Abstract
Leaf-cutting ants combine large-scale herbivory with fungus farming to sustain advanced societies. Their stratified colonies are major evolutionary achievements and serious agricultural pests, but the crucial adaptations that allowed this mutualism to become the prime herbivorous component of neotropical ecosystems has remained elusive. Here we show how coevolutionary adaptation of a specific enzyme in the fungal symbiont has helped leaf-cutting ants overcome plant defensive phenolic compounds. We identify nine putative laccase-coding genes in the fungal genome of Leucocoprinus gongylophorus cultivated by the leaf-cutting ant Acromyrmex echinatior. One of these laccases (LgLcc1) is highly expressed in the specialized hyphal tips (gongylidia) that the ants preferentially eat, and we confirm that these ingested laccase molecules pass through the ant guts and remain active when defecated on the leaf pulp that the ants add to their gardens. This accurate deposition ensures that laccase activity is highest where new leaf material enters the fungus garden, but where fungal mycelium is too sparse to produce extracellular enzymes in sufficient quantities to detoxify phenolic compounds. Phylogenetic analysis of LgLcc1 ortholog sequences from symbiotic and free-living fungi revealed significant positive selection in the ancestral lineage that gave rise to the gongylidia-producing symbionts of leaf-cutting ants and their non–leaf-cutting ant sister group. Our results are consistent with fungal preadaptation and subsequent modification of a particular laccase enzyme for the detoxification of secondary plant compounds during the transition to active herbivory in the ancestor of leaf-cutting ants between 8 and 12 Mya.
Keywords: gene cooption, polyphenols
Large-scale grazing offers rich ecological niches, provided herbivores can overcome the constraints associated with low protein content of plant food. This challenge is particularly acute because plants often respond to herbivory by evolving secondary compounds that reduce protein availability even further (1). Many plant eaters have therefore become specialists, which has allowed them to evolve counteradaptations against specific plant defenses (2). Rather few herbivores have in fact evolved large enough body sizes and population densities to leave substantial ecological footprints as generalist grazers or browsers (2). In vertebrates, the most conspicuous examples are the ungulates and their marsupial analogs, many of whom rely on complex communities of bacterial gut symbionts for making this lifestyle productive (3). Some insects can also be serious defoliators, but normally only as ephemeral plagues (e.g., migratory locusts and cyclical outbreaks of winter moths) (4). The only known insects that cause consistent year-round reductions in live plant biomass comparable to those imposed by ungulates are the Central and South American leaf-cutting ants (5).
Phenolic compounds such as tannins and flavonoids are among the most general and widespread plant defenses against large-scale defoliation (1). They are primarily present in the waxy leaf cuticle and cell vacuoles and, provided sufficient plant resources are available, particularly abundant in the fast-growing plant parts that many large grazers and browsers prefer (6). Phenolic compounds damage the gastrointestinal mucosa and epithelium of mammals and increase metabolic decomposition costs by precipitating digestive enzymes. Mammalian herbivores often neutralize phenolic compounds by increasing gastrointestinal mucus production, by enlisting the degradation services of gut microorganisms, and/or by secreting phenol-binding proteins in the saliva (7). Insect larvae almost invariably experience reduced growth rates when exposed to phenolic compounds despite compensating adaptations (8), but this does not seem to apply to leaf-cutting ants as long as they obtain their forage from a sufficient diversity of plants (9, 10). Leaf-cutting ants owe their status as prime New World pests to what has been described as an “unholy alliance” with a fungus-garden symbiont (Leucocoprinus gongylophorus) that does most of the leaf decomposition (11).
Leaf-cutting ants originated only between 8 and 12 Mya and represent a major evolutionary transition that coincided with an increase in mature colony size (approximately 500–100,000 workers in the genus Acromyrmex and several million in the genus Atta), increased worker size dimorphism, genetically more variable colonies because of queen multiple mating, and higher investment in prophylactic metapleural gland secretions (12–14). This transition was the culmination of tens of millions of years of obligate fungus farming in the attine clade that radiated into 15 currently recognized genera with more than 230 described species, of which 45 are leaf-cutting ants (14). Species in the other genera have significantly smaller mature colony sizes (approximately 50–3,000), monomorphic workers, singly mated queens, and they primarily collect dry leaf litter, caterpillar frass, fallen fruits, and shed flowers (14). Recent research has thus shed considerable light on the evolution of the attine ants, but the fungal adaptations that facilitated the various farming transitions and ultimately the adoption of active herbivory are still poorly understood.
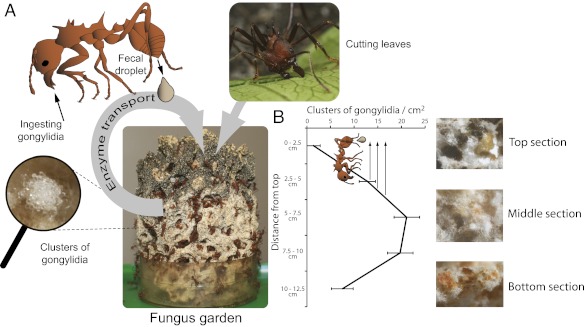
Ungulate stomachs have frequent peristaltic contractions to mix fodder with digestive enzymes, but the gardens of leaf-cutting ants are static. This implies that fungal decomposition enzymes are produced in the central part of fungus gardens where fungal/plant biomass ratios are optimal (Fig. 1A), whereas detoxification is primarily needed at first hyphal encounter with phenolic compounds in the top section of gardens where the ants deposit new substrate in the form of finely fragmented leaf pulp (15). This spatial mismatch of supply and demand has induced consistent selection for carbohydrate-active enzymes that the ants ingest but not digest, so they can be passed on via the fecal fluid that is deposited as droplets with the leaf pulp in the top of gardens (16) (Fig. 1A). This remarkable practice has been known since the 1970s (17), but its molecular mechanisms are only just beginning to be unraveled. A recent study (18) showed that all six pectinolytic enzymes whose protein sequences can be obtained from fecal fluid of Acromyrmex leaf-cutting ants are of fungal origin and that five of them are up-regulated in the special fungal structures (gongylidia: inflated hyphal tips) that fungus gardens produce as food for the farming ants (Fig. 1B).
Fig. 1.
Diagram illustrating the dynamics of fungal enzyme transfer and substrate processing in leaf-cutting ant fungus gardens, which grow at the top or periphery and are cropped and discarded by the ants at the bottom (19). (A) Ants deposit chewed-up fresh leaf fragments combined with fecal fluid at the top of their garden (green shade). Fecal fluid vectoring of fungal enzymes resolves the problem that low fungal biomass in the top sections of gardens would delay substrate decomposition. (B) Fungal gongylidia (Inset Left) concentrate fungal enzymes to be eaten by the ants, but are most abundant in the central layer of the garden where fungal growth is most vigorous [mean number of gongylidia clusters (staphylae)/cm2 ± SE from four A. echinatior colonies]. Leaf-pulp decomposition rate is therefore enhanced when the farming ants transfer fungal enzymes from the middle to the top of a garden by deposition of fecal droplets that vector these gongylidia-produced enzymes.
The transport of pectinolytic enzymes to the top of fungus gardens significantly enhances the hyphae’s access to proteins and sugars inside the plant cells of the leaf-pulp substrate (18, 19), analogous to saliva enzymes facilitating food degradation in the gastrointestinal tract of mammalian herbivores. This intricate form of enzyme transport suggests there has been consistent selection for substrate-processing efficiency ever since leaf-cutting herbivory arose ca. 10 mya (14), because sister groups of attine fungi are saprophytic and thus unlikely to have evolved specialized enzymatic machinery to target live plant tissue, a feature that is characteristic of phytopathogenic fungi (18). However, that leaves unanswered the more fundamental question of how the leaf-cutting ant–fungus symbiosis resolved the challenge of increased leaf-phenol exposure that followed in the wake of herbivory.
Phytophagous fungi interacting with plant-feeding Hemiptera have been shown to neutralize phenolic plant defenses with polyphenol oxidases (20), and leaf-cutting ant cultivars are known to produce phenol-oxidizing enzymes (21, 22) of the laccase subgroup of polyphenoloxidases (benzendiol oxygen oxidoreductase; synonyms: urishiol oxidase, p-diphenol oxidase, EC 1.10.3.2) (23). It was also shown experimentally that high concentrations of phenolic compounds discourage leaf cutting by Atta foragers and inhibit the growth of their L. gongylophorus gardens (21, 22), making it a reasonable conjecture that rapid and efficient phenol detoxification has been an important enabling factor for leaf-cutting ants to become dominant terrestrial herbivores in Central and South America. To assess how leaf-cutting ants overcome phenolic plant defenses, we therefore set out to clarify the molecular structure and function of the laccase enzymes expressed by L. gongylophorus. This included measuring laccase activity in laboratory fungus gardens of leaf-cutting ants that were fed with phenol-containing bramble leaves (Rubus spp.), screening 700 Mb of genomic sequence fragments of L. gongylophorus for laccase genes, and obtaining the fecal droplet proteome of Acromyrmex echinatior workers with MALDI tandem mass spectrometry. We also measured specific gene expression and fecal transport of laccase enzymes in fungus gardens and estimated the likelihood of positive selection on a single relevant laccase gene in the fungi cultivated by leaf-cutting ants and two phylogenetically more basal attine genera, after verifying laccase activity of the gene by heterologous expression in yeast.
Results and Discussion
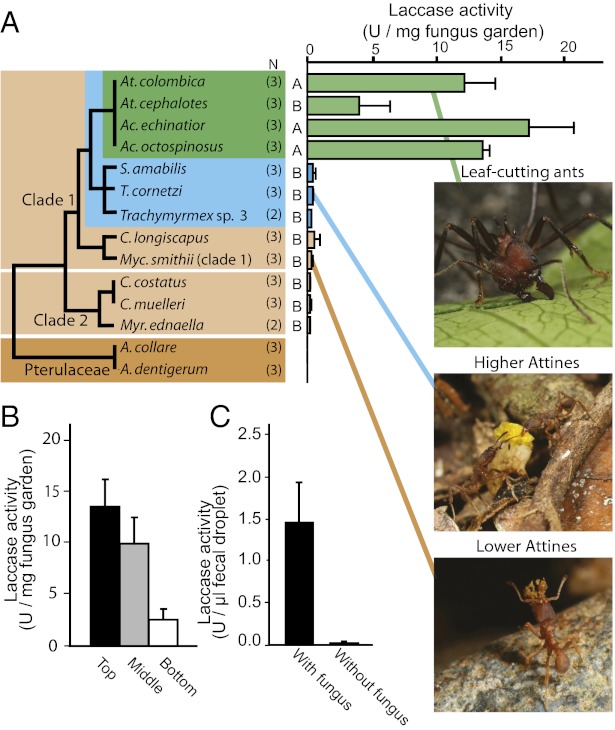
We measured fungus garden laccase activity in representative species of fungus-growing ants and found greatly enhanced laccase activity in leaf-cutting ant gardens (Fig. 2A), consistent with their diet of mainly fresh leaves requiring more phenol detoxification. Further measurements revealed that laccase activity was highest in the top section where new leaf material is positioned (Fig. 2B and SI Appendix, Fig S2), as expected when laccase activity is most important upon first encounter with fresh plant tissue. After a previous study had indicated that a laccase enzyme of fungal origin was active in the ant fecal fluid (23), we verified this by measuring laccase activity in fecal droplets of large workers of A. echinatior kept with and without a fungus garden. This showed that only workers that were allowed to eat fungus garden material had laccase activity in their fecal droplets (Fig. 2C).
Fig. 2.
Fungus garden laccase activity in units per milligram of fungus garden (units = unit oxidizing 1 nmol syringaldazine per minute) + SE for representative attine ants from Panama. (A) Simplified fungal phylogeny showing laccase activity in newly built sections of laboratory fungus gardens of 14 ant species representing 8 of the 15 attine ant genera. Dark brown: pterulaceous fungi cultivated by debris-foraging Apterostigma (almost no activity); light brown: lower-attine ants (Myrmicocrypta, Mycocepurus, and Cyphomyrmex) cultivating either clade 1 or clade 2 leucocoprinaceous fungi on a substrate of mostly dead plant material (very low activity); blue: higher attine ants (Trachymyrmex and Sericomyrmex) cultivating fungi with gongylidia on a substrate of mostly flowers and soft shed leaves (very low activity); green: leaf-cutting ants (Acromyrmex and Atta) cultivating L. gongylophorus (high activity)(n = number of colonies; A and B indicate significantly different means in post hoc tests following ANOVA, F13,39 = 19.48, P < 0.0001). (B) Laccase activity (units per milligram + SE) in the top, middle, and bottom sections of laboratory fungus gardens of three A. echinatior colonies (see SI Appendix, Fig. S2 for similar data on other leaf-cutting ants) showing that laccase activity decreases from top to bottom (ANOVA: F2,6 = 6.33, P = 0.0332). (C) Fecal droplet laccase activity (units per microliter + SE) of medium-sized A. echinatior workers from laboratory colonies with a fungus garden and from workers kept on sugar water for 20 d without a fungus garden (ANOVA: F1,4 = 8.93, P = 0.0404, five workers measured from each of three A. echinatior colonies).
To identify the specific laccase enzymes, we analyzed the fecal droplet proteome of A. echinatior with MALDI tandem mass spectrometry (SI Appendix), which provided partial amino acid sequences. We also identified all putative laccase-coding sequences in the fungal symbiont’s genome by analyzing an unassembled set of 454-generated sequences of genomic DNA extracted from the L. gongylophorus symbiont of another A. echinatior colony (SI Appendix). This revealed nine putative laccase-coding genes that were verified by cDNA sequencing and translated to the corresponding amino acid sequences (SI Appendix, Tables S4 and S5). Alignments of these nine laccase-coding genes with the fecal droplet proteome data showed that a single laccase enzyme (LgLcc1) had a sequence 96% identical to the fecal droplet protein fragments (Fig. 3A), and was markedly different from the remaining eight laccase genes in amino acid sequence and phylogenetic placement in the gene tree (SI Appendix, Fig. S3–S5). The observed discrepancy between peptide mass spectra and cDNA sequences (2 of 51 amino acids, Fig. 3A) is most likely due to biological variation between the respective samples used for the fecal droplet proteome and cDNA analysis. Heterologous expression in yeast further verified that LgLcc1 has laccase activity and is functionally active (Fig. 3B).
Fig. 3.
Laccase gene expression and protein/cDNA sequence comparison demonstrating the transfer of active laccases via the ant gut. (A) Partial alignment of fecal droplet protein sequences and the corresponding translated cDNA of LgLcc1 from gongylidia showing 96% sequence identity (see SI Appendix for details). Numbers refer to amino acid positions in LgLcc1. (B) Recombinant expression of the LgLcc1 gene in yeast (INVSc1) transformed with the vector pYES2 and grown in media supplemented with 20 mM gallic acid. Dark brown staining indicates that LgLcc1 was expressed, secreted into the medium, and able to oxidize a phenolic compound such as gallic acid. Left and Right pictures are identical, but illustrate contrasts with black and white background, respectively. (C) Ratio of laccase gene expression (mean ± SE) (normalized relative to housekeeping genes) between gongylidia and mycelium, measured for nine laccase-coding genes from A. echinatior fungus gardens (green) and the LgLcc1 homolog TcLcc1 from Trachymyrmex cornetzi fungus gardens (blue), a sympatric non–leaf-cutting ant sister group representative. Values above 1 show an up-regulation of laccase gene expression in gongylidia, whereas values below 1 show a down-regulation of laccase gene expression in gongylidia relative to undifferentiated mycelium (based on 10,000 random permutations in reallocation tests of four A. echinatior and three T. cornetzi fungus gardens, respectively). (D) Absolute gene expression (mean number of transcripts + SE per nanogram of total RNA) of TcLcc1 and LgLcc1 in gongylidia and undifferentiated mycelium from three T. cornetzi and four A. echinatior fungus gardens, respectively. (E) Unrooted maximum likelihood phylogeny of ant-cultivated fungal laccase sequences showing positive selection on the branch leading to the clade of gongylidia-producing fungi (L. gongylophorus and Leucocoprinus sp. “T. cornetzi”). Branch-site tests identified the branch (highlighted in bold) containing codon positions that are significantly positively selected (2ΔlnL = 7.44, P = 0.025). The orthologous laccase from the free-living fungi Cyathus bulleri (ABW75771) and Lac2 from Coprinus comatus (JQ228449) are included as outgroups to the fungus-growing ant cultivars. dN/dS ratios (ω) and branch lengths (substitutions per codon) are given above and below each branch, respectively. The nonsignificant ω = 4.204 for the branch leading to ClLcc1 is due to very few synonymous substitutions (see SI Appendix for details).
Phenolic compounds are not only present in live plant tissue, but to a lesser extent also in the shed leaves and leaf litter (24) collected by most other fungus-growing ants, suggesting that polyphenol-oxidase activity was an inherent property of ant fungus farming before active leaf cutting arose. Ant-cultivated leucocoprinaceous fungi are closely related to saprophytic basidiomycetes that mostly decompose leaf litter (25), and laccase enzymes are known from the gardens of leaf-litter collecting fungus-growing termites (26), indicating the inherent need for phenol degradation when using leaf litter as fungal substrate. The genomes of saprophytic fungi normally contain multiple laccase-coding genes (27, 28), so that the actual number of laccases that we found in L. gongylophorus may merely represent the enzyme spectrum of the last common ancestor of the gongylidia-bearing cultivars (14, 29). However, our measurements of gene expression of the nine laccase genes from A. echinatior fungus gardens showed that only the LgLcc1 coding gene was significantly up-regulated in the fungal gongylidia relative to undifferentiated fungal mycelium (Fig. 3C). This single laccase gene (LgLcc1) thus appears to have been selected to enable ingestion and nondigestive passage through the gut of the ants so the enzyme that it codes for can be transported to the top outermost edges of fungus gardens via fecal droplets (Fig. 1 A and B). Similar to the previously identified pectinase enzymes (18), this ant-vectoring mechanism for LgLcc1 underlines that the ants eat substantial amounts of gongylidia. How crucial this feeding is for ant performance remains to be established, but it seems clearly adaptive for the symbiosis. It ensures that this fungal laccase is most abundant and active at the locations where fresh phenol-containing plant material is added to the fungus garden, but where the low biomass of incipient hyphal growth without gongylidia cannot deliver adequate quantities of laccase enzyme (Figs. 1 and 2B). An additional advantage of LgLcc1 ingestion may be that it helps neutralize phenolic compounds in plant sap that foraging workers may drink when cutting leaves (30).
The increased expression of a single laccase gene suggests that LgLcc1 evolved a novel function by direct cooption (31) of an ancestral laccase-coding gene, which prompted us to investigate whether LgLcc1 has undergone structural changes during its putative coevolution with the leaf-cutting ants. We therefore analyzed the LgLcc1 sequence for molecular signatures of positive selection and identified the homologous sequences of LgLcc1 in the closely related gongylidia-producing fungal cultivar of the (non–leaf-cutting) higher attine ant Trachymyrmex cornetzi (TcLcc1), and in the fungal cultivar of the lower attine ant Cyphomyrmex longiscapus (ClLcc1). The set of orthologous laccase sequences that we obtained was analyzed for positive selection acting on specific enzyme residues during ant-cultivar evolution using likelihood models that allow dN/dS (ω) to vary across codons for each lineage in the alignment (SI Appendix). This revealed character changes with clear signatures of positive selection (2ΔlnL = 7.44, P = 0.025, Fig. 3E and SI Appendix, Tables S6 and S7) that mapped onto the branch subtending LgLcc1 in Acromyrmex cultivars and TcLcc1 in Trachymyrmex cultivars, a result that remained robust no matter what phylogenetic outgroups we used (SI Appendix, Table S9).
The fact that positive selection was determined on the branch subtending the clade LgLcc1 in Acromyrmex cultivars and TcLcc1 in Trachymyrmex cultivars suggests that the laccase enzyme was already modified in conjunction with the evolution of gongylidia ca. 20 Mya (29). This event coincided with the symbiosis becoming obligate for both parties and the fungal cultivar losing its gene flow with free-living relatives (14, 29). Based on the foraging behaviors of extant fungus-growing ants (32), substrate use by ancestral higher-attine ants was likely a combination of plant debris, insect frass, flower parts, and possibly soft fresh leaves. Selection for structural modifications of the ancestral Lcc1 laccase may thus initially have been variable in intensity and direction, consistent with the relatively weak signals of directional selection that we find for the branch leading to TcLcc1 (SI Appendix, Tables S7 and S9). However, as positive selection on the branch leading to LgLcc1 + TcLcc1 was consistently significant (SI Appendix, Table S9), it seems reasonable to infer that early structural changes in TcLcc1 may have served as preadaptations for more substantial modifications when active leaf cutting evolved 8–10 My later.
Gene expression analysis of T. cornetzi fungus gardens showed that TcLcc1 was significantly up-regulated in the gongylidia (Fig. 3C), but that the number of laccase (Lcc1) gene transcripts remained an order of magnitude lower than in gongylidia of the A. echinatior cultivar L. gongylophorus (Fig. 3D). Fig. 3D gives a ratio of 2–3 for LgLcc1 overexpression after normalizing relative to the total amount of RNA, whereas Fig. 3C suggests an up-regulation ratio of about 16 when normalizing relative to the expression of housekeeping genes, which we believe is a more reliable comparison. C. longiscapus cultivates a more distantly related garden symbiont without gongylidia (29) so that differential tissue-specific gene expression could not be measured for this symbiont. Highly elevated gene transcription is energetically costly and thus normally constrained by natural selection (33). These results are therefore consistent with adaptive benefits of elevated Lcc1 gene expression in leaf-cutting ant fungus gardens.
Codons on the positively selected branch shared between LgLcc1 in Acromyrmex cultivars and TcLcc1 in Trachymyrmex cultivars were predicted using Bayes-empirical-Bayes analysis, producing four structural amino acid substitutions with a posterior probability >90% (SI Appendix, Table S8). However, none of these amino acid substitutions were located in active-site loop regions (SI Appendix, Fig. S7) that are known to alter reaction specificity in other enzymes when they are artificially modified (34). The positively selected codons in the LgLcc1 and TcLcc1 genes therefore do not appear to be associated with fungus garden laccase activity per se, because the Trachymyrmex fungus gardens did not have increased laccase activity (Fig. 2A). We hypothesize that the positively selected codons may instead be associated with the ability of this laccase enzyme to avoid digestion in the ant guts, as the up-regulation of Lcc1 in the gongylidia indicates similar functionality in gardens of both Trachymyrmex and Acromyrmex. The combination of likely adaptations in LgLcc1 and TcLcc1 may have constrained gene duplication, which is otherwise often observed for genes that provide adaptive benefits (35) and may thus have selected for further modifications in transcription regulation that would explain the up-regulations that we observed.
Studies so far have emphasized that the attine transition to active herbivory gave access to an abundant novel resource base (11, 14, 15) and allowed the leaf-cutting ants to reach colony sizes two to three orders of magnitude larger than their Trachymyrmex sister clade that also cultivate fungi with gongylidia, but on a substrate of mostly shed flowers and wilted leaves and with slower garden growth rates (14, 29). Our present study shows that coevolutionary selection for laccase enzyme activity in the fungal cultivar may have greatly facilitated the transition to active herbivory by alleviating a crucial digestion challenge, analogous to the fitness advantages that herding human populations obtained from the disappearance of lactose intolerance (36).
Materials and Methods
Detailed materials and methods with references are described in SI Appendix.
Biological Material.
Fungus-growing ant colonies from 12 species (Acromyrmex echinatior, Ac. octospinosus, Atta colombica, At. cephalotes, Sericomyrmex amabilis, Trachymyrmex cornetzi, Trachymyrmex sp. 3, Cyphomyrmex longiscapus, C. muelleri, C. costatus, Mycocepurus smithii, Myrmicocrypta ednaella, Apterostigma collare, and A. dentigerum) were collected in Parque Nacional Soberanía, Panama (the Gamboa area and forest along Pipeline Road) during 2005–2008. Colonies were transported to the University of Copenhagen, Denmark, and maintained as laboratory colonies.
Measurement of Laccase Activity.
Proteins were extracted in duplicate by homogenizing 50 mg laboratory fungus garden material in 0.1 M citric acid–sodium citrate buffer (pH = 5.5). Laccase activity was measured by recording the change in absorbance at 530 nm per minute during the linear phase using syringaldazine as enzymatic substrate. Fecal droplets of A. echinatior medium-sized workers were obtained by gently squeezing the thorax and abdomen with forceps until an approximately 0.1–0.2 μL drop was expelled and 4.8 μL ddH2O was immediately added to prevent evaporation. Laccase activity was measured in each of five fecal droplets, from five workers each of three colonies, kept with and without a fungus garden, respectively.
Peptide Sequencing.
To analyze the fecal droplet proteome, fecal droplets from 50 A. echinatior workers were combined and separated on a 1D SDS/PAGE gel. Individual bands were excised from the gel, loaded directly onto a matrix-assisted laser desorption ionization (MALDI) target plate, and peptide mass spectra obtained with tandem mass spectrometry (SI Appendix, Fig. S8). The MS/MS spectra were analyzed manually with the AminoCalc program (Protana A/S) to find the distance between fragment ions and obtain the amino acid sequences (SI Appendix).
Gene Identification and Expression Analysis.
Genes encoding laccase enzymes in the fungal symbiont were identified by searching an unassembled set of 700 Mb sequence data (estimated 7× coverage), which represents a draft heterokaryotic genome of L. gongylophorus obtained from another A. echinatior colony collected in Gamboa (Ae322). This sequence set was generated by shotgun sequencing of gDNA extracted from axenic fungal isolates using Roche 454 pyrosequencing (SI Appendix). The sequence set was variable, indicating that the fungal genome was polyploid, which in combination with the lack of paired-end sequencing data precluded any further assembly. However, it recovered up to 97% of a set of approximately 35 protein sequences in fecal droplets that we have independently obtained (partly published in ref. 18), indicating that a very high proportion of coding sequences had been recovered. We therefore proceeded with searching for putative laccase-coding genes with an iterative psi-tBLASTn search strategy based on conserved laccase signature regions (SI Appendix). The specific expressions of the identified putative laccase-coding genes were measured with real-time qPCR of cDNA generated from RNA extracted from gongylidia and undifferentiated mycelium, respectively, collected directly from laboratory colonies.
Positive Selection Analysis.
Maximum likelihood estimates of the dN/dS ratio (ω) for each site (codon) along a protein were used to detect signatures of positive natural selection among cultivar laccase sequences using the program codeml implemented in PAML 4.4. Each lineage (branch) was tested independently for positive selection (ω > 1) on particular lineages and sites by applying a neutral model that allows ω to vary between 0–1 and a selection model that also incorporates sites with ω > 1. Statistical significance was determined with a likelihood ratio test of these two models for each lineage and adjusted for multiple testing with Bonferroni correction.
Supplementary Material
Acknowledgments
We thank Morten Rasmussen for assistance with obtaining the fungal 454 sequences; Duur Aanen, David Nash, and Michael Poulsen for comments on a previous version of the manuscript; the Smithsonian Tropical Research Institute for providing logistic help and facilities to work in Gamboa; and the Autoridad Nacional del Ambiente y el Mar for permission to sample ants in Panama and to export them to Denmark. H.H.D.F.L., M.S., S.N., and J.J.B. were supported by the Danish National Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (ENA) (accession no. ERP001825) and the GenBank database (accession nos. JQ307223–JQ307233).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212709110/-/DCSupplemental.
References
- 1.Coley PD, Bryant JP, Chapin FS., 3rd Resource availability and plant antiherbivore defense. Science. 1985;230(4728):895–899. doi: 10.1126/science.230.4728.895. [DOI] [PubMed] [Google Scholar]
- 2.Shipley LA, Forbey JS, Moore BD. Revisiting the dietary niche: When is a mammalian herbivore a specialist? Integr Comp Biol. 2009;49(3):274–290. doi: 10.1093/icb/icp051. [DOI] [PubMed] [Google Scholar]
- 3.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berryman AA. What causes population cycles of forest Lepidoptera? Trends Ecol Evol. 1996;11(1):28–32. doi: 10.1016/0169-5347(96)81066-4. [DOI] [PubMed] [Google Scholar]
- 5.Wirth R, Herz H, Ryel RJ, Beyschlag W, Hölldobler B. 2003. Herbivory of Leaf-Cutting Ants: A Case Study on Atta colombica in the Tropical Rainforest of Panama, eds Baldwin IT, et al., Ecological studies (Springer, Berlin), Vol. 164.
- 6.Endara MJ, Coley PD. The resource availability hypothesis revisited: A meta-analysis. Funct Ecol. 2011;25:389–398. [Google Scholar]
- 7.Dearing MD, Foley WJ, McLean S. The influence of plant secondary metabolites on the nutritional ecology of herbivorous terrestrial vertebrates. Annu Rev Ecol Syst. 2005;36:169–189. [Google Scholar]
- 8.Dowd PF. In: Microbial Mediation of Plant-Herbivore Interactions. Barbosa P, Krischik VA, Jones CG, editors. New York: Wiley; 1991. pp. 411–440. [Google Scholar]
- 9.Howard JJ. Leafcutting ant diet selection: The role of nutrients, water, and secondary chemistry. Ecology. 1987;68:503–515. [Google Scholar]
- 10.Bucher EH, Marchesini V. Herbivory by leaf-cutting ants: Nutrient balance between harvested and refuse material. Biotropica. 2004;36:327–332. [Google Scholar]
- 11.Cherrett JM, Powell RJ, Stradling DJ. In: Insect-Fungus Interactions. Wilding N, Collins NM, Hammond PM, Webber JF, editors. London: Academic; 1989. pp. 93–120. [Google Scholar]
- 12.Villesen P, Murakami T, Schultz TR, Boomsma JJ. Identifying the transition between single and multiple mating of queens in fungus-growing ants. Proc Biol Sci. 2002;269(1500):1541–1548. doi: 10.1098/rspb.2002.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes WO, Pagliarini R, Madsen HB, Dijkstra MB, Boomsma JJ. Antimicrobial defense shows an abrupt evolutionary transition in the fungus-growing ants. Evolution. 2008;62(5):1252–1257. doi: 10.1111/j.1558-5646.2008.00347.x. [DOI] [PubMed] [Google Scholar]
- 14.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci USA. 2008;105(14):5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Fine Licht HH, Schiøtt M, Mueller UG, Boomsma JJ. Evolutionary transitions in enzyme activity of ant fungus gardens. Evolution. 2010;64(7):2055–2069. doi: 10.1111/j.1558-5646.2010.00948.x. [DOI] [PubMed] [Google Scholar]
- 16.Schiøtt M, De Fine Licht HH, Lange L, Boomsma JJ. Towards a molecular understanding of symbiont function: Identification of a fungal gene for the degradation of xylan in the fungus gardens of leaf-cutting ants. BMC Microbiol. 2008;8:40. doi: 10.1186/1471-2180-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd ND, Martin MM. Faecal proteinases of the fungus-growing ant, Atta texana: Properties, significance and possible origin. Insect Biochem. 1975;5:619–635. [Google Scholar]
- 18.Schiøtt M, Rogowska-Wrzesinska A, Roepstorff P, Boomsma JJ. Leaf-cutting ant fungi produce cell wall degrading pectinase complexes reminiscent of phytopathogenic fungi. BMC Biol. 2010;8:156. doi: 10.1186/1741-7007-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moller IE, De Fine Licht HH, Harholt J, Willats WGT, Boomsma JJ. The dynamics of plant cell-wall polysaccharide decomposition in leaf-cutting ant fungus gardens. PLoS ONE. 2011;6(3):e17506. doi: 10.1371/journal.pone.0017506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles PW. Interaction of plant phenols and salivary phenolases in relationship between plants and Hemiptera. Entomol Exp Appl. 1969;12:736–744. [Google Scholar]
- 21.Nichols-Orians C. Condensed tannins, attine ants, and the performance of a symbiotic fungus. J Chem Ecol. 1991;17:1177–1195. doi: 10.1007/BF01402942. [DOI] [PubMed] [Google Scholar]
- 22.Nichols-Orians C. Differential-effects of condensed and hydrolyzable tannin on polyphenol oxidase activity of attine symbiotic fungus. J Chem Ecol. 1991;17:1811–1819. doi: 10.1007/BF00993730. [DOI] [PubMed] [Google Scholar]
- 23.Rønhede S, Boomsma JJ, Rosendahl S. Fungal enzymes transferred by leaf-cutting ants in their fungus gardens. Mycol Res. 2004;108(Pt 1):101–106. doi: 10.1017/s0953756203008931. [DOI] [PubMed] [Google Scholar]
- 24.Kurokawa H, Nakashizuka T. Leaf herbivory and decomposability in a Malaysian tropical rain forest. Ecology. 2008;89(9):2645–2656. doi: 10.1890/07-1352.1. [DOI] [PubMed] [Google Scholar]
- 25.Vellinga EC. Ecology and distribution of Lepiotaceous fungi (Agaricaceae) Nova Hedwigia. 2004;78:273–299. [Google Scholar]
- 26.Taprab Y, et al. Symbiotic fungi produce laccases potentially involved in phenol degradation in fungus combs of fungus-growing termites in Thailand. Appl Environ Microbiol. 2005;71(12):7696–7704. doi: 10.1128/AEM.71.12.7696-7704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Courty PE, et al. Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol. 2009;182(3):736–750. doi: 10.1111/j.1469-8137.2009.02774.x. [DOI] [PubMed] [Google Scholar]
- 28.Kilaru S, Hoegger PJ, Kües U. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr Genet. 2006;50(1):45–60. doi: 10.1007/s00294-006-0074-1. [DOI] [PubMed] [Google Scholar]
- 29.Mikheyev AS, Mueller UG, Abbot P. Comparative dating of attine ant and lepiotaceous cultivar phylogenies reveals coevolutionary synchrony and discord. Am Nat. 2010;175(6):E126–E133. doi: 10.1086/652472. [DOI] [PubMed] [Google Scholar]
- 30.Littledyke M, Cherrett JM. Direct ingestion of plant sap from cut leaves by the leaf-cutting ants Atta cephalotes (L.) and Acromyrmex octospinosus (Reich) (Formicidae: Attini) Bull Entomol Res. 1976;66:205–217. [Google Scholar]
- 31.True JR, Carroll SB. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol. 2002;18:53–80. doi: 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]
- 32.De Fine Licht HH, Boomsma JJ. Forage collection, substrate preparation and diet composition in fungus-growing ants. Ecol Entomol. 2010;35:259–269. [Google Scholar]
- 33.Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22(6):1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- 34.Park H-S, et al. Design and evolution of new catalytic activity with an existing protein scaffold. Science. 2006;311(5760):535–538. doi: 10.1126/science.1118953. [DOI] [PubMed] [Google Scholar]
- 35.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 36.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39(1):31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.