotechnol. 2003;21(6):697–700. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- 5.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402(6757):83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 6.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 7.Hishigaki H, Nakai K, Ono T, Tanigami A, Takagi T. Assessment of prediction accuracy of protein function from protein—Protein interaction data. Yeast. 2001;18(6):523–531. doi: 10.1002/yea.706. [DOI] [PubMed] [Google Scholar]
- 8.Shah I, Hunter L. Predicting enzyme function from sequence: A systematic appraisal. Proc Int Conf Intell Syst Mol Biol. 1997;5:276–283. [PMC free article] [PubMed] [Google Scholar]
- 9.Raes J, Harrington ED, Singh AH, Bork P. Protein function space: Viewing the limits or limited by our view? Curr Opin Struct Biol. 2007;17(3):362–369. doi: 10.1016/j.sbi.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96(6):2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci USA. 2002;99(9):5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson CA, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol. 2000;297(1):233–249. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- 13.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 14.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6(1):91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y-C, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods. 2010;7(9):667–668, author reply 668. doi: 10.1038/nmeth0910-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerber D, Maerkl SJ, Quake SR. An in vitro microfluidic approach to generating protein-interaction networks. Nat Methods. 2009;6(1):71–74. doi: 10.1038/nmeth.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melin J, Quake SR. Microfluidic large-scale integration: The evolution of design rules for biological automation. Annu Rev Biophys Biomol Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 20.von Mering C, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417(6887):399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 21.Yu NY, et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin MY, Koonin EV. ‘Conserved hypothetical’ proteins: Prioritization of targets for experimental study. Nucleic Acids Res. 2004;32(18):5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishi T, Ikemura T, Kanaya S. GeneLook: A novel ab initio gene identification system suitable for automated annotation of prokaryotic sequences. Gene. 2005;346:115–125. doi: 10.1016/j.gene.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz AS, Yu J, Gardenour KR, Finley RL, Jr, Ideker T. Cost-effective strategies for completing the interactome. Nat Methods. 2009;6(1):55–61. doi: 10.1038/nmeth.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrish JR, et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8(7):R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rain JC, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409(6817):211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 27.Titz B, et al. The binary protein interactome of Treponema pallidum—The syphilis spirochete. PLoS ONE. 2008;3(5):e2292. doi: 10.1371/journal.pone.0002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arifuzzaman M, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16(5):686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato A, et al. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10(5):393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.