Abstract
Posttranscriptional/translational regulation of gene expression is mediated by diverse RNA binding proteins and plays an important role in development and defense processes. Among the RNA-binding proteins, the mammalian Pumilio RNA-binding family (Puf) acts as posttranscriptional and translational repressors. An Arabidopsis Puf mutant, apum5-D, was isolated during a T-DNA insertional mutant screen for mutants with reduced susceptibility to Cucumber mosaic virus (CMV) infection. Interestingly, CMV RNA contained putative Pumilio-homology domain binding motifs in its 3′ untranslated region (UTR) and internal places in its genome. APUM5 directly bound to the 3′ UTR motifs and some internal binding motifs in CMV RNAs in vitro and in vivo. We showed that APUM5 acts as a translational repressor that regulates the 3′ UTR of CMV and affects CMV replication. This study uncovered a unique defense system that Arabidopsis APUM5 specifically regulates CMV infection by the direct binding of CMV RNAs.
Keywords: plant defense, Pumilio RNA binding protein, TuMV
Plant viruses are the obligate pathogens, and host proteins facilitate the multiplication of viruses by affecting processes such as viral replication, cell-to-cell movement, and systemic movement of the virus (1). These processes occur upon the interaction between the virus and host proteins or after the suppression of the host basal defense mechanism and often lead to abnormal phenotypes of virus-infected host plants, such as small, highly branched bushes with deformed leaves, stunting, and reduced apical dominance (2–4). Thus, the reduced growth and developmental changes in the virus-infected plants are typical symptoms of virus infection and signify that the virus has undergone a successful life cycle. To understand the molecular mechanisms underlying viral multiplication and symptoms in plants, it is necessary to identify and characterize the host factors involved in these processes.
To identify novel host factors involved in the multiplication of plant RNA viruses in susceptible plants, T-DNA insertion mutants of Arabidopsis thaliana Col-0 ecotype, in which CMV-Kor multiplication is abrogated (5), were screened. An Arabidopsis Puf mutant, apum5-D, in which Cucumber mosaic virus (CMV) multiplication was affected during viral spreading, was isolated. APUM5 contains the Pumilio-homology domain (PHD) and encodes a putative Puf, which was originally identified in Drosophila melanogaster and Caenorhabditis elegans (6). Pufs are highly conserved in various organisms and work as posttranscriptional and translational repressors (7, 8). PHD has RNA binding activity; there are eight repeats with three alpha helices in each repeat. The inner surface of the PHD binds the RNA. The outer surface of the domain permits protein–protein interactions with diverse proteins, such as deadenylase and general translation factors (9, 10). Arabidopsis Pufs also exhibit RNA-binding activity and have conserved binding motifs (11, 12). However, plant Puf functions have not yet been fully identified or characterized.
Results
apum5-D Mutant Showed Altered Susceptibility to CMV.
To identify novel host factors associated with the multiplication of plant RNA viruses, we screened for Arabidopsis mutants with altered susceptibility to CMV infection, exploiting the fact that CMV-infected plants exhibit visible symptoms that are readily discernible (5, 13) (Fig. S1A). Twenty candidates showed decreased CMV coat protein (CP) levels in inflorescence tissue at 18 d postinoculation (dpi), as measured by ELISA (Fig. S1B). The lines that contained a single T-DNA insertion were further selected by DNA blot analysis, and the T-DNA insertion sites were identified by thermal asymmetric interlaced PCR (TAIL-PCR). At 18 dpi, CMV-inoculated mutant plant #75010 was clearly distinguishable from CMV-inoculated Col-0 plants (Fig. 1A) and was chosen for further analysis (Fig. S1 C–E). Mutant #75010 had a T-DNA insertion in APUM5 encoding a member of the Arabidopsis Puf (APUM) family. APUM5 was found to possess a structurally conserved Pumilio RNA-binding domain and a putative transmembrane sequence in the N-terminal region (Fig. S1 E and F). The T-DNA insertion was located about 650 bp upstream of the translational start codon (Fig. S1D). We determined APUM5 transcript levels in the #75010 mutant. APUM5 expression was about three times greater in the #75010 mutant than in the wild type, as determined by quantitative RT-PCR (Fig. 1B). The mutant was redesignated as apum5-D. CMV CP and CMV RNA levels in the apum5-D mutant were also lower than those in Col-0 plants after infection (Fig. 1 C and D). The reduced level of CP was consistent with reduced susceptibility. The apum5-D mutant showed about a 20% increase over Col-0 plants, both in stem length and fresh weight, at 18 dpi with CMV infection (Fig. 1 E and F). These results indicated that elevated APUM5 expression might confer resistance to CMV infection in the apum5-D mutant.
Fig. 1.
Screening of T-DNA insertion mutant lines for the detection of changes in CMV susceptibility. (A) The mutant #75010 infected with CMV showed a much healthier growth phenotype than the CMV-infected Col-0 plant at 18 dpi. (B) The apum5-D mutant showed an up-regulated gene expression compared with Col-0 in 3-wk-old plants, as shown by qRT-PCR analysis. Significant difference (Student t test; *P < 0.05) is indicated with an asterisk. (C) ELISA for CMV coat protein accumulation detection in Col-0 and apum5-D mutant. Significant difference (Student t test; *P < 0.05) is indicated with an asterisk. (D) RNA blot analysis for CMV detection in Col-0 and apum5-D mutant plants. CMV-inoculated plant RNA was extracted and RNA blot analysis was performed using 3′ UTR probes specific to CMV. rRNA was used as a loading control. The relative band intensity was quantified by using the ImageJ software. (E and F) Analysis of fresh weight and stem length in CMV-inoculated Col-0 and apum5-D mutant plants at 18 dpi. Significant differences (Student t test; *P < 0.05, **P < 0.01) are indicated with asterisks. Error bars show mean ± SD (n = 12).
PHD of APUM5 Binds to the CMV 3′ Untranslated Region Motif and Some Other Internal Motifs in the CMV RNA Genome.
The PHD of Pufs is a conserved region that binds to the sequence-specific motifs in the 3′ untranslated region (UTR) of target genes and the defined nucleotide core motif, UGUA (A/C/U) AUA, is bound by diverse Puf family proteins (14, 15). We hypothesized that direct interactions with sequence-specific motifs of CMV RNAs would contribute to the gain-of-function phenotype of the apum5-D mutant. As expected, the putative binding motif was found in CMV tripartite RNA 3′ UTR regions (Fig. S2A; motif 1C, 2C, and 3C). In addition to the 3′ UTRs, additional putative core binding motifs in the CMV RNA genomes were found (Fig. S2 A and B). Furthermore, CMV group-I, but not group-II, strains contain putative binding motifs in the 3′ UTR (Fig. S2C). Purified GST–APUM5–PHD protein bound effectively to CMV 3′ UTR motifs in the EMSA (Fig. 2A). Furthermore, GST–APUM5–PHD also exhibited a strong binding affinity for hbNRE2 (Fig. 2A). This binding was also confirmed by competition assay (Fig. 2 B and C). The binding specificity of GST–APUM5–PHD was further tested using mutant CMV RNAs. GST–APUM5–PHD did not bind to these mutant CMV RNAs at a concentration of 10 nM (Fig. 2D). Next, we examined whether the other internal putative binding motifs in the CMV RNA genome other than the 3′ UTR motifs could bind to APUM5. Interestingly, APUM5–PHD bound to CMV RNA internal motifs 1B, 3A, and 3B, but not to 1A, 2A, or 2B (Fig. 2E). This result suggested that nearby sequences, in addition to the core motif in the CMV genome, can affect binding in vitro. To further confirm the binding activity and specificity in vivo, an RNA coimmunoprecipitation (co-IP) experiment was performed. In the APUM5–GFP fraction, CMV-positive-strand RNAs were highly enriched compared with the GFP control fraction, but CMV-negative-strand RNAs were not enriched (Fig. 2F). These results indicated that APUM5 binds to CMV-positive-strand RNAs preferentially in vivo. Thus, the putative CMV RNA motif functions as a platform for the binding of APUM5.
Fig. 2.
Analysis of APUM5 binding to CMV RNAs both in vitro and in vivo. (A) The 3′ UTR sequence of CMV RNA 1 (3201–3210 nt), 2 (2884–2913 nt), and 3 (2139–2168 nt) contains the APUM5 binding motif that interacts with GST–APUM5–PHD protein. GST protein was used as a negative control. Drosophila hbNRE2 interacts with GST–APUM5–PHD protein. (B) Competition assay for the CMV 3′ UTR binding motif. The 10 nM GST–APUM–PHD protein was incubated with 32P-labeled CMV 3′ UTR RNA and various amount of unlabeled CMV 3′ UTR RNA were indicated at the top. (C) Competition assay with various amount of unlabeled nonspecific RNA (UCCUGGCCUGGAAAAUCCUGACUUUCGCGU). The relative band intensity was quantified using the ImageJ software. (D) Specific binding analysis of the core binding motif sequence in CMV 3′ UTR. Mutant RNA sequences are indicated at the top of the panel and 10 fM RNA was incubated in each reaction with 10 nM GST–APUM5–PHD protein. (E) GST–APUM5–PHD protein also interacts with some (CMV RNA motif 1B, 3A, and 3B), but not all, putative UGUA-containing sequences in internal CMV RNA genomes. Single and double asterisks indicate RNA-protein complexes and free probes, respectively. (F) RNA coimmunoprecipitation experiment. The 35S–APUM5–GFP and 35S–GFP constructs were agro-infiltrated into the CMV-infected Nicotiana benthamiana leaves. After 5 d, total proteins were extracted and immunoprecipitated with monoclonal GFP antibody. Bound RNAs were recovered. Expression of APUM5–GFP (∼133 kDa) and GFP (26 kDa) in the plant was confirmed by protein blot analysis for each fraction. Recovered RNAs were converted to cDNA and then subjected to qRT-PCR with CMV RNA (+) or (−) strand-specific primers. Error bars indicate mean ± SD (n = 5). Significant differences (Student t test; *P < 0.05, **P < 0.01) are indicated by asterisks.
APUM5 Is Involved in CMV Resistance but Does Not Play a Role in Basal Defense.
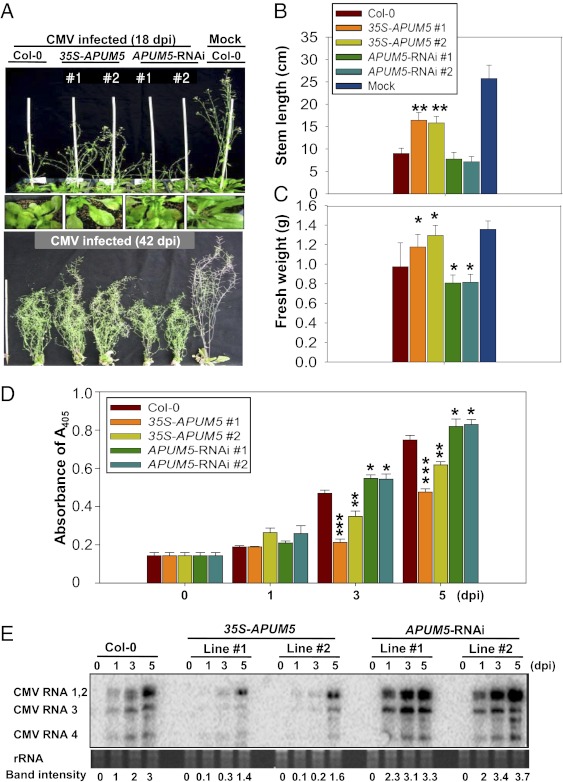
APUM5, but not a close homolog, APUM6, expression was significantly increased by salicylic acid (SA) treatment and CMV inoculation (Fig. S3 A and B). APUM5pro-GUS activities increased in leaves that were inoculated with CMV and strong APUM5pro-GUS activity was observed in systemic leaves (Fig. S3 C and D). To examine further the function of APUM5 in the plant responses to CMV infection, transgenic plants in which APUM5 expression was decreased by RNA interference (RNAi) were generated, as well as plants that overexpressed APUM5 (Fig. S4). APUM5 transgenic plants were inoculated with CMV. At 18 dpi, 35S-APUM5 transgenic plants showed reduced susceptibility, whereas APUM5-RNAi plants showed increased susceptibility to infection compared with CMV-inoculated Col-0 plants (Fig. 3 A–C). At 42 dpi, a much later stage, APUM5-RNAi plants still exhibited dwarfism and were shorter than Col-0 plants (Fig. 3A Lower). CMV CP accumulations and CMV RNA levels were reduced in 35S-APUM5 transgenic plants compared with Col-0 plants, whereas APUM5-RNAi plants exhibited increased accumulation of CMV RNAs and CP at the early stage (Fig. 3 D and E). At 18 dpi, 35S-APUM5 and APUM5-RNAi plants still showed changes in CMV RNA levels and CP accumulations (Fig. S5 A and B). However, APUM5 did not affect the SA-inducible expression of PR genes and reactive oxygen species accumulation (Fig. S6 A and B).
Fig. 3.
Analysis of APUM5 transgenic plants for susceptibility change upon CMV infection. (A) The phenotypes associated with CMV infection in Col-0 and APUM5 transgenic plants at 18 and 42 dpi. Approximately 3.5-wk-old plants were inoculated with CMV. (B) Stem length analysis of CMV-infected Col-0 and APUM5 transgenic plants at 18 dpi. Significant differences (Student t test; *P < 0.05, **P < 0.01) are indicated by asterisks. (C) Fresh weight analysis of CMV-infected Col-0 and APUM5 transgenic plants at 18 dpi. Significant differences (Student t test; *P < 0.05, **P < 0.01) are indicated by asterisks. Error bars show the mean ± SD (n = 36). (D) ELISA for CMV coat protein accumulation in Col-0 and APUM5 transgenic plants. Protein samples were collected on the indicated days and used for ELISA. Significant differences (Student t test; *P < 0.05, **P < 0.01, ***P < 0.001) are indicated by asterisks. (E) CMV RNA accumulation analysis in Col-0 and APUM5 transgenic plants. RNA was extracted from plants on the indicated days, and RNA gel blot analysis was performed with CMV-specific 3′ UTR probes. The relative band intensity was quantified using the ImageJ software.
CMV genomes contain tRNA-like structure (TLS) but do not have poly(A) tail in their 3′ UTRs. On the other hand, Turnip mosaic virus (TuMV) is a single-stranded Arabidopsis-infecting RNA virus that does have a poly(A) tail in its 3′ UTR. TuMV also contains putative Pumilio-binding core motifs in its genome and 3′ UTR (16). In TuMV-UK1 3′ UTR, putative “UGUA” core sequences were found (Fig. S5E). When the level of TuMV viral RNA accumulation was examined, 35S-APUM5 transgenic plants exhibited reduced TuMV RNA levels, whereas APUM5-RNAi plants showed increased RNA levels compared with wild-type plants (Fig. S5C). As for long-term symptoms, 35S-APUM5 transgenic plants showed an attenuated infection phenotype compared with wild-type plants (Fig. S5D). These results suggest that APUM5 also acts to repress TuMV viral RNA accumulation in the initial stages of infection and affects symptom developments.
APUM5 Regulates the CMV 3′ UTR at the Translational Level and Inhibits the CMV Replication in Protoplast.
To better understand the function of APUM5, a modified reporter system consisting of a GFP vector containing the full CMV 3′ UTR sequence or endochitinase 3′ UTR (17) as a negative control was used (Fig. 4A) (18). Confocal microscopy analysis showed that the GFP reporter containing the endochitinase 3′ UTR resulted in a normal GFP expression pattern in Col-0 and 35S-APUM5 transgenic protoplasts (Fig. 4B). However, CMV 3′ UTR reporter signals were reduced to ∼55% in the 35S-APUM5 transgenic protoplasts compared with the Col-0 protoplasts (Fig. 4 B and D). The protein levels of the GFP CMV 3′ UTR reporter were reduced in the 35S-APUM5 protoplasts compared with Col-0 (Fig. 4 C and D). However, APUM5 overexpression did not affect GFP reporter mRNA levels (Fig. 4C). Thus, mRNA stability was not changed. To confirm the specific role of the CMV 3′ UTR motif further, a mutant CMV 3′ UTR reporter was generated by site-directed mutagenesis (Fig. 4A). As a result, the GFP signal intensity of the CMV mutant 3′ UTR reporter was not changed in 35S-APUM5 transgenic protoplasts compared with Col-0 protoplasts (Fig. 4 E and G). Immunoblot analysis also showed that GFP protein levels of the CMV mutant 3′ UTR reporter expressed in Col-0 and 35S-APUM5 protoplasts were quite similar (Fig. 4 F and G). In APUM5-RNAi transgenic protoplasts, we did not find any change of reporter GFP protein or mRNA level compared with Col-0 protoplasts (Fig. S7 A–C). These results were consistent with the suggestion that APUM5 might act as a translational repressor in CMV infection. We also checked whether APUM5 affects CMV replication via CMV 3′ UTR binding. The “UGUACUUCUA” motif of CMV 3′ UTRs of CMV RNA 1, RNA 2, and RNA 3 was changed to “AAAACUUCUA” and in vitro transcripts were transformed into Col-0 and 35S-APUM5 protoplasts. Mutant CMV in vitro transcripts were normally replicated in both Col-0 and 35S-APUM5 protoplasts. However, wild-type CMV in vitro transcripts exhibited significantly reduced replication in 35S-APUM5 protoplasts compared with Col-0 protoplasts (Fig. 4H). In APUM5-RNAi transgenic protoplasts, we also tested whether down-regulation of APUM5 affects CMV replication. However, wild-type CMV in vitro transcripts were normally replicated in both Col-0 and APUM5-RNAi protoplasts (Fig. S7D). Thus, APUM5 may function as a translational repressor via direct binding to the CMV 3′ UTR motifs, although we did not show the effect of APUM5 on the other internal putative binding sites.
Fig. 4.
APUM5 repressed the CMV 3′ UTR reporter at the translational level and inhibited CMV replication in protoplast. (A) Schematic diagrams of GFP-fused reporter constructs. Endochitinase 3′ UTR was used as a negative control reporter. CMV mutant 3′ UTR reporter was generated by mutating the Pumilio binding core motif of the wild-type CMV 3′ UTR. (B) GFP-fused reporter constructs with endochitinase 3′ UTR or CMV 3′ UTR were transformed into protoplasts of Col-0 and 35S-APUM5 transgenic plants by the polyethylene glycol-mediated transformation method. Next, the GFP reporter signal was detected by LSM 700 confocal microscope (Carl Zeiss). (C) Western blot analysis was performed with 5 μg of total protoplast protein. Luciferase (LUC) was used as an internal control. Rubisco protein was used as an equal loading control. Total cellular mRNA was extracted and GFP mRNA levels were determined by RT-PCR. Actin7 mRNA level was monitored for equal loading control. (D) GFP signal intensities were quantified by LSM 700 ZEN software (Carl Zeiss) and the ImageJ program. Significant difference (Student t test; *P < 0.05, ***P < 0.001) is indicated by asterisks. Quantification of Western blot analysis was performed by Multi Gauge V3.0 (Fujifilm). Significant difference (Student t test; *P < 0.05) is indicated by an asterisk. (E) A GFP-fused CMV mutant 3′ UTR reporter construct was transformed into protoplasts of Col-0 and 35S-APUM5 transgenic plants. Next, the GFP reporter signal was detected by confocal microscopy. (F) Protein blot analysis of the GFP-fused CMV mutant 3′ UTR reporter was performed with 5 μg of total protoplast protein. LUC was used as an internal control. Rubisco protein was used as an equal loading control. Total cellular mRNA was extracted, and GFP mRNA levels were determined by RT-PCR. (G) GFP signal intensities were quantified by LSM 700 ZEN software and the ImageJ program. Quantification of protein blots was performed by Multi Gauge V3.0 software. (H) Wild-type and mutant-type CMV RNA 1, 2, and 3 in vitro transcripts were cotransformed into Col-0 and 35S-APUM5 transgenic protoplasts. After 24 h, total RNAs were extracted and RNA blot analysis was performed. For quantification of CMV replication in Col-0 and 35S-APUM5 transgenic protoplasts, signal intensities of RNA blots were quantified using the ImageJ program. Significant difference (Student t test; *P < 0.05) is indicated by an asterisk.
Recently, another repression mechanism involving Puf binding to a target mRNA was identified. Xenopus Pum2 directly interacts with the 5′ m7G cap structure, thereby blocking the assembly of the initiation complex, whereas Pum2 does not interact with the eIF4E protein (24). This suggests that Puf can affect the posttranscriptional regulation step both at the level of the 3′ UTR and the 5′ cap of mRNA. According to this model, Pum2 recognizes the m7G cap structure through its tryptophan 344 residue, which is conserved in the Pum2 of several organisms, such as human Pum2, mouse Pum2, and Xenopus Pum2. Then, Pum2 represses the translation of target mRNA by competing with eIF4E to bind to the m7G cap structure. At first we checked whether the tryptophan amino acid was conserved in APUM5, as in vertebrates. However, the tryptophan residue did not exist in APUM5 (Fig. S8 A and B). Furthermore, the APUM5 N-terminal region is not well conserved compared with mammalian or other plant Pufs (Fig. S8B). Nonetheless, we examined whether the N-terminal region of APUM5 interacted with the m7G cap structure. As expected, N-t APUM5 did not interact with the cap structure in vitro (Fig. S8C). These results imply that several Pufs can repress the posttranscriptional step in different ways.
Discussion
We identified an Arabidopsis Puf, APUM5, as a mutant with reduced susceptibility to CMV infection. The apum5-D mutant showed approximately threefold up-regulated expression of APUM5 and behaved as a gain-of-function mutant (Fig. 1). The 35S-APUM5 transgenic plants exhibited phenotypes similar to those of the apum5-D mutant (Fig. 3). The 35S-APUM5 protoplasts affected CMV replication via CMV 3′ UTR binding in vitro replication assays (Fig. 4H). Thus, APUM5 affects the translational step of CMV infection by binding to Pumilio-binding motifs in the CMV 3′ UTR. Interestingly, CMV group-I strains contain Pumilio-binding motifs at the 3′ UTR and these motifs are highly conserved. However, CMV group II strains do not have the motifs at the 3′ UTR (Fig. S2C). Thus, APUM5 could function in CMV group-I strains but not group-II strains. We also showed decreased infection phenotype change in the 35S-APUM5 transgenic plants upon TuMV inoculation, which contains putative Pumilio-binding core motifs in its genome and 3′ UTR (Fig. S5 C–E) (16). These results suggest that Pumilio binding motif of plant RNA viruses could be a rather broad target for this resistance mechanism, although we did not carry out a binding assay with the TuMV motif. A specific interaction between viral RNAs and host RNA-binding proteins affects viral RNA multiplication (19, 20, 21). BTR1, which encodes three K-homology RNA-binding domains, negatively regulates Tomato mosaic virus (ToMV) multiplication in Arabidopsis by interacting with the 5′ terminal region of ToMV genomic RNA (20).
APUM5 binds to the putative Pumilio-binding motifs in the CMV RNAs and to hbNRE2 (Fig. 2A). Drosophila Pum binds to hbNRE2 mRNA and represses its ability to regulate embryo development via deadenlyase-dependent and -independent pathways (22, 23). This indicates that APUM5 might be associated with deadenylase-dependent and -independent pathways to suppress CMV RNAs, even though CMV has a TLS instead of a poly(A) tail at its 3′ end. Furthermore, the host plant might have evolved a mechanism by which APUM5 recognizes putative Pumilio-binding motifs of CMV RNAs to repress CMV RNAs. Indeed, host protein Y-box binding protein-1 directly interacts with Dengue virus 3′ UTR and represses the translation of Dengue virus, and the virus lacks a 3′ poly(A) tail but has tRNA-like structure (21).
Xenopus Pum2 directly binds to the 5′ m7G cap structure for blocking the assembly of the initiation complex (24). However, N-t APUM5 did not bind to the cap structure in vitro (Fig. S8). These results imply that APUM5 affects CMV replication and symptoms via direct binding to CMV RNA, but not through competition with the eIF4E. The function of APUM5 is still not clear with respect to the nature of its endogenous mRNA targets and de novo interacting partners.
Materials and Methods
Plant Material, Growth Conditions, and Transgenic Plants.
Arabidopsis wild-type and transgenic plants had the ecotype Columbia-0 background and were grown in a 16 h light/8 h dark photoperiod at 23 °C in soil. For the constitutive expression of APUM5 (At3g20250), a modified pCAMBIA2300 vector was used (25), and the APUM5 ORF was amplified using Pfu DNA polymerase (Promega) and cloned. For the repression of APUM5 expression, the RNAi technique was exploited using the pHANNIVAL vector system, which can transcribe self-complementary hairpin RNA molecules (26). Specific fragments of ∼300 nucleotide sequences were chosen from the 3′ UTR of APUM5 and APUM6 (At4g25880) mRNA and amplified using Ex-Taq (TaKaRa). The pHANNIBAL cassettes containing APUM5 and APUM6 RNAi constructs were cloned into the pART27 binary vector. To prepare for the APUM5 promoter-GUS construct, the 1.3-kb promoter region was amplified using Ex-Taq and then cloned into the pBI101 vector system (Clontech). Arabidopsis plants were transformed according to the floral dip method (27) using Agrobacterium tumefaciens strain GV3101, and T3 homozygotes were obtained by antibiotic selection.
CMV and TuMV Inoculation and Analysis of Infected Plants.
Col-0, 35S-APUM5, and APUM5-RNAi transgenic plants were dusted with carborundum (Hayashi Chemical) at ∼3.5 wk of age and gently rubbed with phosphate buffer (20 mM, pH 7.0) containing CMV-Kor using brushes as described (28). At 18 or 42 dpi, stem length and fresh weight were measured, and the experiments were performed at least three times. TuMV-GFP inoculation assay was as described (29).
RNA co-IP Experiment.
To perform the in vitro and in vivo protein–RNA interaction, co-IP and qRT-PCR were performed using a slightly modified method (30). Briefly, 35S-APUM5-GFP and 35S-GFP constructs were agro-infiltrated into the CMV-inoculated Nicotiana benthamiana leaves. After 5 d, protein samples were extracted and immunopreciptated with monoclonal GFP antibody (Invitrogen). The protein–RNA complexes were washed with washing buffer [10 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 1 mM MgCl2, and 0.5% Nonidet P-40]. The collected pellet was treated with 2× PK buffer [0.2 M Tris⋅HCl (pH 7.5), 25 mM EDTA, 0.3 M NaCl, and 2% (vol/vol) SDS] containing 80 μg of proteinase K and incubated at 50 °C for 30 min. Bound RNAs were recovered by a phenol extraction method and then qRT-PCR was performed. NtAcitn was used as internal control for normalization. The primers used for the qRT-PCR are listed in Table S1.
Reporter Assay.
Protoplasts of Col-0 and APUM5 transgenic plants were isolated according to a previously described protocol, with several modifications (18). To generate reporter constructs, the full wild-type CMV 3′ UTR and mutated CMV 3′ UTR fragments were ligated into the N terminus of the modified 326-GFP3G vector, from which the nos-terminator region was removed by digestion with XhoI and EcoRI. The modified 326-GFP3G reporter containing endochitinase 3′ UTR was used as a negative reporter control and 35S-LUC in pUC vector was used as an internal control. Reporter constructs were introduced into protoplasts by polyethylene glycol-mediated transformation (18). GFP signal intensities were quantified by LSM 700 ZEN software (Carl Zeiss) and the ImageJ program (http://rsbweb.nih.gov/ij/).
Details of RNA, DNA, protein works and reporter assays, full methods, and associated references are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Hyun Kyu Song (Korea University) for providing the modified pET-GST vector, Byung-Cheon Jeong (Korea University) for technical assistance, Prof. James C. Carrington (Donald Danforth Plant Science Center) for providing the TuMV-GFP clone, Prof. Yoon Ki Kim (Korea University) for providing a cDNA clone of cap-binding protein, and Drs. Young Jin Kim and Gil-Je Lee (Korea University) for helpful discussions. This work was supported by Science Research Center-Engineering Research Center Program (Plant Signaling Network Research Center) of the Ministry of Education, Science and Technology Grant R11-2003-008-02001-0 and Wujangchoon Project Grant PJ007850 from the Rural Development Administration, Republic of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214287110/-/DCSupplemental.
References
- 1.Maule A, Leh V, Lederer C. The dialogue between viruses and hosts in compatible interactions. Curr Opin Plant Biol. 2002;5(4):279–284. doi: 10.1016/s1369-5266(02)00272-8. [DOI] [PubMed] [Google Scholar]
- 2.Bucher GL, et al. Local expression of enzymatically active class I beta-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 2001;28(3):361–369. doi: 10.1046/j.1365-313x.2001.01181.x. [DOI] [PubMed] [Google Scholar]
- 3.Yamanaka T, et al. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA. 2000;97(18):10107–10112. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsushita Y, Deguchi M, Youda M, Nishiguchi M, Nyunoya H. The tomato mosaic tobamovirus movement protein interacts with a putative transcriptional coactivator KELP. Mol Cells. 2001;12(1):57–66. [PubMed] [Google Scholar]
- 5.Kim MJ, Kim HR, Paek KH. Arabidopsis tonoplast proteins TIP1 and TIP2 interact with the cucumber mosaic virus 1a replication protein. J Gen Virol. 2006;87(Pt 11):3425–3431. doi: 10.1099/vir.0.82252-0. [DOI] [PubMed] [Google Scholar]
- 6.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann R, Nüsslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119(2):402–417. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- 8.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15(6):762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22(8):1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Droll D, et al. The trypanosome Pumilio-domain protein PUF7 associates with a nuclear cyclophilin and is involved in ribosomal RNA maturation. FEBS Lett. 2010;584(6):1156–1162. doi: 10.1016/j.febslet.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francischini CW, Quaggio RB. Molecular characterization of Arabidopsis thaliana PUF proteins—binding specificity and target candidates. FEBS J. 2009;276(19):5456–5470. doi: 10.1111/j.1742-4658.2009.07230.x. [DOI] [PubMed] [Google Scholar]
- 12.Tam PP, et al. The Puf family of RNA-binding proteins in plants: Phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010;10:44. doi: 10.1186/1471-2229-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palukaitis P, García-Arenal F. Cucumoviruses. Adv Virus Res. 2003;62:241–323. doi: 10.1016/s0065-3527(03)62005-1. [DOI] [PubMed] [Google Scholar]
- 14.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2(3):E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotton S, et al. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J Virol. 2009;83(20):10460–10471. doi: 10.1128/JVI.00819-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin HH, Huang LF, Su HC, Jeng ST. Effects of the multiple polyadenylation signal AAUAAA on mRNA 3′-end formation and gene expression. Planta. 2009;230(4):699–712. doi: 10.1007/s00425-009-0977-4. [DOI] [PubMed] [Google Scholar]
- 18.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 19.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA. 2002;8(3):265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujisaki K, Ishikawa M. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology. 2008;380(2):402–411. doi: 10.1016/j.virol.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 21.Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282(42):30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- 22.Chagnovich D, Lehmann R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci USA. 2001;98(20):11359–11364. doi: 10.1073/pnas.201284398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124(15):3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 24.Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16(1):221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ham BK, et al. Tobacco Tsip1, a DnaJ-type Zn finger protein, is recruited to and potentiates Tsi1-mediated transcriptional activation. Plant Cell. 2006;18(8):2005–2020. doi: 10.1105/tpc.106.043158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wesley SV, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27(6):581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1(2):641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 28.Kim MJ, Huh SU, Ham BK, Paek KH. A novel methyltransferase methylates Cucumber mosaic virus 1a protein and promotes systemic spread. J Virol. 2008;82(10):4823–4833. doi: 10.1128/JVI.02518-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Ruiz H, et al. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell. 2010;22(2):481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez M, Galy B, Hentze MW, Muckenthaler MU. Identification of target mRNAs of regulatory RNA-binding proteins using mRNP immunopurification and microarrays. Nat Protoc. 2007;2(8):2033–2042. doi: 10.1038/nprot.2007.293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.