Abstract
With the aim of enhancing G-quadruplex binding activity, two new glucosaminosides (16, 18) of penta-methylated epigallocatechin were synthesized by chemical glycosylation. Subsequent ESI-TOF-MS analysis demonstrated that these two glucosaminoside derivatives exhibit much stronger binding activity to human telomeric DNA and RNA G-quadruplexes than their parent structure (i.e., methylated EGC) (14) as well as natural epigallocatechin (EGC, 6). The DNA G-quadruplex binding activity of 16 and 18 is even more potent than strong G-quadruplex binder quercetin, which has a more planar structure. These two synthetic compounds also showed a higher binding strength to human telomeric RNA G-quadruplex than its DNA counterpart. Analysis of the structure-activity relationship revealed that the more basic compound, 16, has a higher binding capacity with DNA and RNA G-quadruplexes than its N-acetyl derivative, 18, suggesting the importance of the basicity of the aminoglycoside for G-quadruplex binding activity. Molecular docking simulation predicted that the aromatic ring of 16 π-stacks with the aromatic ring of guanine nucleotides, with the glucosamine moiety residing in the groove of G-quadruplex. This research indicates that glycosylation of natural products with aminosugar can significantly enhance their G-quadruplex binding activities, thus is an effective way to generate small molecules targeting G-quadruplexes in nucleic acids. In addition, this is the first report that green tea catechin can bind to nucleic acid G-quadruplex structures.
Introduction
Nucleic acid G-quadruplexes, four-stranded helical structures held together by a core of guanine tetrads, are secondary structures formed in particular G-rich sequences. Potential nucleic acid G-quadruplex structures have been identified in telomeric DNA and RNA sequences [1]–[5] as well as non-telomeric chromosomal promoters [6]–[9] of biological significance. These higher-order structures in nucleic acids represent a new class of molecular targets for selective DNA- and RNA-interacting compounds; in view of the fact that cancer cells have high telomerase activity and abnormal overexpression of oncogenes relative to normal cells, they are promising targets for cancer drug discovery [10]. In addition, numerous compounds have been designed to inhibit telomerase or to inactivate the transcription of oncogenes, such as c-Myc, c-kit, and Bcl-2 [6], [9], [11]–[13], suggesting that the design of drugs targeting telomere or promoter G-quadruplexes is a rational and promising approach for generating new anticancer agents [14]. While recognition of G-quadruplex has mostly been achieved with the use of planar aromatic ligands through stacking interactions with the G-tetrad [14], grooves and negatively charged phosphate residues in G-quadruplexes are alternative binding sites to consider in the design of G-quadruplex stabilizing ligands [15]–[22].
Green tea catechins, the main biologically-active constituents of green tea, have gained significant recognition as cancer preventive agents. Green tea catechins are composed of four major polyphenols: (−)-epigallocatechin gallate (EGCG), (−)-epigallocatechin (EGC), (−)-epicatechin gallate (ECG), and (−)-epicatechin (EC) [23]. They show a variety of pharmacological activities, including cancer-preventive, antioxidant, anti-cancer [24], anti-angiogenesis activities [25], as well as inhibiting the fibrillogenesis of amyloid β peptide [26]–[27], anti-mutagenic and anti-viral activities [28]–[31]. It has been reported that catechins affect DNA replication, DNA repair, and transcription [32]–[34]. A recent study revealed that nucleic acids are binding targets of green tea catechins: nucleic acids extracted from EGCG-treated human cancer cells were catechin-colored, and direct binding of catechins with single-stranded and double-stranded DNA/RNA was observed by cold spray ionization-mass spectrometry [34]. However, a molecular docking study indicated that catechins, including EGC, are poor DNA G-quadruplex-stabilizing ligands compared with the more planar compound quercetin [35]. Therefore, structural modification of EGC is necessary for enhancing its G-quadruplex binding affinity.
It has been discovered that aminosugar moieties play an essential role in both the in vitro and in vivo antitumor activity of anthracyclines based on a DNA-binding mechanism [36]. It was also found that the N-acetyl glucosamine moiety seems to enhance the cytotoxic activity of the saponin julibroside III towards KB cancer cells [37]. A recent study found that non-planar aminoglycosides, such as neomycin and paromomycin, recognize the wide groove of Oxytricha nova telomeric G-quadruplex DNA [15]. These findings led to a prediction that the coupling of aminosugars with ligands that bind to G-quadruplex through stacking interactions may lead to enhanced G-quadruplex stabilizing properties. In our previous study, it was demonstrated that glycosylation of shikonin/alkannin with N-acetyl glucosamine is an effective way to generate a potent G-quadruplex DNA ligand [38]. Based on these observations, in this study we herein designed and synthesized two new glucosaminosides of EGC (16, 18) and subsequently examined their binding affinities with both telomeric DNA and RNA G-quadruplexes by ESI-TOF-MS. Furthermore, the binding of these two glucosaminosides (16, 18) with oncogene G-quadruplexes was also explored. Finally, the binding mode of 16 with human telomeric DNA G-quadruplex was investigated by computational docking experiments.
Results
Synthesis of EGC Glucosaminosides
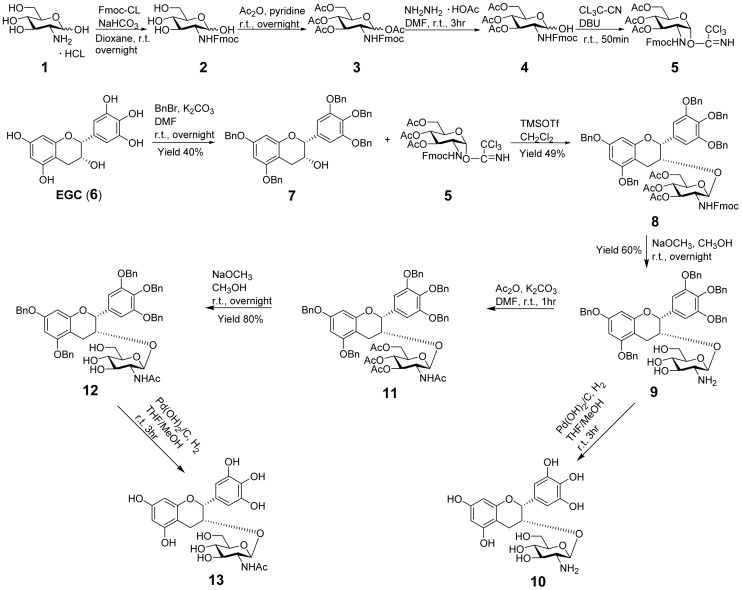
Glycosylation is an effective method for connecting saccharide units to natural products in order to obtain biologically active glycosides [39]–[40]. Many glycosylated natural products have been reported to show high activity against a variety of human tumors [36]–[37]. In this study, chemical glycosylation was employed as a key approach to acquiring EGC glucosaminosides. As illustrated in Figure 1, our initial efforts were focused on the design and synthesis of EGC-3-O-β-glucosaminoside (10) and its N-acetyl derivative (13), starting from the readily available (−)-EGC and D-(+)-glucosamine hydrochloride. The glycosyl donor 5 was prepared according to the method previously described in the literature [38], [41]–[42]. The amino group of glucosamine was firstly blocked by 9-fluorenylmethoxycarbonyl chloride (Fmoc-Cl) and followed by acetylation of the hydroxyl groups. After selective deprotection, the anomeric hydroxyl group was transformed to trichloroacetimidate and thus activated for glycosylation [38].Trimethylsilyl triflate (TMSOTf) was used as a catalyst for the glycosylation of the penta-benzyl ether of EGC (7). After deprotection and acetylation of the amino group, EGC glucosaminoside [10, ESI-TOF-MS m/z (C21H26NO11)+: calcd 468.1500, found 468.1489] and its N-acetyl product 13 [ESI-TOF-MS m/z (C23H28NO12)+: calcd 510.1606, found 510.1599] were synthesized. Unfortunately, we failed to purify these two products due to their instability during the course of column chromatographic purification.
Figure 1. Figure.
1. Synthesis of glucosaminosides of EGC.
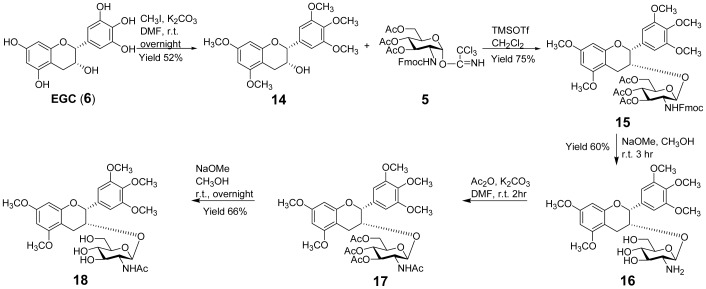
To avoid this instability, we slightly modified our target products by blocking the active phenolic hydroxyl groups of EGC with methyl groups. The penta-methylated EGC (14) was subsequently glycosylated with glycosyl donor 5 by the action of TMSOTf, followed by deprotection to give 16. Its N-acetyl derivative 18 was synthesized by further acetylation of 16 and subsequent deacetylation (Figure 2). Compounds 16 and 18 were characterized to be 5, 7, 3′, 4′, 5′-penta-O-methyl epigallocatechin β-D-glucosaminoside and 5, 7, 3′, 4′, 5′-penta-O-methyl epigallocatechin N-acetyl β-D-glucosaminoside, respectively, on the basis of 1H-NMR, 13C-NMR and high resolution mass spectroscopic evidence (Figure S1, S2, S3, S4, S5, S6, S7, S8, S9) [43]–[44].
Figure 2. Synthesis of glucosaminosides of penta-methylated EGC.
Analysis of Human Telomeric G-quadruplex DNA Binding by ESI-TOF-MS
Mass spectrometry coupled with the source of electrospray ionization (ESI), a soft ionization method, has played a more active role in the investigation of noncovalent complexes of nucleic acids with small organic molecules. It has the advantages of direct assignment of the stoichiometry and gives an indication of the relative amounts of different species of complexes [45]–[46]. Mass spectrometry, combined with techniques of ion mobility and molecular dynamics, has demonstrated that DNA G-quadruplexes in telomeric repeats are conserved in a solvent-free environment [47].
The G-quadruplex DNA-binding activities of glucosaminosides of penta-methylated EGC (16, 18), as well as their aglycone (14) and natural EGC (6), were examined with a 27 nt human telomeric sequence d[(TTAGGG)4TTA] which forms an intramolecular G-quadruplex, by ESI-TOF-MS. Quercetin, a flavonoid with a similar but more planar structure than EGC, was used as a reference compound for the comparison of G-quadruplex binding activity of the natural and synthetic compounds, since it was reported to be stacked with terminal tetrads of monomeric G-quadruplexes [48].
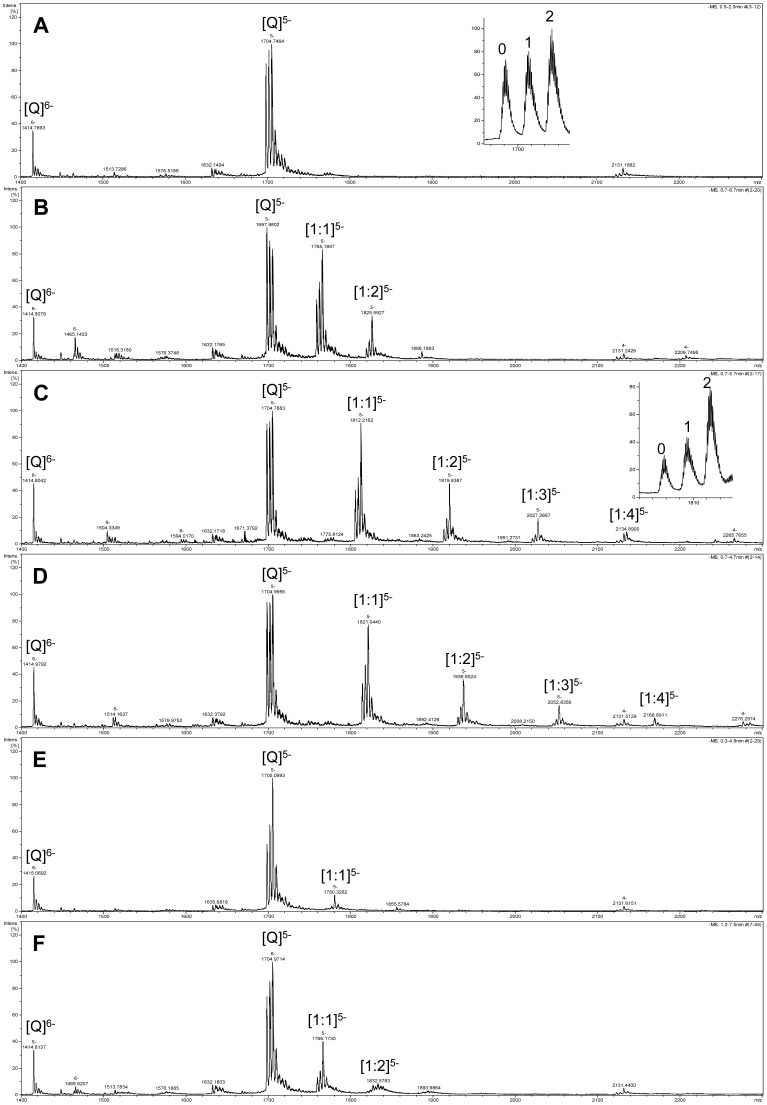
The ESI-TOF-MS spectrum of telomeric DNA showed that the addition of the NH4OAc buffer facilitated the detection of quadruplex (Q5− in Figure 3A) [49] in the −5 charge state ions at m/z 1697.9, 1701.3, and 1704.7. These three ions correspond to the lone oligodeoxynucleotide and the oligodeoxynucleotides with one and two NH4 + ion adducts, respectively. When the drug was added to DNA, the complex peaks with two NH4 + ion adducts became more predominant than those with one NH4 + ion adduct or none when a molar ratio of DNA/drug of 1∶1 was used (Figure 3B–F). This indicated that the G-quadruplex structure stabilized by drugs holds NH4 + ions inserted between G-tetrads more tightly than free G-quadruplex in the course of being introduced into the gas phase. In other words, drug-bound G-quadruplex is more stable than when it is unbound. In order to compare the stabilization effect of different molecules on DNA G-quadruplex, the peak area ratio of all [complex]5− to [quadruplex]5− was used to evaluate the relative binding affinities (Figure 4) [49]–[51].
Figure 3. ESI-TOF-MS spectra of telomeric DNA d[(TTAGGG)4TTA] (Q) in the absence and presence of drugs.
Negative ESI-TOF-MS spectra of human telomeric DNA sequence d[(TTAGGG)4TTA] were recorded under conditions of (A) without drug, (B) with quercetin, (C) with compound 16, (D) with compound 18, (E) with compound 14 and (F) with EGC. The inserts in spectra A and C are the enlargements of free G-quadruplex and complex ions with 1∶1 binding stoichiometry, respectively. The numbers in the inserts represent the number of ammonium ion adducts. Spectra were recorded with a molar ratio of DNA/drug of 1∶1 (C = 50 µM) in 50 mM ammonium acetate buffer (pH 7.6) containing 50% methanol.
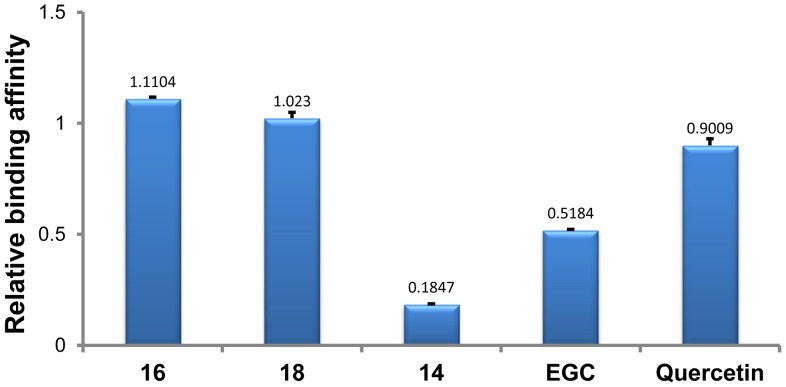
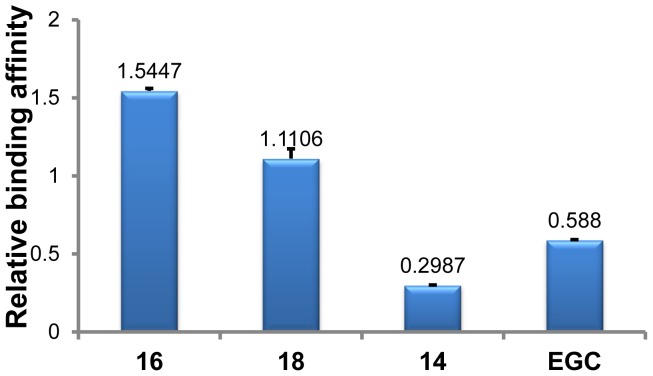
Figure 4. Relative binding affinities of drugs with intramolecular human telomeric DNA G-quadruplex.
The numbers indicated on the top of each column showed the mean values from two determinations.
As illustrated in Figure 4, the relative binding affinities of all tested drugs with intramolecular human telomeric DNA G-quadruplex followed the descending order of 16>18> Quercetin>EGC >14. Two synthetic glucosaminosides (16 and 18) demonstrated a more strong stabilizing effect on intramolecular human telomeric DNA G-quadruplex than their parent structure, methylated EGC (14). These two glucosaminosides also showed higher relative binding affinities than the natural catechin EGC and the even more planar flavonol quercetin. This indicated that the introduction of a glucosamine moiety into penta-methylated EGC (14), the weakest G-quadruplex binder among all tested compounds, resulted in a largely enhanced G-quadruplex stabilizing ability. This finding was further supported by the results of UV-melting study [15]–[22] that the melting temperature (T m) of dAGGG(TTAGGG)3 was increased 3.92, 1.86, 0.34 and 0°C?by the presence of 50 µM of 16, 18, EGC and 14, respectively. On the basis of the above results, the following structure-activity relationships can be summarized. First, the more basic compound 16 demonstrated more potent G-quadruplex DNA-binding capacity than compound 18, suggesting the importance of basicity of the aminoglycoside in G-quadruplex DNA-binding activity. Secondly, the distinct binding behaviors of EGC and its penta-methylated derivative (14) to G-quadruplex DNA indicated that the hydroxyls in EGC are essential groups for its G-quadruplex DNA-binding activity.
It was also found that both a glucosaminoside of methylated EGC (16) and its acetyl-N derivative (18) bind to the intramolecular human telomeric DNA G-quadruplex with 1∶1, 1∶2, 1∶3 and 1∶4 stoichiometries when a molar ratio of DNA/drug of 1∶1 is used. However, their aglycone 14 only shows 1∶1 binding stoichiometry under the same condition. The multiple stoichiometries of 16 and 18 binding with intramolecular human telomeric DNA G-quadruplex suggested that, unlike their aglycone, these two glucosaminosides of penta-methylated EGC bind to multiple sites on the human telomeric DNA G-quadruplex.
Molecular Modeling of Methylated EGC Glucosaminoside Derivatives Binding with Human Telomeric DNA G-quadruplex
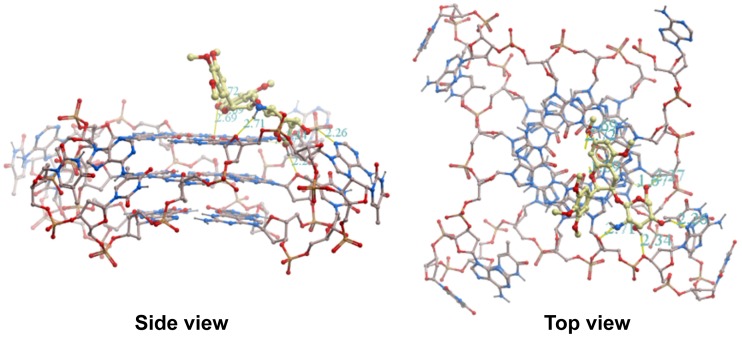
Using 16 as a model compound, a molecular modeling study was performed on its binding with intramolecular human telomeric G-quadruplex (PDB code: 1KF1 [52]) to provide insight into the binding mode of aminoglucosides of methylated EGC. As illustrated in Figure 5, the aglycone moiety of 16 is predicted to bind to the 5′ terminal face of the G-quadruplex through stacking interactions between the aromatic rings of methylated EGC and guanine nucleotides. The part of the glucosamine moiety of 16 is predicted to reside in the grooves of the G-quadruplex through hydrogen bonding interactions between donors in the G-quadruplex and hydrogens in both the amino and hydroxyl groups of 16. The amino hydrogen of 16 forms a hydrogen bond with the oxygen atom of the deoxyribose in the phosphate backbone. The hydroxyl hydrogens of 16 form hydrogen bonds with oxygen atoms from the phosphate sugars and adenine residues of the G-quadruplex. This kind of binding mode, towards the top of the 5′ terminus of the G-quadruplex, is the most favorable binding interaction, with a binding energy of −35.99 kJ/mol.
Figure 5. Molecular modeling of 16 binding with the human intramolecular telomeric G-quadruplex (PDB code: 1KF1).
Oxygen atoms are highlighted in red, nitrogen atoms in blue, phosphorus atoms in yellow and carbon atoms in beige.
Analysis of Binding with Oncogene G-quadruplex DNA by ESI-TOF-MS
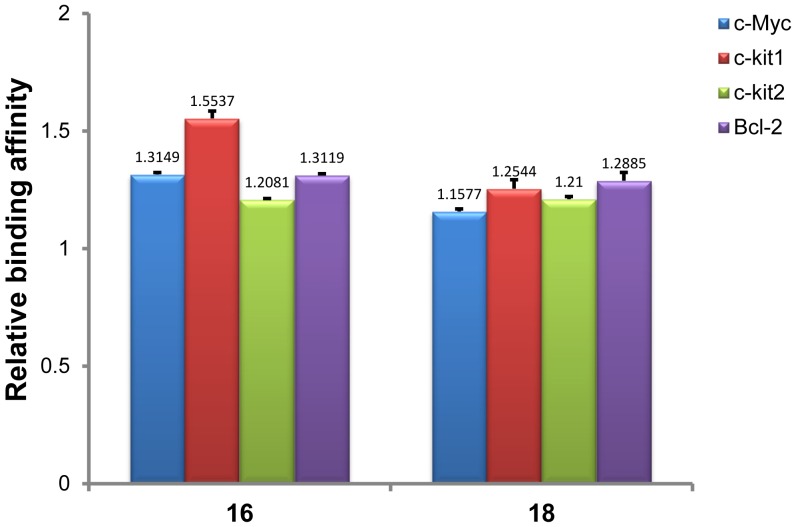
In addition to human telomeric G-quadruplex DNA, the binding of two synthetic glucosaminosides of methylated EGC (16, 18) with oncogene G-quadruplexes derived from the sequences of c-Myc, c-kit1, c-kit2 and Bcl-2 was further investigated by the same ESI-TOF-MS technique. All the oncogene sequences displayed the ability to form G-quadruplex structures in NH4OAc buffer, as ESI-TOF-MS spectra revealed that the major ion in −5 charge state of each sequence corresponds to the m/z value of the oligodeoxynucleotide with two NH4 + ions adduct (Figure S10). As with telomeric DNA G-quadruplex, 16 and 18 also showed multiple binding stoichiometries with all oncogene DNA G-quadruplexes. The relative binding affinities presented in Figure 6 show that 16 demonstrated comparative binding strength with c-Myc, c-kit1, c-kit2, and Bcl-2 DNA G-quadruplexes. Compound 18 exhibited almost equivalent binding capacity with all oncogene G-quadruplexes. These results demonstrate that glucosaminosides (16, 18) of methylated EGC exhibit binding capacity to different oncogene G-quadruplexes, though without obvious G-quadruplex selectivity.
Figure 6. Relative binding affinities of 16 and 18 with different intramolecular oncogene G-quadruplexes.
The experiments were conducted with a molar ratio of DNA/drug of 1∶1 (100 µM:100 µM) in 100 mM ammonium acetate (pH 7.6) containing 50% methanol. The numbers indicated on the top of each column showed the mean values from two determinations.
Analysis of Interaction with Human Telomeric RNA G-quadruplex by ESI-TOF-MS
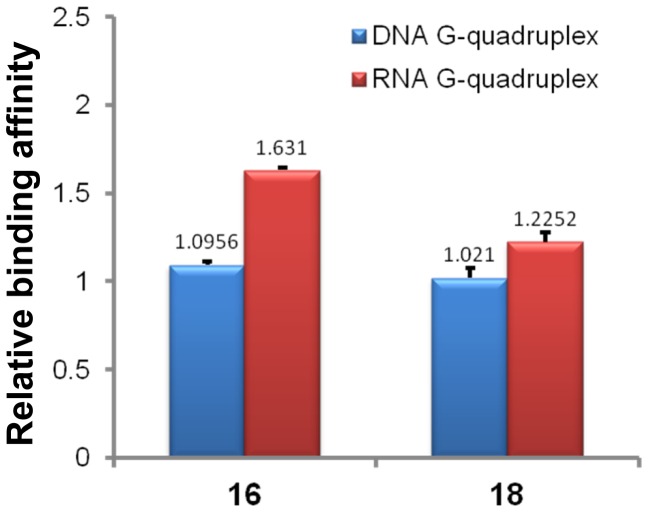
A recent finding demonstrated that telomere DNA is transcribed into telomeric repeat-containing RNA in mammalian cells. The telomeric repeat-containing RNA sequence, r(UUAGGG)4, folds into a parallel G-quadruplex in solution that is more stable than its DNA counterpart [53]–[56]. The binding of synthetic glucosaminosides (16, 18) of penta-methylated EGC, penta-methylated EGC (14) and EGC (6) were therefore studied with a 27 nt human telomeric RNA sequence, r[(UUAGGG)4UUA], under the same ESI-TOF-MS conditions as used for its DNA counterpart (Figure S11) [55]–[56]. As shown in Figure 7, the RNA G-quadruplex binding strength of these four compounds also followed the same descending trend of 16>18> EGC >14 as seen with a DNA G-quadruplex. By Comparing with the DNA G-quadruplex binding results (Figure 4), it was found that the relative affinity of each compound for the RNA G-quadruplex was slightly higher. In order to confirm this result, competitive binding experiments were carried out for two methylated EGC glucosaminosides (16, 18) with both DNA and RNA G-quadruplexes to further elucidate the DNA and RNA G-quadruplex binding selectivity of each compound (Figure S12). In each competition experiment, human telomeric DNA and RNA G-quadruplexes were mixed with each compound to give a final molar ratio of DNA/RNA/compound of 1∶1∶2. The peak area ratio of drug-bound complex to free nucleic acid of each species of G-quadruplex was calculated to give the value of relative affinity (Figure 8), which confirmed that both 16 and 18 show stronger binding affinity with RNA G-quadruplex than DNA G-quadruplex. To provide further evidence to support the ESI-TOF-MS results of the above binding studies of these aminoglucosaminosides, we undertook UV-melting experiments [15]–[22]. It was demonstrated that 16 and 18 increased melting temperature of rAGGG(UUAGGG)3 6.08 and 3.26°C, respectively, under the same tested condition with that of DNA analogue. The larger ΔT m value for RNA G-quadruplex further confirmed that 16 and 18 exhibited stronger stabilizing effects on RNA G-quadruplex than DNA counterpart. However, there are no obvious increases for the melting temperature of a double-stranded oligodeoxynucleotide (ds-DNA) 5′-AGGGTTAGGGT-3′/3′-TCCCAATCCCA-5′ in the presence of 16 (0.22°C)?and 18 (−0.15°C), indicating the selectivity of two synthetic compounds to RNA and DNA G-quadruplexex over duplex oligonucleotide (Figure S13).
Figure 7. Relative binding affinities of drugs with intramolecular human telomeric RNA G-quadruplex.
The numbers indicated on the top of each column showed the mean values from two determinations.
Figure 8. Relative binding affinities of drugs with DNA and RNA G-quadruplex in competitive binding experiments.
The numbers indicated on the top of each column showed the mean values from two determinations.
Discussion and Conclusion
In this study, two glucosaminosides of methylated EGC, compounds 16 and 18, have been successfully synthesized for the first time. Both the DNA and RNA G-quadruplex binding activity of these two glucosaminosides has been evaluated and compared with that of green tea catechin EGC by ESI-TOF-MS analysis. The DNA and RNA G-quadruplex binding capacity of both synthetic compounds and natural EGC with human telomeric DNA and RNA sequences both followed the order of 16>18> EGC >>14. This finding indicated that introduction of a glucosamine moiety to penta-methylated EGC (14), the weakest G-quadruplex binder among all tested compounds, resulted in much stronger G-quadruplex stabilizing ability that exceeds natural EGC. In addition, they exhibited stronger DNA G-quadruplex binding activity than the more planar structure quercetin. Analysis of the structure-activity relationship revealed that the more basic glucosaminoside 16 generally showed more potent G-quadruplex binding capacity than that of 18, indicating the importance of basicity of aminoglycoside in G-quadruplex binding activity. Furthermore, it was demonstrated that both glucosaminosides of penta-methylated EGC had a greater binding affinity with the RNA G-quadruplex than its DNA counterpart. Additionally, it has been shown that glucosaminosides 16 and 18 can also bind to different oncogene G-quadruplexes, although without obvious G-quadruplex selectivity. Taken together, these results demonstrate that aminoglycosylation of natural products is an effective way to design and synthesize small molecules targeting G-quadruplexes in nucleic acids.
ESI-TOF-MS analysis revealed that glucosaminosides 16 and 18 demonstrated more binding stoichiometries with an intramolecular human telomeric G-quadruplex than EGC under a molar ratio of DNA/drug of 1∶1, suggesting that there are more binding sites for 16 and 18 in the intramolecular human telomeric G-quadruplex than the natural tea catechin EGC and their aglycone (14). Subsequent molecular docking simulation predicted that the aromatic ring of compound 16 π-stacked with the aromatic ring of guanine nucleotides, with the glucosamine moiety residing in the groove of G-quadruplex. This prediction is consistent with the binding mode of aminoglycosides neomycin and paramonomycin [15]. This kind of binding mode is also in agreement with the multiple binding stoichiometries of 16 with the 27 nt human telomeric G-quadruplex detected by ESI-TOF-MS.
Although green tea catechins were proven to bind to normal (single-stranded and double-stranded) DNA and RNA [28]–[31], this is the first time they were also found to bind to DNA and RNA G-quadruplex structures. The distinct binding behaviors of EGC and its penta-methylated derivative, compound 14, to DNA and RNA G-quadruplexes suggested that the hydroxyl groups in EGC are essential for stabilizing DNA and RNA G-quadruplexes. The binding of nucleic acid G-quadruplexes by green tea catechins may be in part responsible for their cancer-preventive activities.
Materials and Methods
General
The trichloroacetimidate method was employed to conduct glycosylation reaction, which was performed in the presence of TMSOTf at −40°C under an atmosphere of argon, followed by deprotection of sugar moiety with sodium methoxide in methanol and further catalytic hydrogenation with palladium hydroxide on carbon powder. Unless otherwise noted, all reactions were conducted in oven-dried glassware. Column chromatography for product purification was performed on DAVISIL® chromatographic silica gel LC60A (40–63 micron, GRACE Davison, Germany), and Chromatorex ODS (100–200 mesh, Fuji Silysia Chemical Ltd., Japan). Purity checking of product was accomplished on an ultra performance LC system (Acquity™, Waters) equipped with photodiode array detector (Waters) and a Bruker micrOTOF ESI-TOF mass spectrometer by using an Acquity UPLC® BEH C18 column (17 µM, 2.1×100 mm, part No. 186002352, Ireland). 1H and 13C NMR were recorded at room temperature on a Bruker 400 MHz NMR Avance-III (Switzerland) or a Varian 400 MHz NMR Inova 400 (USA) spectrometers operating at 400 MHz (1H) and 100 MHz (13C). Coupling constants were given in Hz and chemical shifts were represented in δ (ppm) relative to Me4Si as internal standard. HR-ESI-MS was performed on a Q-TOF mass spectrometer (Bruker Daltonics, MA, U.S.A). Optical rotation was measured on a JASCO P-1010 polarimeter (Japan) with a 1 dm cell (C given in g/100 mL).
3,4,5-tri-O-acetyl-2-deoxy-2-(9-Fluorenylmethoxycarbonylamino)-α-D-glucopyranosyl Trichloroacetimidate (5)
To a solution of glucosamine chloride (8.15 g, 37.8 mmol) in water (50 mL), NaHCO3 (4.76 g, 56.7 mmol) and 9-fluorenylmethoxycarbonylchloride (11.7 g, 45.4 mmol) in dioxane (50 mL) were added and the reaction stirred at room temperature until silica gel TLC (CHCL3-EtOAc-MeOH 7∶2∶3) showed complete conversion of the starting material to N-protected glucosamine (2). The reaction solution was slowly poured into ice water (900 mL) and filtered under decreased pressure after stirring at room temperature for 2 h. The residue was washed by water and dried at 50°C to afford crude 2 (13.8 g). The N-protected glucosamine (2, 13.8 g, 34.6 mmol) in pyridine (50 mL) was acetylated with acetic anhydride (18 mL, 173 mmol) by stirring overnight at room temperature to produce tetra-O-acetyl carbohydrate (3). The reaction solution was poured into ice water and filtered under decreased pressure, followed by silica gel column chromatography (PE-acetone 7∶3) to give 3 (18.5 g). The anomeric acetyl group of 3 (8.65 g, 15.2 mmol) in DMF (50 mL) was then deprotected with hydrazine acetate (1.54 g, 16.72 mmol) by stirring at room temperature for 5 h. The reaction solution was then diluted with EtOAc (300 mL), then washed with water (100 mL×4). The organic layer was dried with anhydrous Na2SO4 and concentrated to give 4 (7.3 g). To a solution of 4 (1.844 g, 3.50 mmol) in anhydrous CH2Cl2 (60 mL), CCl3-CN (2.91 mL, 13.97 mmol) and DBU (58.2 µL, 0.35 mmol) was added. After stirring at room temperature for 50 minutes, the reaction mixture was directly loaded to silica gel chromatography column eluted with n-hexane-EtOAc (6∶4) containing 0.5% triethylamine to afford 5 (2.25 g, 96% yield).
(−)-5, 7, 3′, 4′, 5′-Penta-O-methyl Epigallocatechin (14)
To a solution of epigallocatechin (612 mg, 2.0 mmol) in DMF (20 mL), methyl iodide (1.0 mL, 16.0 mmol) and K2CO3 (2.21 g, 16.0 mmol) was added. The mixture was stirred at room temperature for 15 h. 180 mL of water was poured into the reaction solution, followed by extraction with ethyl acetate (100 mL × 4). The organic solutions was dried with anhydrous Na2SO4 and subjected to silica gel chromatography (n-hexane-ethyl acetate 9∶1 to 5∶5) to afford 14 (388 mg, 52% yield) as a white amorphous powder. [α]D 20 −76.16 (C = 0.44, EtOAc). HRMS (ESI) m/z (C20H25O7)+: calcd 377.1595, found 377.1583. 1H-NMR (400 MHz, CDCl3) δ: 4.94 (s, H-2), 4.30 (m, H-3), 2.98 (dd, J = 1.6, 17.3 Hz, H-4β), 2.90 (dd, J = 4.3, 17.3 Hz, H-4α), 6.21 (d, J = 2.3, H-6), 6.13 (d, J = 2.3 Hz, H-8), 6.75 (2H, s, H-2′, 6′), 3.86 (3H, s, 4′-OCH3), 3.80, 3.78 (3H each, s, 5, 7-OCH3), 3.90 (6H, s, 3′, 5′-OCH3), 1.78 (d, J = 5.6 Hz, 3-OH). 13C-NMR (100 MHz, DMSO) δ: 78.4 (C-2), 64.4 (C-3), 28.6 (C-4), 101.0 (C-4a), 158.9, 158.7 (C-5, C-7), 93.4 (C-6), 91.4 (C-8), 155.4 (C-8a), 135.2 (C-1′), 104.4 (C-2′, C-6′), 152.5 (C-3′, C-5′), 136.7 (C-4′), 55.4, 55.2 (5, 7-OCH3), 55.9 (3′, 5′-OCH3), 60.0 (4′-OCH3).
Compound 15
A heterogeneous mixture of compound 14 (113 mg, 0.30 mmol) and trichloroacetimidate 5 (336 mg, 0.50 mmol) in anhydrous CH2Cl2 (6 mL) with 4 Å molecular sieves (500 mg) was stirred at room temperature for 30 min under an argon atmosphere and then was cooled to −40°C. TMSOTf (600 µL, 0.06 M in dry CH2CL2) was added quickly to the precooled reaction mixture and the resulting mixture was allowed to warm to room temperature over 3 h. Triethylamine (24 µL) was added to quench the reaction. The organic solution was then directly loaded to silica gel chromatograph column eluted with n-hexane-ethyl acetate (from 9∶1 to 5∶5) to give 15 (199.1 mg, 75% yield). HRMS (ESI) m/z (C47H52NO16)+: calcd 886.3281, found 886.3275.
(−)-5, 7, 3′, 4′, 5′-penta-O-methyl Epigallocatechin β-D-glucosaminoside (16)
Compound 15 (177 mg, 0.20 mmol) was dissolved in methanol (14 mL) and sodium methoxide (40 mg, 0.74 mmol) was added. After 3 h of stirring at room temperature, the reaction solution was filtered and evaporated under reduced pressure. The residue was purified by silica gel chromatography eluted with chloroform-methanol-water (from 90∶10∶1 to 80∶20∶2) to afford 16 (64 mg, 60% yield) as a white amorphous powder. Optical rotation [α]D 20 −30.48 (C = 0.44, MeOH). HRMS (ESI) m/z (C26H36NO11)+: calcd 538.2283, found 538.2271. 1H-NMR (400 MHz, DMSO and D2O) δ: 5.14 (s, H-2), 4.50 (m, H-3), 2.72 (dd, J = 3.6, 17.4 Hz, H-4a), 2.67 (dd, J = 4.0, 17.4 Hz, H-4b), 6.12 (d, J = 2.3 Hz, H-6), 6.09 (d, J = 2.3 Hz, H-8), 6.84 (2H, s, H-2′, 6′), 3.71, 3.65 (3H each, s, 5, 7-OCH3), 3.72 (6H, s, 3′, 5′-OCH3), 3.74 (3H, s, 4′-OCH3), 4.26 (d, J = 7.8 Hz, glc-1), 2.31 (dd, J = 8.0, 9.0 Hz, glc-2), 3.04 (t, J = 9.0 Hz, glc-3), 2.96 (t, J = 9.0 Hz, glc-4), 3.12 (m, glc-5), 3.68 (m, overlapped with H2O peak, glc-6a), 3.34 (dd, J = 6.4, 11.6 Hz, glc-6b). 13C-NMR (100 MHz, DMSO) δ: 77.5 (C-2), 70.1 (C-3), 23.9 (C-4), 101.3 (C-4a), 159.6, 158.9 (C-5, 7), 93.6 (C-6), 91.9 (C-8), 155.6 (C-8a), 134.2 (C-1′), 105.6 (C-2′, 6′), 152.6 (C-3′, 5′), 137.1 (C-4′), 100.9 (glc-1), 57.5 (glc-2), 76.4 (glc-3), 70.6 (glc-4), 77.4 (glc-5), 61.6 (glc-6), 55.9, 55.6 (5, 7-OCH3), 56.2 (3′, 5′-OCH3), 60.5 (4′-OCH3).
Compound 17
To a solution of compound 16 (16 mg, 0.03 mmol) in DMF (2 mL), acetic anhydride (40 µL, 0.39 mmol) and K2CO3 (12 mg, 0.09 mmol) was added at room temperature. After 2 hours of stirring, ice water (20 mL) was poured into the mixture solution to stop reaction. Then ethyl acetate (10 mL × 3) was used to extract the target product. The organic solution was dried with anhydrous Na2SO4 and evaporated to give 17 (15.6 mg, 74% yiled). HRMS (ESI) m/z (C34H44NO15)+: calcd 706.2705, found 706.2695.
(−) 5, 7, 3′, 4′, 5′-penta-O-methyl Epigallocatechin N-acetyl β-D-glucosaminoside (18)
Sodium methoxide (6.06 mg, 0.11 mmol) was added to a solution of 17 (15.6 mg, 0.022 mmol) in methanol (3 mL). The mixture was allowed to stir at room temperature overnight. After neutralization by formic acid (100 µL), the reaction solution was loaded to an ODS column eluted with 70% methanol to give 18 (8.5 mg, 66% yield) as a white amorphous powder. [α]D 20 −47.12 (C = 0.42, MeOH). HRMS (ESI) m/z (C28H38NO12)+: calcd 580.2389, found 580.2377. 1H-NMR (400 MHz, CD3OD) δ: 5.04 (s, H-2), 4.49 (m, H-3), 2.92 (dd, J = 3.6, 17.3 Hz, H-4a), 2.70 (dd, J = 3.9, 17.3 Hz, H-4b), 6.13 (d, J = 2.3 Hz, H-6), 6.10 (d, J = 2.3 Hz, H-8), 6.91 (2H, s, H-2′, 6′), 3.77, 3.74 (3H each, s, 5, 7-OCH3), 3.85 (6H, s, 3′, 5′-OCH3), 3.80 (3H, s, 4′-OCH3), 4.58 (d, J = 7.8 Hz, glc-1), 3.82 (m, glc-2), 3.45–3.58 (3H, m, glc-6a, 6b, 3), 3.21–3.24 (2H, m, glc-4, 5), 1.58 (3H, s, -COCH3). 13C-NMR (100 MHz, CD3OD) δ: 79.7 (C-2), 71.9 (C-3), 24.6 (C-4), 101.8 (C-4a), 161.2, 160.5 (C-5, 7), 94.6 (C-6), 92.4 (C-8), 157.3 (C-8a), 136.1 (C-1′), 106.8 (C-2′, 6′), 153.9 (C-3′, 5′), 138.5 (C-4′), 99.7 (glc-1), 57.7 (glc-2), 75.6 (glc-3), 72.5 (glc-4), 78.0 (glc-5), 63.1 (glc-6), 56.1, 56.0 (5, 7-OCH3), 56.9 (3′, 5′-OCH3), 61.2 (4′-OCH3), 173.7 (C = O in acetyl group), 22.7 (CH3 in acetyl group).
Mass Spectrometry
All ESI-MS experiments were carried out on a Bruker MicrOTOFQ mass spectrometer in negative ion mode, with the capillary voltage set to +3500 V, the dry N2 gas flow set to 4.0 L/min at 100 celcius, and injection flow rate of sample set to 3 µL/min. Data processing was performed by the software Bruker Daltonics DataAnalysis. All the nucleic acids oligomers (desalted grade) were purchased from Invitrogen and used without further purification. The stock solutions of all nucleic acid oligomers were prepared in milli Q water at the concentration of 1 mM, and further diluted by 1 M NH4OAc buffer (pH 7.6) to the desired concentration. All stock solutions of drugs were prepared in methanol at a concentration of 400 µM. For the analysis of the noncovalent complex of oncogene DNA G-quadruplex with drug, the samples were prepared at a final concentration of 100 µM DNA and 100 µM drug in 100 mM NH4OAc (pH 7.6) containing 50% methanol. For the analysis of human telomeric DNA and RNA G-quadruplexes, the samples were injected at a final strand concentration of 50 µM oligomer and 50 µM drug in 50 mM NH4OAc (pH 7.6) containing 50% methanol. Each sample of nucleic acid-drug complex solution was prepared in duplicate.
The oligonucleotide sequences are shown as the following:
human telomeric DNA: 5′-TTAGGGTTAGGGTTAGGGTTAGGGTTA-3′ (M = 8496.6241 Da)
c-myc DNA: 5′-TGGGGAGGGTGGGGAGGGTGGGGAAGG-3′ (M = 8687.7137 Da)
c-kit 1 DNA: 5′-AGAGGGAGGGCGCTGGGAGGAGGGGCT-3′ (M = 8576.6545 Da)
c-kit 2 DNA: 5′-CCCGGGCGGGCGCGAGGGAGGGGAGGT-3′ (M = 8513.5935 Da)
truncated bcl-2 DNA: 5′-CGGGCGCGGGAGGAAGGGGGCGGGAGC-3′ (M = 8562.6312 Da)
human telomeric RNA: 5′-UUAGGGUUAGGGUUAGGGUUAGGGUUA-3′ (M = 8789.3336 Da)
Molecular Modeling
A computer model to study the binding of 16 with human telomeric DNA G-quadruplex was performed by using the literature method [57]. Molecular modeling was performed using the ICM-Pro 3.4-8a program (Molsoft). The X-ray crystal structure of the intramolecular G-quadruplex DNA was obtained from the Protein Data Bank (PDB code: 1KF1) and used as the model to perform molecular modeling [57].
UV-melting Study
UV-melting profiles were recorded by using a Beckman Coulter DU800® spectrophotometer equipped with a high performance temperature controller. The absorbance was monitored at 295 nm for G-quadruplex in 25 mM Tris-HCl buffer (pH 7.0) containing 5 mM KCl and 1% DMSO, and at 260 nm for duplex oligonucleotides in 25 mM Tris-HCl buffer (pH 7.0) containing 1% DMSO. The concentration of all oligonucleotides was 5 µM for the ΔT m measurement in the absence and presence of compounds (50 µM).
Supporting Information
Positive ESI-TOF-MS spectrum of compound 14.
(TIF)
1H-NMR spectrum of compound 14. The spectrum was taken in CDCl3 at room temperature.
(TIF)
13C-NMR spectrum of compound 14. The spectrum was taken in DMSO-d6 at room temperature.
(TIF)
Positive ESI-TOF-MS spectrum of compound 16.
(TIF)
1H-NMR spectrum of compound 16. The spectrum was taken in the mixture of DMSO and D2O at room temperature.
(TIF)
13C-NMR spectrum of compound 16. The spectrum was taken in the mixture of DMSO and D2O at room temperature.
(TIF)
Positive ESI-TOF-MS spectrum of compound 18.
(TIF)
1H-NMR spectrum of compound 18. The spectrum was taken in CD3OD at room temperature.
(TIF)
13C-NMR spectrum of compound 18. The spectrum was taken in CD3OD at room temperature.
(TIF)
Negative ESI-TOF-MS spectra of oncogene G-rich sequences. (A–B) c-Myc sequence d[(TG4AG3)2TG4A2G2] with compound 16 and compound 18. (C–D) c-kit1 sequence d[AG(AG3)2CGCTG3AG2AG4CT] with compound 16 and compound 18. (E–F) c-kit2 sequence d[C3G3CG3(CG)2AG3AG4AG2T] with compound 16 and compound 18. (G–H) truncated Bcl-2 sequence d[CG3CGCG3AG2A2G5CG3AGC] with compound 16 and compound 18. Q represents quadruplex oligodeoxynucleotides. Spectra were recorded with 1∶1 DNA-to-drug molar ratio (C = 100 µM) in 50 mM ammonium acetate buffer (pH 7.6) containing 50% methanol.
(TIF)
Negative ESI-TOF-MS spectra of human telomeric RNA sequence r[(UUAGGG)4UUA] (Q). (A) without drug, (B) with compound 16, (C) with compound 18, (D) with compound 14, and (E) with EGC. Spectra were recorded with 1∶1 DNA-to-drug molar ratio (C = 50 µM) in 50 mM ammonium acetate buffer (pH 7.6) containing 50% methanol.
(TIF)
ESI-TOF-MS spectra of human telomeric DNA and RNA G-quadruplex in competitive binding experiments. (A) an equal molar mixture of human telomeric DNA d[(TTAGGG)4TTA] (Q) and RNA r[(UUAGGG)4UUA] (Q) without drug, (B) with compound 16, and (C) with compound 18. Spectra were recorded with 1∶1∶2 molar ratio of DNA:RNA:drug (25 µM: 25 µM: 50 µM) in 50 mM ammonium acetate buffer (pH 7.6) containing 50% methanol.
(TIF)
Thermal denaturation profiles of oligonucleotides in the absence and presence of 16. (A) UV-melting profiles of rAGGG(UUAGGG)3 (5 µM) in the absence and presence (50 µM) of compound 16 in 25 mM Tris-HCl buffer (pH 7.0) containing 5 mM KCl and 1% DMSO, (B) UV-melting profiles of dAGGG(TTAGGG)3 (5 µM) in the absence and presence (50 µM) of compound 16 in 25 mM Tris-HCl buffer (pH 7.0) containing 5 mM KCl and 1% DMSO, (C) UV-melting profiles of double-stranded oligodeoxynucleotide (Ds-DNA) 5′-AGGGTTAGGGT-3′/3′-TCCCAATCCCA-5′ (5 µM) in the absence and presence (50 µM) of compound 16 in 25 mM Tris-HCl buffer (pH 7.0) containing 1% DMSO.
(TIF)
Funding Statement
This work was financially supported by a grant from the Macao Foundation (grant numbers 0205 and 0199). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lipps HJ, Rhodes D (2009) G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 19: 414–422. [DOI] [PubMed] [Google Scholar]
- 2. Neidle S, Parkinson GN (2003) The structure of telomeric DNA. Curr Opin Struct Biol 13: 275–283. [DOI] [PubMed] [Google Scholar]
- 3. Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801. [DOI] [PubMed] [Google Scholar]
- 4. Schoeftner S, Blasco MA (2009) A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luke B, Lingner J (2009) TERRA: telomeric repeat-containing RNA. EMBO J 28: 2503–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maizels N (2006) Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol 13: 1055–1059. [DOI] [PubMed] [Google Scholar]
- 7. Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ (2005) Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol 12: 847–854. [DOI] [PubMed] [Google Scholar]
- 8. Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA 99: 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dexheimer TS, Sun D, Hurley LH (2006) Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl2 P1 promoter. J Am Chem Soc 128: 5404–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hurley LH (2002) DNA and associated processes as targets for cancer therapy. Nat Rev Cancer 2: 188–200. [DOI] [PubMed] [Google Scholar]
- 11. Cuesta J, Read MA, Neidle S (2003) The design of G-quadruplex ligands as telomerase inhibitors. Mini-Rev Med Chem 3: 11–21. [DOI] [PubMed] [Google Scholar]
- 12. Todd AK, Haider SM, Parkinson GN, Neidle S (2007) Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res 35: 5799–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arola A, Vilar R (2008) Stabilisation of G-quadruplex DNA by small molecules. Curr Top Med Chem 8: 1405–1415. [DOI] [PubMed] [Google Scholar]
- 14. Tan J-H, Gu L-Q, Wu J-Y (2008) Design of selective G-quadruplex ligands as potential anticancer agents. Mini-Rev Med Chem 8: 1163–1178. [DOI] [PubMed] [Google Scholar]
- 15. Ranjan N, Andreasen KF, Kumar S, Hyde-Volpe D, Arya DP (2010) Aminoglycoside binding to Oxytricha nova telomeric DNA. Biochemistry 49: 9891–9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arya DP (2011) New approaches toward recognition of nucleic acid triple helices. Acc Chem Res 44: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue L, Ranjan N, Arya DP (2011) Synthesis and spectroscopic studies of the aminoglycoside (neomycin)-perylene conjugate binding to human telomeric DNA. Biochemistry 50: 2838–2849. [DOI] [PubMed] [Google Scholar]
- 18. Xue L, Xi H, Kumar S, Gray D, Davis E, et al. (2010) Probing the recognition surface of a DNA triplex: binding studies with intercalator-neomycin conjugates. Biochemistry 49: 5540–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Willis B, Arya DP (2010) Triple recognition of B-DNA by a neomycin-hoechst 33258-pyrene conjugate. Biochemistry 49: 452–469. [DOI] [PubMed] [Google Scholar]
- 20. Shaw NN, Xi H, Arya DP (2008) Molecular recognition of a DNA:RNA hybrid: sub-nanomolar binding by a neomycin-methidium conjugate. Bioorg Med Chem Lett 18: 4142–4145. [DOI] [PubMed] [Google Scholar]
- 21.Xue L, Charles I, Arya DP (2002) Pyrene-neomycin conjugate: dual recognition of a DNA triple helix. Chem Comm 70–71. [DOI] [PubMed]
- 22. Arya DP, Xue L, Tennant P (2003) Combining the best in triplex recognition: synthesis and nucleic acid binding of a BQQ-neomycin conjugate. J Am Chem Soc 125: 8070–8071. [DOI] [PubMed] [Google Scholar]
- 23. Vuong QV, Stathopoulos CE, Nguyen MH, Golding JB, Roach PD (2011) Isolation of green tea catechins and their utilization in the food industry. Food Rev Int 27: 227–247. [Google Scholar]
- 24. Naghma K, Hasan M (2008) Multitargeted therapy of cancer by green tea polyphenols. Cancer Lett 269: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao Y, Cao R (1999) Angiogenesis inhibited by drinking tea. Nature 398: 381. [DOI] [PubMed] [Google Scholar]
- 26. Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, et al. (2008) EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol 15: 558–566. [DOI] [PubMed] [Google Scholar]
- 27. Zhang H, Wang J-R, Yau L, Ho H, Chan C, et al. (2012) Cellular Lipidomic study on the Aβ-induced neurotoxicity and neuroprotective Effect of EGCG by using UPLC/MS based glycerolipid profiling and multivariate analysis. Mol Biosyst 8: 3208–3215. [DOI] [PubMed] [Google Scholar]
- 28. Yang CS, Wang X (2010) Green Tea and Cancer Prevention. Nutr Cancer 62: 931–937. [DOI] [PubMed] [Google Scholar]
- 29. Rahman MA, Amin ARMR, Shin DM (2010) Chemopreventive potential of natural compounds in head and neck cancer. Nutr Cancer 62: 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lambert JD, Elias RJ (2010) The antioxidant and pro-oxidant activities of green tea polyphenols: A role in cancer prevention. Arch Biochem Biophys 501: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song JM, Lee KH, Seong BL (2005) Antiviral effect of catechins in green tea on influenza virus. Antivir Res 68: 66–74. [DOI] [PubMed] [Google Scholar]
- 32. Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, et al. (2003) Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res 63: 7563–7570. [PubMed] [Google Scholar]
- 33. Balasubramanian S, Eckert R (2004) Green tea polyphenol and curcumin inversely regulate human involucrin promoter activity via opposing effects on CCAAT/enhancer-binding protein function. J Biol Chem 279: 24007–24014. [DOI] [PubMed] [Google Scholar]
- 34. Kuzuhara T, Sei Y, Yamaguchi K, Suganuma M, Fujiki H (2006) DNA and RNA as new binding targets of green tea catechins. J Biol Chem 281: 17446–17456. [DOI] [PubMed] [Google Scholar]
- 35. Phosrithong N, Ungwitayatorn J (2010) Molecular docking study on anticancer activity of plant-derived natural products. Med Chem Res 19: 817–835. [Google Scholar]
- 36. Ferla BL, Airoldi C, Zona C, Orsato A, Cardona F, et al. (2011) Natural glycoconjugates with antitumor activity. Nat Prod Rep 28: 630–648. [DOI] [PubMed] [Google Scholar]
- 37. Ikeda T, Fujiwara S, Araki K, Kinjo J, Nohara T, et al. (1997) Cytotoxic glycosides from Albizia julibrissin . J Nat Prod 60: 102–107. [DOI] [PubMed] [Google Scholar]
- 38. He H, Bai L-P, Jiang Z-H (2012) Synthesis and human telomeric G-quadruplex DNA-binding activity of glucosaminosides of shikonin/alkannin. Bioorg Med Chem Lett 22: 1582–1586. [DOI] [PubMed] [Google Scholar]
- 39. Deng S, Yu B, Xie J, Hui Y (1999) Highly efficient glycosylation of sapogenins. J Org Chem 64: 7265–7266. [Google Scholar]
- 40. Liu L, Yi L, Yang X, Yu Z, Wen X, et al. (2008) Sythesis and spectroscopic characterization of binaphthol aminosugars for stimulation of DNA strand slippage synthesis. Tetrahedron 64: 5885–5890. [Google Scholar]
- 41. Wrodnigg TM, Lundt I, Stütz AE (2006) Synthesis of N-protected galactosamine building blocks from D-tagatose via the Heyns rearrangement. J Carbohydr Chem 25: 33–41. [Google Scholar]
- 42. Xi Z, Hwang G-S, Goldberg I H, Harris JL, Pennington WT, et al. (2002) Targeting DNA bulged microenvironments with synthetic agents: lessons from a natural product. Chem Biol 9: 925–931. [DOI] [PubMed] [Google Scholar]
- 43. Gunaherath GMKB, Gunatilaka AAL, Sultanbawa MUS (1981) Dulcitol and (−)-4′-O-methylepigallocatechin from Kokoona zeylanica . J Nat Prod 45: 140–142. [Google Scholar]
- 44. Schmidt CA, Murillo R, Bruhn T, Bringmann G, Goettert M, et al. (2010) Catechin derivatives from Parapiptadenia rigida with in vitro wound-healing properties. J Nat Prod 73: 2035–2041. [DOI] [PubMed] [Google Scholar]
- 45. Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R (2002) Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int J Mass Spectrom 216: 1–27. [Google Scholar]
- 46. Gavelica V, De Pauw E, Rosu F (1999) Interaction between antitumor drugs and a double-stranded oligonucleotide studies by electrospray ionization mass spectrometry. J Mass Spectrom 34: 1328–1337. [DOI] [PubMed] [Google Scholar]
- 47. Baker ES, Bernstein SL, Gabelica V, De Pauw E, Bowers MT (2006) G-quadruplexes in telomeric repeats are conserved in a solvent-free environment. Int J Mass Spectrom 253: 225–237. [Google Scholar]
- 48. Sun HX, Tang YL, Xiang JF, Xu GZ, Zhang YZ, et al. (2006) Spectroscopic studies of the interaction between quercetin and G-quadruplex DNA. Bioorg Med Chem Lett 16: 3586–3589. [DOI] [PubMed] [Google Scholar]
- 49. Bai L-P, Hagihara M, Jiang Z-H, Nakatani K (2008) Ligand binding to tandem G quadruplexes from human telomeric DNA. ChemBioChem 9: 2583–2587. [DOI] [PubMed] [Google Scholar]
- 50. Wan KX, Shibue T, Gross ML (2000) Non-covalent complexes between DNA-binding drugs and double-stranded oligodeoxynucleotides: a study by ESI ion-trap mass spectrometry. J Am Chem Soc 122: 300–307. [DOI] [PubMed] [Google Scholar]
- 51. Bai L-P, Cai Z, Zhao Z-Z, Nakatani K, Jiang Z-H (2008) Site-specific binding of chelerythrine and sanguinarine to single pyrimidine bulges in hairpin DNA. Anal Bioanal Chem 392: 709–716. [DOI] [PubMed] [Google Scholar]
- 52. Parkinson GN, Lee MPH, Neidle S (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417: 876–880. [DOI] [PubMed] [Google Scholar]
- 53. Xu Y, Suzuki Y, Ito K, Komiyama M (2010) Telomeric repeat-containing RNA structure in living cells. PNAS 107: 14579–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Phan AT (2010) Human telomeric G-quadruplex: structure of DNA and RNA sequences. FEBS J 277: 1107–1117. [DOI] [PubMed] [Google Scholar]
- 55.Collie G, Reszka AP, Haider SM, Gabelica V, Parkinson GN, et al.. (2009) Selectivity in small molecule binding to human telomeric RNA and DNA quadruplexes. Chem Commun 7482–7484. [DOI] [PubMed]
- 56. Collie GW, Parkinson GN, Neidle S, Rosu F, De Pauw E, et al. (2010) Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures. J Am Chem Soc 132: 9328–9334. [DOI] [PubMed] [Google Scholar]
- 57. Ma D-L, Lai T-S, Chan F-Y, Chung W-H, Abagyan R, et al. (2008) Discovery of a drug-like G-quadruplex binding ligand by high-throughput docking. ChemMedChem 3: 881–884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Positive ESI-TOF-MS spectrum of compound 14.
(TIF)
1H-NMR spectrum of compound 14. The spectrum was taken in CDCl3 at room temperature.
(TIF)
13C-NMR spectrum of compound 14. The spectrum was taken in DMSO-d6 at room temperature.
(TIF)
Positive ESI-TOF-MS spectrum of compound 16.
(TIF)
1H-NMR spectrum of compound 16. The spectrum was taken in the mixture of DMSO and D2O at room temperature.
(TIF)
13C-NMR spectrum of compound 16. The spectrum was taken in the mixture of DMSO and D2O at room temperature.
(TIF)
Positive ESI-TOF-MS spectrum of compound 18.
(TIF)
1H-NMR spectrum of compound 18. The spectrum was taken in CD3OD at room temperature.
(TIF)
13C-NMR spectrum of compound 18. The spectrum was taken in CD3OD at room temperature.
(TIF)
Negative ESI-TOF-MS spectra of oncogene G-rich sequences. (A–B) c-Myc sequence d[(TG4AG3)2TG4A2G2] with compound 16 and compound 18. (C–D) c-kit1 sequence d[AG(AG3)2CGCTG3AG2AG4CT] with compound 16 and compound 18. (E–F) c-kit2 sequence d[C3G3CG3(CG)2AG3AG4AG2T] with compound 16 and compound 18. (G–H) truncated Bcl-2 sequence d[CG3CGCG3AG2A2G5CG3AGC] with compound 16 and compound 18. Q represents quadruplex oligodeoxynucleotides. Spectra were recorded with 1∶1 DNA-to-drug molar ratio (C = 100 µM) in 50 mM ammonium acetate buffer (pH 7.6) containing 50% methanol.
(TIF)
Negative ESI-TOF-MS spectra of human telomeric RNA sequence r[(UUAGGG)4UUA] (Q). (A) without drug, (B) with compound 16, (C) with compound 18, (D) with compound 14, and (E) with EGC. Spectra were recorded with 1∶1 DNA-to-drug molar ratio (C = 50 µM) in 50 mM ammonium acetate buffer (pH 7.6) containing 50% methanol.
(TIF)
ESI-TOF-MS spectra of human telomeric DNA and RNA G-quadruplex in competitive binding experiments. (A) an equal molar mixture of human telomeric DNA d[(TTAGGG)4TTA] (Q) and RNA r[(UUAGGG)4UUA] (Q) without drug, (B) with compound 16, and (C) with compound 18. Spectra were recorded with 1∶1∶2 molar ratio of DNA:RNA:drug (25 µM: 25 µM: 50 µM) in 50 mM ammonium acetate buffer (pH 7.6) containing 50% methanol.
(TIF)
Thermal denaturation profiles of oligonucleotides in the absence and presence of 16. (A) UV-melting profiles of rAGGG(UUAGGG)3 (5 µM) in the absence and presence (50 µM) of compound 16 in 25 mM Tris-HCl buffer (pH 7.0) containing 5 mM KCl and 1% DMSO, (B) UV-melting profiles of dAGGG(TTAGGG)3 (5 µM) in the absence and presence (50 µM) of compound 16 in 25 mM Tris-HCl buffer (pH 7.0) containing 5 mM KCl and 1% DMSO, (C) UV-melting profiles of double-stranded oligodeoxynucleotide (Ds-DNA) 5′-AGGGTTAGGGT-3′/3′-TCCCAATCCCA-5′ (5 µM) in the absence and presence (50 µM) of compound 16 in 25 mM Tris-HCl buffer (pH 7.0) containing 1% DMSO.
(TIF)