Abstract
The extensive carbohydrate coat, the variability of protein structural features on HIV-1 envelope glycoproteins (Env), and the steric constraints of the virus-cell interface during infection, present challenges to the elicitation of effective full-length (∼150 kDa), neutralizing antibodies against HIV. These hurdles have motivated the engineering of smaller antibody derivatives that can bind Env and neutralize the virus. To further understand the mechanisms by which these proteins neutralize HIV-1, we carried out cryoelectron tomography of native HIV-1 BaL virions complexed separately to two small (∼15 kDa) HIV-neutralizing proteins: A12, which binds the CD4-binding site on Env, and m36, whose binding to Env is enhanced by CD4 binding. We show that despite their small size, the presence of these proteins and their effects on the quaternary conformation of trimeric Env can be visualized in molecular structures derived by cryoelectron tomography combined with subvolume averaging. Binding of Env to A12 results in a conformational change that is comparable to changes observed upon its binding to the CD4-binding site antibody, b12. In contrast, binding of Env to m36 results in an “open” quaternary conformation similar to that seen with binding of soluble CD4 or the CD4i antibody, 17b. Because these small neutralizing proteins are less sterically hindered than full-length antibodies at zones of virus-cell contact, the finding that their binding has the same structural consequences as that of other broadly neutralizing antibodies highlights their potential for use in therapeutic applications.
Keywords: gp120, gp41, cryoelectron microscopy, AIDS vaccine, virus entry
The HIV-1 envelope glycoprotein (Env) is anchored in the viral membrane and facilitates infection through its interaction with T-cell membrane proteins. Env is a trimer of dimers composed of gp120 and gp41 polypeptides, which associate noncovalently on the surface of the virus. Three copies of this heterodimer assemble to form a functional Env trimer “spike” that is visible on the viral surface in electron micrographs of purified virus particles. HIV entry into the cell is initiated when gp120 makes contact with the cell surface receptor CD4. The quaternary molecular structures of Env and the associated conformational changes that result from its binding to CD4 and numerous monoclonal antibodies have been analyzed by cryoelectron tomography (1–5). These studies have identified three distinct quaternary conformations of trimeric Env. A “closed” conformation, defined by the close positioning of adjacent gp120 V1/V2 loops at the apex of the spike, is observed when trimeric Env is unliganded and when it is bound to the CD4-binding site-directed neutralizing antibodies VRC01, VRC02, or VRC03 (2, 3). A second, “partially open” conformation is found when Env is bound by the CD4-binding site antibody, b12, and is characterized by a slight outward and rotational displacement of the gp120 monomers with respect to the central axis of the spike (2). Finally, a third, “open” Env structure featuring a quaternary conformation with large rearrangements of gp120 and gp41 is observed upon binding of soluble CD4 or the CD4-induced (CD4i) antibody, 17b (1–3).
Successful protein engineering efforts have yielded an array of small proteins and single domain antibody derivatives that are capable of neutralizing HIV-1 (6-11). Single domain antibody derivatives (sdAb), whether engineered or extracted from full-length antibodies, correspond to the smallest independently folded antibody domain that retains specificity for a target epitope (Fig. 1A). Antibody derivatives are being used in a variety of applications including treatment of autoimmune diseases and the diagnosis and treatment of cancers, with many derivatives either FDA approved or in preclinical or clinical trials (12, 13). Derivatives targeting HIV-1 have been engineered, and a subset of these proteins has been shown to have neutralization potency comparable to or better than that of full-size IgG antibodies, despite having a 10-fold smaller size (∼15 kDa versus ∼150 kDa) (6, 14). These smaller proteins can also have additional advantages, such as greater thermal and pH stability, better biocompatibility and tissue permeability, lower cost, and increased ease of production of recombinant protein in aqueous and lyophilized formulations (7).
Fig. 1.
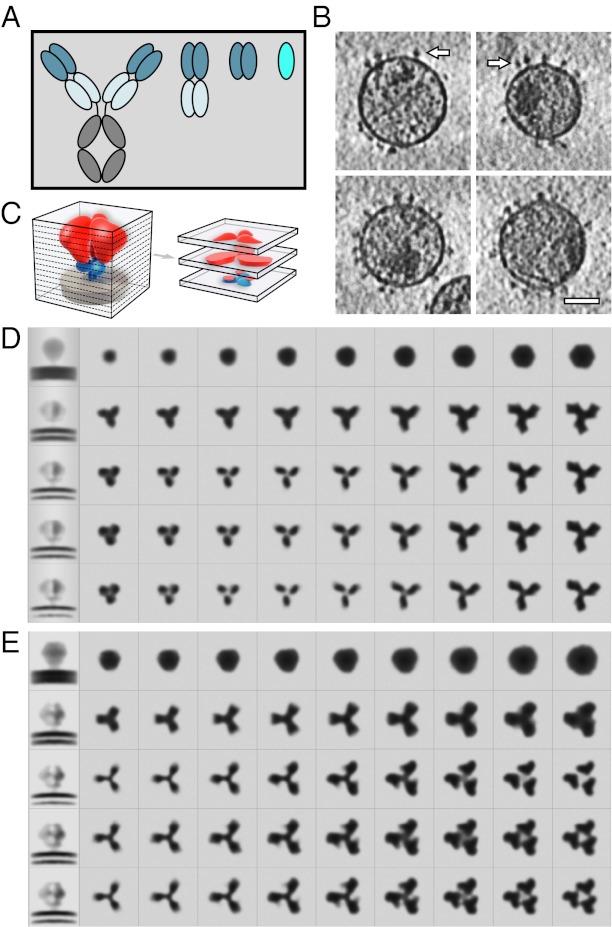
Schematic representation of domain antibody architecture and steps in structure determination. (A) Full-size IgG antibody and its derivatives. From left to right: IgG (∼150 kDa), Fab (∼50 kDa), scFv (∼25 kDa), and a single domain (∼15 kDa). (B) Slices through representative tomograms illustrating the presence of envelope glycoprotein spikes (shown with white arrows) on the surface of the viral membrane. (Scale bar: 50 nm.) (C) Schematic illustrating Env trimer structure represented as a series of horizontal slices through the structure volume. Env is shown with gp120 in red, gp41 in blue, and the viral membrane in gray. (D and E) Slices through the 3D structure of Env in the unbound (D) and A12-bound (E) states, showing progressive improvement in structure with successive rounds of refinement. The first column in each row shows a central, vertical slice through the volume. The remaining columns show successive horizontal slices through the Env volume beginning at the viral membrane (second column) and ending near the apex of the trimer (final column).
One class of such molecules is derived from llama antibodies. Like humans and most other mammals, llamas produce antibodies containing light chains and heavy chains with both variable and constant domains. However, llamas and other members of the Camelidae family also produce a subset of antibodies that have heavy chains but lack light chains (15). The variable region of these antibodies is ∼15 kDa, comprising a single domain. Three such constructs of llama heavy chain-only antibodies (termed “VHH”) were recently isolated and shown to target gp120 with picomolar dissociation constants. Each VHH construct displays low IC90 values in neutralization assays, and neutralizes HIV-1 subtypes B and C in a manner similar to that seen with other broadly neutralizing antibodies (10). These antibodies were identified by immunizing llamas with recombinant gp120, selecting the resulting antibody repertoire, and then using directed evolution via phage display to refine the affinity for gp120. Biochemical studies of three VHH proteins (D7, C8, and A12) showed that these proteins target the CD4-binding site of gp120. Inspection of the crystal structure of the complex of monomeric gp120 with A12 [Protein Data Bank (PDB) ID code: 3RJQ], the most potent of these constructs, confirms this prediction. Another class of molecules produced through directed evolution of the complementarity-determining region of the variable portion of a human IgG heavy chain is domain antibody m36, which can neutralize HIV-1 primary isolates from clades A to D at low nanomolar concentrations (6). The binding affinity of m36 and its neutralization efficacy are enhanced by the presence of soluble CD4 (sCD4), placing m36 in the CD4i category of HIV-1 neutralizing proteins (16).
Understanding the structural aspects of the interaction of proteins such as A12 and m36 with Env on intact viruses is important for understanding their function in virus neutralization. Here, we have used cryoelectron tomography to determine structures of A12, m36, or m36/sCD4 complexed to trimeric Env displayed on intact HIV-1 BaL virus. By applying emerging techniques for subvolume averaging and 3D image classification (3, 17), we obtained density maps for these complexes at ∼20 Å resolution. These structures identify the locations of the bound proteins on gp120 and reveal the ligands’ effects on the conformation of trimeric Env. By fitting crystallographic structures of subcomponents of the protein complex into our maps, we derived trimeric molecular models of the A12-Env, m36-Env, and m36/sCD4-Env complexes. We then compared these models to previously determined structures of trimeric Env bound to other ligands to interpret the structural effects of small ligand interactions with Env.
Results and Discussion
Quaternary Structure of Trimeric Env Bound to Llama Antibody Fragment VHH A12.
Subvolume averaging and iterative classification methods allow determination of the 3D structure of trimeric Env displayed on the surface of the virus. These methods have been used for structural analysis of trimeric Env bound to a variety of antibodies and ligands (1–5). To resolve these structures, HIV-1 BaL viruses displaying native Env alone or in complex with added ligand were imaged in 3D by using cryoelectron tomography (Fig. 1B, Env is indicated by white arrows). From the resulting 3D images (tomograms), the individual Env subtomograms were extracted and subjected to iterative alignment and classification. The averaged 3D Env structures were progressively refined, providing visualization of increasing structural detail with successive steps (Fig. 1 C–E). Slices through the final 3D averages of unbound and A12-bound Env (second through final columns of Fig. 1D and Fig. 1E, respectively) show that in comparison with unliganded trimeric Env, the gp120 protomers in the A12-bound state are slightly farther apart at the apex and feature additional density protruding from each protomer. The intensity distribution in these maps reflects the averaged density from structurally homogeneous and well-ordered elements in the protein. N-linked oligosaccharides on the surface, which are likely to be diffuse and partially disordered, contribute much less to the overall density in proportion to ordered polypeptide regions with the same mass.
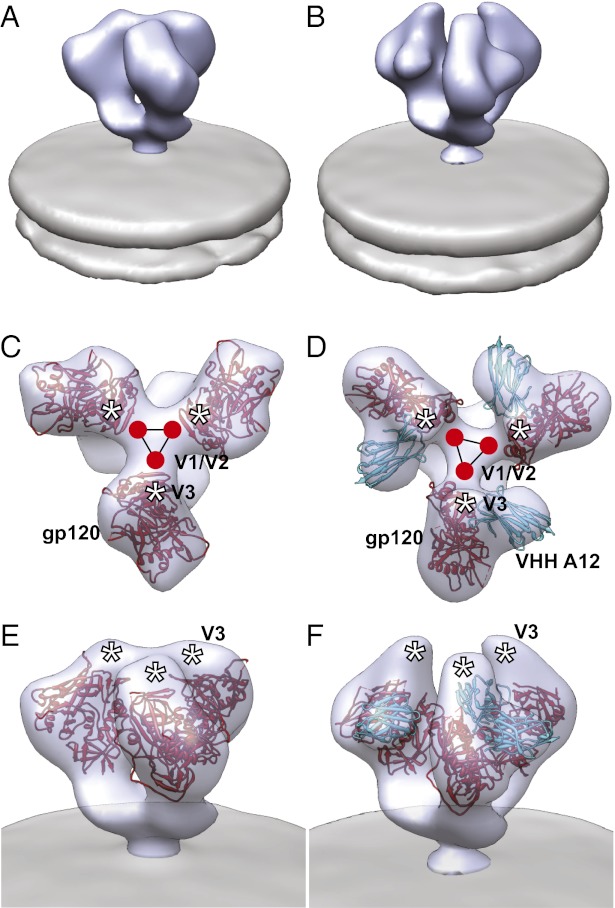
Visualization of the averaged density maps as isosurfaces also clearly shows the presence of additional density in A12-bound Env compared with the unliganded state (Fig. 2 A and B). Automated fitting of three copies of the structure of A12-liganded gp120 derived by X-ray crystallography into the density map generates a model for the molecular architecture of this trimeric complex (Fig. 2 D and F). The A12 coordinates overlap precisely with the location of the additional density observed in the density maps, highlighting the differences in quaternary structure of Env between unliganded (Fig. 2 A, C, and E) and A12-bound states (Fig. 2 B, D, and F). A12 binding is also observed to result in a ∼20–25° rotation of each gp120 protomer around an axis parallel to the central threefold axis, repositioning the V1/V2 loop regions away from the apex of the spike. This rotation of each protomer is similar to that seen with b12 binding and may be required to avoid steric interaction of bound A12 with neighboring protomers as in the case of b12.
Fig. 2.
A12-bound Env trimer displays partially open conformation. (A and B) Perspective views of density maps for Env in the unbound (A) and A12-bound (B) states. Env density is shown as a solid blue contour with the viral membrane in gray. (C–F) Top (C and D) and perspective (E and F) views of the density map and molecular architecture of unbound (C and E) and A12-bound (D and F) trimeric Env. Env density is shown as a transparent blue isosurface, and the viral membrane is shown in gray. Maps for unbound (C and E) and A12-bound (D and F) Env were fitted with the trimeric molecular model for gp120 in the unbound state (fitted with the molecular trimer model for unbound Env, PDB ID code 3DNN, displayed with the more recent gp120 protomer coordinates from PDB ID code 3NGB) and cocrystal coordinates for the gp120–A12 complex (PDB ID 3RJQ), respectively. Coordinates show gp120 (red) and VHH A12 (cyan). The estimated locations of the V1/V2 and V3 loop stumps are highlighted by the red circles and white asterisks, respectively, and the black triangle is shown as a visual guide to follow the rotations of the three gp120 protomers with A12 binding.
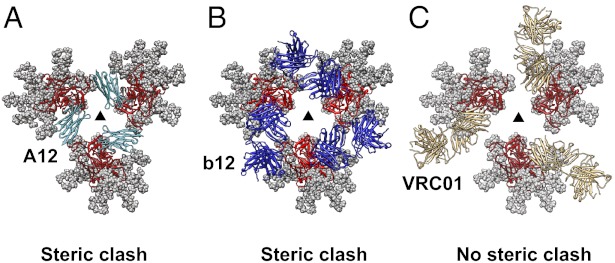
To further interpret the structural impact of A12 binding in the context of other antibody-bound Env structures, we compared the A12-bound Env structure with those for b12 and VRC01, because all three ligands bind gp120 at the CD4 binding site. The rotation in gp120 observed with A12 binding is closely comparable to that observed with binding of b12, in contrast to the effect of VRC01 binding, which causes no significant change in Env quaternary structure. To describe these differences in more detail, we created models for the molecular architecture of each of these antibody-complexed Env trimers in which the three gp120 protomers are maintained in the same quaternary conformation as that observed for unliganded trimeric Env (Fig. 3). The modeling suggests that A12 is unable to bind gp120 if Env remains in the closed quaternary conformation because, in this state, the distal end of bound A12 would be too close to the neighboring gp120 protomer (Fig. 3A). Thus, although A12, b12, and VRC01 bind very similar regions of the CD4 binding site on gp120, the steric interactions at the distal ends of the bound antibody moieties are likely to play a role in determining whether the binding requires small rotations in gp120, as in the case of A12 (Fig. 3A) and b12 (Fig. 3B) or can be accommodated without any quaternary structural change, as in the case of VRC01 (Fig. 3C).
Fig. 3.
In contrast to VRC01, A12 or b12 cannot be accommodated on native trimeric Env without steric clash. (A) Top view of a hypothetical, glycosylated model for A12-bound Env if no conformational changes occurred in the trimer upon A12 binding. Three copies of the complex of A12 with monomeric gp120 (PDB ID code 3RJQ) were aligned to the trimeric molecular model for gp120 in the unliganded state (PDB ID code 3DNN). (B and C) Top views of a model for glycosylated trimeric gp120 in the unliganded state, as in A, but with either b12 (B) or VRC01 (C) Fab bound. The models show that steric clashes would occur if A12 or b12 were to bind the native state without conformational rearrangements. Coordinates show gp120 (red), VHH A12 (cyan, PDB ID: 3RJQ), b12 Fab (blue, PDB ID: 2NY7), and VRC01 Fab (gold, PDB ID: 3NGB).
Quaternary Structure of Trimeric Env Bound to m36 With and Without sCD4.
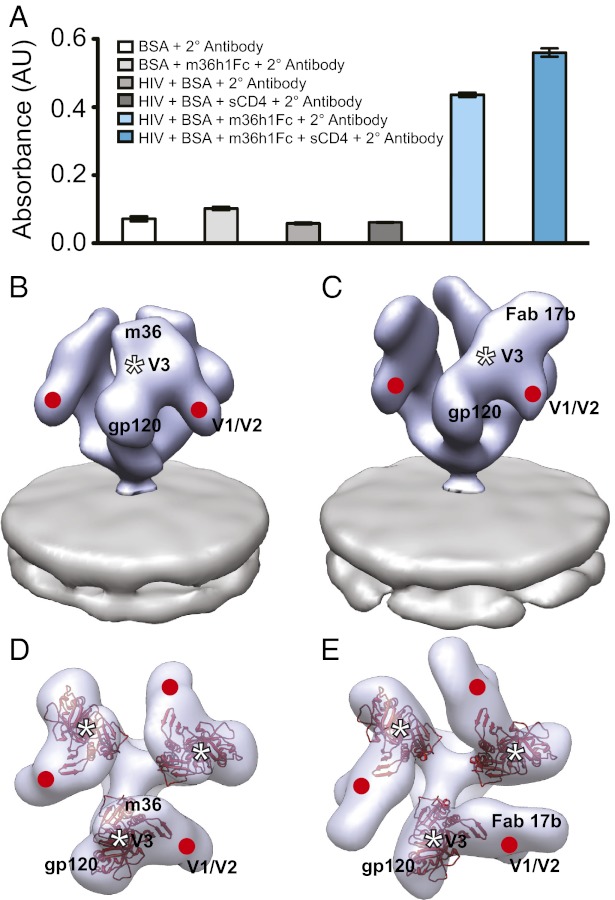
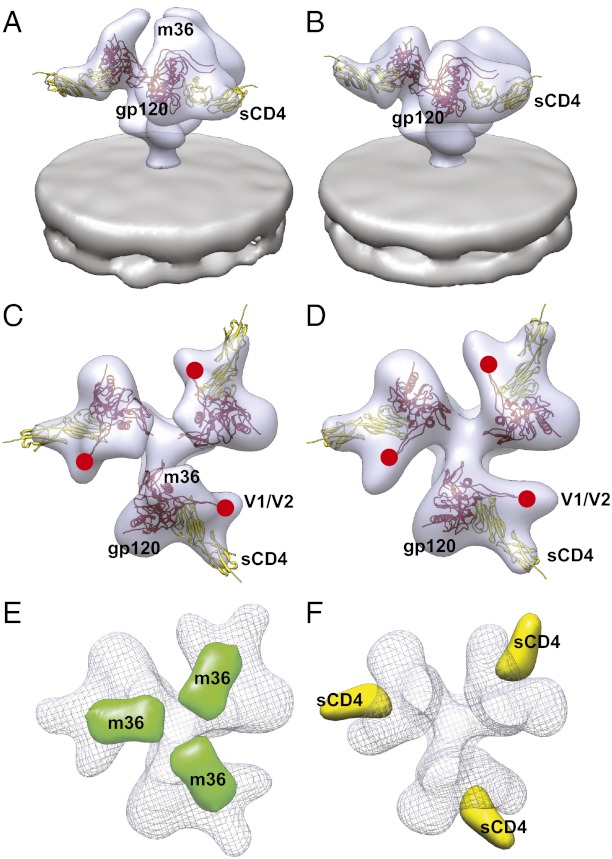
Unlike the A12 domain antibody, which binds the CD4 binding site, m36 is reported to bind a region of gp120 made more accessible by the binding of CD4 (6, 18). To establish whether m36 can also bind gp120 in the context of native Env, we measured m36 binding to HIV-1 BaL in the presence and absence of sCD4. Binding was measured by using an ELISA adapted for use with whole HIV or SIV virions (5). The ELISA method uses a construct with an antibody Fc region fused to m36 (m36h1Fc). The Fc region facilitates binding of a secondary antibody carrying a reporter enzyme (6). ELISA experiments show that m36 displays binding to native trimeric Env that is measurably increased in the presence of sCD4 (Fig. 4A). In this respect, m36 features binding properties similar to those seen with 17b, a well-characterized coreceptor-targeted CD4i monoclonal antibody (3). Determination of the 3D structure of m36 complexed to trimeric Env by subvolume averaging reveals that the m36-bound conformation is dramatically different from that of unliganded trimeric Env (Fig. 4 B and D), with changes similar to that seen in the 17b-bound conformation (Fig. 4 C and E). In each case, the binding results in a large displacement of the density corresponding to the V1/V2 loops away from the center of the spike, along with an out-of-plane rotation of each gp120 protomer. m36-bound Env thus appears to display the same open quaternary conformation as that observed after binding of 17b, 17b/sCD4, and sCD4 alone (3).
Fig. 4.
m36 binding results in formation of activated, open conformation of trimeric Env. (A) Binding of m36h1Fc and m36h1Fc with sCD4 to Env on whole virions as measured by enzyme-linked immunosorbance assay (ELISA). The first four bars in the graph give absorbance measurements for negative controls of nonspecific interactions between experimental constituents (HIV, BSA, secondary antibody, m36h1Fc, and sCD4). The final two bars give absorbance measurements for m36h1Fc bound to native Env both in the absence and presence of sCD4. (B–E) Perspective (B and C) and top (D and E) views of density maps of Env bound by m36 (B and D) or 17b (C and E) (3). Env density is shown in blue with one solid contour (B and C) or one transparent contour (D and E), and the viral membrane is shown in gray. Because crystallographic models are not available for the gp120–m36 complex, top views for both m36-bound (D) and 17b-bound (E) Env are fitted with the molecular model for gp120 (red) in the open conformation originally derived for the sCD4/17b conformation of trimeric Env (PDB ID code 3DNO, displayed with the more recent gp120 protomer coordinates from PDB ID code 3NGB). The estimated locations of the V1/V2 and V3 loop stumps are highlighted by the red circles and white asterisks, respectively.
The structure of m36-bound Env shows an area of extra density at the apex of the spike, near the coreceptor binding site (Fig. 4B). This extra density is apparent compared with the structure of unliganded Env (Fig. 2A). An atomic model for the structure of m36, either alone or in complex with monomeric gp120, is not yet available. To aid in definitively localizing the site on gp120 where m36 is bound, we therefore determined the structure of the complex of native trimeric Env complexed simultaneously to m36 and sCD4 (Fig. 5 A and C). To confirm the location of m36 binding, we generated a difference map (Fig. 5E) comparing the structure of this m36/sCD4 ternary complex (Fig. 5 A and C) with the structure of trimeric Env complexed to sCD4 alone (Fig. 5 B and D) (3). This map reveals a single peak on each gp120 protomer corresponding to the presence of m36 (Fig. 5E). The footprint of m36 binding on gp120 is near the base of the V3 loop on gp120 in a location similar to that of antibodies 17b and X5 (3, 19, 20). To assess the similarity between the sCD4-bound state and the m36/sCD4-bound state, we generated a second difference map. In this case, the m36-bound Env map was subtracted from the m36/sCD4-bound Env map. Inspection of this difference shows a single set of peaks that correspond to the presence of sCD4 (Fig. 5F). The fact that only a single set of peaks are observed in these two difference maps provides unambiguous evidence that m36 binding, both with and without sCD4, results in the same open conformation that is observed with binding of soluble sCD4 alone. These maps also localize m36 binding to the V3-loop region of native trimeric Env.
Fig. 5.
Difference maps reveal location of m36 binding site on trimeric Env. (A–D) Perspective (A and B) and top (C and D) views of the density maps and molecular models for Env complexed to m36 and sCD4 (A and C) or sCD4 alone (B and D). Because no cocrystal structure is available for gp120 with m36 and sCD4, the m36/sCD4-bound Env density map was fitted with coordinates for the gp120–sCD4 complex (subset from PDB ID code 1GC1); the density map for sCD4-bound Env (B and D) was also fitted with the same coordinates. Red circles mark the estimated locations of the V1/V2 loop stumps. (E and F) Top views of the difference maps that result from subtracting either the Env-sCD4 density map (B and D) or Env-m36 density map (Fig. 4 B and D) from the density map for the Env-m36/sCD4 complex (A and C). Subtracting Env-sCD4 from Env-m36/sCD4 reveals the location of the m36 ligand density (E, green). The analogous subtraction of Env-m36 from Env-m36/sCD4 reveals the sCD4 ligand density (F, yellow). Wire mesh density maps for Env-sCD4 and Env-m36 are shown in E and F, respectively, to provide spatial context for the difference maps.
Three Distinct Conformational States of Trimeric Env.
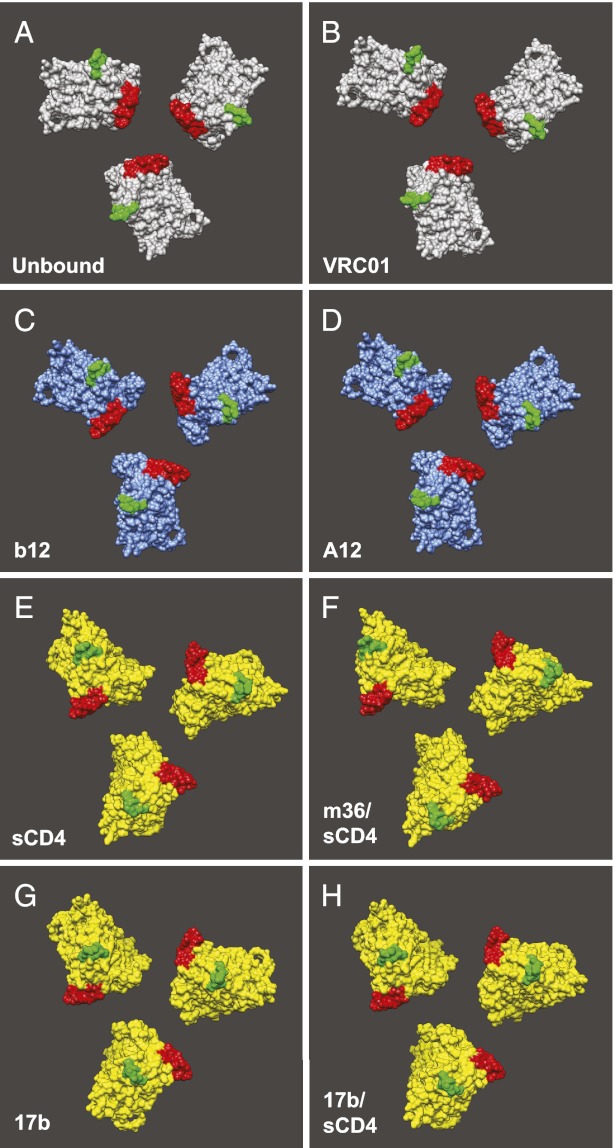
The structural characterization we have presented of native, trimeric Env bound to either A12 or m36 shows that these small antibody derivatives can modulate the quaternary conformation of Env in a manner similar to full-length IgG molecules, resulting in the generation of either partially or fully open Env states. A12 is targeted to the CD4-binding site, whereas m36 recognizes a site on gp120 that is similar to that expected to be recognized by the antibody 17b and coreceptors on the target membrane. Taken together with previous tomographic studies, the present study further confirms the spectrum of distinct quaternary states that can be sampled by native trimeric Env in the context of viral entry and neutralization (summarized in Fig. 6). The consequence of A12 binding is most comparable to that of b12 in that they both cause a small conformational change in Env, which we refer to as a partially open conformation. This partially open state is distinct from the fully open state generated by binding of m36, sCD4, or 17b, in which much larger Env conformational changes are observed, and which we have described recently as an “activated” conformation of Env (3). The structure of the open quaternary conformation of 17b-bound trimeric Env at ∼9 Å resolution provides a glimpse of the significance of the open conformation. In this open state, the three N-terminal helices of gp41 appear to have sprung open into the central portion of trimeric Env that is largely a cavity in the unliganded state. The fact that coreceptor mimics such as m36 and 17b can bind and generate the same types of conformational changes observed with sCD4 binding provides useful insights into early events in the mechanism of viral entry and is in agreement with data for CD4-independent entry of some HIV-1 isolates (21–23). It is likely that the open structures we observe are the conformational prerequisites needed to initiate further conformational changes that ultimately lead to fusion of the viral and plasma cell membranes.
Fig. 6.
Three distinct quaternary conformations of trimeric Env. Trimeric molecular models derived from fitting atomic coordinates into Env density maps. Model configurations are in the closed (gray), partially open (blue), or activated open (yellow) states. Native, unliganded (A) and VRC01-bound (B) structures are found in the closed Env configuration; b12- (C) and A12-bound (D) structures are in the partially open state; sCD4- (E), m36/sCD4- (F), 17b- (G), or 17b/sCD4-bound (H) structures are in the open state. In all models, the V1/V2 and V3 loop stumps are highlighted in red and green, respectively.
The size of m36 is comparable to that of the CD4 binding domain (domain 1) and to the extracellular portion of coreceptors CCR5 and CXCR4; both m36 and the first domain of CD4 that binds to gp120 have the same Ig fold. The three variable loops of m36 that are likely involved in binding to gp120 could also engage in interactions similar to those likely to be formed by the three variable loops of the chemokine receptors. Therefore, one could speculate that this structural similarity of m36 with both CD4 and coreceptors represents a type of functional mimicry for inducing the observed open conformation. At the present resolution, the CD4-, 17b-, and m36-bound structures appear similar and can be fit by the same coordinates, although as the resolution of these structures improves, subtle differences in these structures are expected to appear. Further increases in resolution should also provide additional information on the conformational changes in gp41 that lead to the formation of the activated and prehairpin intermediates that occur before membrane fusion.
Using cryoelectron tomography, we have described the quaternary conformations of a number of trimeric Env complexes displayed on intact viruses and as soluble ectodomains (1–5). When trimeric Env is in the unliganded state, or when it is bound to VRC01, VRC02, or VRC03 antibodies, it is in a closed conformation with the V1/V2 loops located close to the apex of the spike. When trimeric gp120 is bound to CD4, 17b, or m36, it transitions from the closed state to an open state in which the three gp120 monomers undergo a major structural rearrangement involving large rotations of each gp120 monomer. When trimeric Env is bound to b12 or A12, there is a partial opening of the trimer, with a slight rearrangement of each gp120 monomer. One way to interpret these conformational transitions is that the formation of the partially open or fully open conformations is an irreversible, triggered change, driven by the binding of the respective ligands. An alternative way to interpret these observations is to postulate that trimeric spikes are in a dynamic equilibrium between closed, partially open, or open states, and the role of ligand binding is to shift the equilibrium in favor of one of these states. It could also be that the closed and partially open states are in a state of dynamic equilibrium and that the formation of the open state is an irreversible, triggered change. Which of these scenarios is more accurate? Does binding induce the opening, or does the opening precede binding? Our experiments do not allow us to distinguish between these possibilities and, indeed, it may be difficult to classify the changes into one or other extreme scenario. However, as structural details of these conformations begin to be discerned at progressively higher resolutions by using cryoelectron microscopy and other complementary structural and computational approaches, our understanding of these changes can be expected to improve.
Materials and Methods
Sample Preparation.
Quantifoil Multi A Holey Carbon Film grids (Quantifoil Micro Tools) were cleaned in a Solarus plasma cleaner (Gatan). All sample solutions were prepared by premixing HIV-1 BaL virions (Biological Products Core of the AIDS and Cancer Virus Program, SAIC Frederick, Inc.; ∼1011 virions per mL, aldrithiol-2 treated) with saturating amounts of VHH A12, sdAb m36, or sdAb m36 and sCD4, and 10-nm–sized protein-A gold colloid (Utrecht University). Vitrified sample grids were prepared by adding ∼2 μL of sample solution to a grid and immediately blotting at 22 °C and 100% humidity, and plunge freezing in liquid ethane (approximately −180 °C) by using a Mark III Vitrobot (FEI Company).
Cryoelectron Tomography.
Tomographic tilt series were collected by using a Tecnai G2 Polara transmission electron microscope (FEI Company) operated at 200 kV and equipped with an energy filter and 2K × 2K postenergy filter CCD (Gatan). The specimens were maintained at a temperature of approximately −182 °C throughout all steps in imaging. Tilt series were acquired over a ±60° angular range at 2° tilt intervals to yield 61 projections per series. Each projection image in the series was acquired at a nominal magnification of 34,000× (effective 4.1 Å pixel size at the specimen plane) with an average underfocus value of 2.5 μm, and a dose of 1–2 e− per Å2. The tilt series were aligned and reconstructed in IMOD to yield tomograms (24, 25).
Subtomogram Analysis.
Virion subtomograms were manually segmented, down-sampled by a factor of four, denoised by using edge-enhancing anisotropic diffusion as implemented in IMOD, and subjected to unsupervised membrane segmentation by using the energy-based 3D approach described (26). Automated identification of envelope glycoprotein spikes on virion surfaces began by assigning, to each position on the segmented virion surface, the cross-correlation score between a cylindrically symmetric phantom and the local voxel intensities. Automated spike segmentation proceeded by designating those tomogram subvolumes with local cross-correlation maxima, provided the maxima were above a given threshold. Designated subvolumes were segmented from virion surfaces without denoising or binning and had final dimensions of 100 × 100 × 100 voxels. Subvolumes were classified and averaged iteratively by using a collaborative alignment and clustering algorithm based on the concept of minimizing the matrix rank and its convex surrogate, the nuclear norm (27). Refinement was done until class convergence, typically between 8 and 20 iterations.
X-Ray Coordinate Fitting.
Tomographic map visualization was done by using Bsoft, EMAN2, and UCSF Chimera software packages at different stages of data processing (28–30). X-ray coordinate fitting to tomographic maps was done by using the steepest-ascent local optimization utility in UCSF Chimera. Coordinates were initially placed in random orientations in the density maps, and the local maxima of the sum of pointwise products between the coordinates and the density map were computed repeatedly until convergence (typically 4–5 repeats per fit). Fitting was done until convergence was achieved. Coordinates for the gp120 core were extracted from the cocrystal structure for gp120 with VRC01 (PDB ID code 3NGB). Coordinates for gp120 with sCD4 were extracted from the ternary crystal structure for gp120 with 17b and sCD4 (PDB ID code 1GC1). Because no cocrystal structures for m36 with gp120 were available, the molecular model for m36/sCD4-bound Env was built by using the extracted coordinates for gp120/sCD4. Despite the absence of m36 in the coordinates, sCD4 provided sufficient constraint for proper density map fitting. Coordinates for gp120 with VHH A12 (PDB ID code 3RJQ), b12 Fab (PDB ID code 2NY7), and VRC01 Fab (PDB ID code 3NGB) were used as deposited in the PDB. Carbohydrates were modeled onto the gp120 sequence by using the GLYPROT module at the publicly available website, www.glycosciences.de.
Whole Virion ELISA.
Whole, aldrithiol-2–treated HIV-1 BaL virions in buffer TNE (100 mM Tris, 150 mM NaCl, and 1 mM EDTA at pH 7.5) were incubated overnight at 4 °C in the wells of a clear, flat-bottom 96-well plate (Corning; product no. 3596). Unless otherwise specified, all subsequent steps were done at 4 °C with washes done with TNE. After incubation, wells were washed once, blocked with cold 1% BSA for 1 h, and washed three more times. Primary antibody (m36h1Fc fusion construct) was added to virus-coated wells and incubated for 1 h before washing wells three times. Soluble sCD4 was added with m36h1Fc for the corresponding experiment. Alkaline phosphatase-conjugated secondary antibody against IgG Fc (Sigma-Aldrich; product no. A9544) was diluted in buffer TNE in accordance with manufacturer instruction, added to primary antibody wells, and incubated for 1 h before washing wells three times. A solution of 4-nitrophenyl phosphate (Sigma-Aldrich; product no. N2765, 1 mg/mL) in phosphatase substrate buffer (50 mM NaHCO3 and 1 mM MgCl2 at pH 9.8) was added to wells, and the plate was rocked in the dark at 25 °C. Absorbance was measured at 405 nm (Molecular Devices; SpectraMax 250). Experiments and controls were done in triplicate, and controls against nonspecific interactions of primary and secondary antibodies were performed in parallel.
Acknowledgments
We thank Steven Fellini, Susan Chacko, and colleagues for support with our use of the high-performance computational capabilities of the Biowulf Linux cluster at National Institutes of Health (NIH) (http://biowulf.nih.gov); Dr. Dan Shi and Robert Mueller for assistance with electron microscopy; Dr. Kedar Narayan for assistance with whole virion ELISA experiments; Dr. Lesley Earl for helpful comments; Ethan Tyler for assistance with preparing figures; and the NIH AIDS reagent program for providing sCD4. This work was supported by funds from the NIH Intramural AIDS Targeted Antiviral Program and Center for Cancer Research at the National Cancer Institute (to S.S.), the NIH-Cambridge Scholars Program and an NIH Intramural AIDS Research Fellowship (to J.R.M.), and National Cancer Institute, National Institutes of Health Contract HHSN261200800001E (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Harris A, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci USA. 2011;108(28):11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran EEH, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8(7):e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White TA, et al. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol. 2011;85(23):12114–12123. doi: 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: Strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6(12):e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Zhu Z, Feng Y, Dimitrov DS. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc Natl Acad Sci USA. 2008;105(44):17121–17126. doi: 10.1073/pnas.0805297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorlani A, et al. Llama antibody fragments have good potential for application as HIV type 1 topical microbicides. AIDS Res Hum Retroviruses. 2012;28(2):198–205. doi: 10.1089/aid.2011.0133. [DOI] [PubMed] [Google Scholar]
- 8.McCoy LE, et al. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J Exp Med. 2012;209(6):1091–1103. doi: 10.1084/jem.20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Dimitrov DS. Human monoclonal antibodies and engineered antibody domains as HIV-1 entry inhibitors. Curr Opin HIV AIDS. 2009;4(2):112–117. doi: 10.1097/COH.0b013e328322f95e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsman A, et al. Llama antibody fragments with cross-subtype human immunodeficiency virus type 1 (HIV-1)-neutralizing properties and high affinity for HIV-1 gp120. J Virol. 2008;82(24):12069–12081. doi: 10.1128/JVI.01379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moulaei T, et al. Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure. 2010;18(9):1104–1115. doi: 10.1016/j.str.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 13.Hudson PJ, Souriau C. Engineered antibodies. Nat Med. 2003;9(1):129–134. doi: 10.1038/nm0103-129. [DOI] [PubMed] [Google Scholar]
- 14.Strokappe N, et al. Llama antibody fragments recognizing various epitopes of the CD4bs neutralize a broad range of HIV-1 subtypes A, B and C. PLoS ONE. 2012;7(3):e33298. doi: 10.1371/journal.pone.0033298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 16.Labrijn AF, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77(19):10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank GA, et al. Computational separation of conformational heterogeneity using cryo-electron tomography and 3D sub-volume averaging. J Struct Biol. 2012;178(2):165–176. doi: 10.1016/j.jsb.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Xiao X, Wang Y, Zhu Z, Dimitrov DS. Bifunctional fusion proteins of the human engineered antibody domain m36 with human soluble CD4 are potent inhibitors of diverse HIV-1 isolates. Antiviral Res. 2010;88(1):107–115. doi: 10.1016/j.antiviral.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317(5846):1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBranche CC, et al. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol. 1999;73(12):10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dumonceaux J, et al. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72(1):512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang PF, et al. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol. 2002;76(2):644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 25.Amat F, et al. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161(3):260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Bartesaghi A, Sapiro G, Subramaniam S. An energy-based three-dimensional segmentation approach for the quantitative interpretation of electron tomograms. IEEE Trans Image Process. 2005;14(9):1314–1323. doi: 10.1109/TIP.2005.852467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuybeda O, et al. 2012. A collaborative framework for 3D alignment and classification of heterogeneous subvolumes in cryo-electron tomography. J Struct Biol, 10.1016/j.jsb.2012.10.010.
- 28.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157(1):3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]