Abstract
Cross-contamination between cell lines is a longstanding and frequent cause of scientific misrepresentation. Estimates from national testing services indicate that up to 36% of cell lines are of a different origin or species to that claimed. To test a standard method of cell line authentication, 253 human cell lines from banks and research institutes worldwide were analyzed by short tandem repeat profiling. The short tandem repeat profile is a simple numerical code that is reproducible between laboratories, is inexpensive, and can provide an international reference standard for every cell line. If DNA profiling of cell lines is accepted and demanded internationally, scientific misrepresentation because of cross-contamination can be largely eliminated.
Human cell lines are widely used in laboratory-based research. A significant proportion of this research is misleading, because the cell lines are of a different origin to that being claimed (1). Cross-contamination of cell lines is a longstanding problem and a repeated and frequent cause of scientific misrepresentation. This paper highlights the problem and describes a method for its detection.
The first continuous cell line derived from a human cancer, HeLa, was described in 1952 (2). Interspecies cross-contamination was soon described with HeLa (3, 4). Intraspecies cross-contamination became detectable by 1967 with the development of genetic markers, and it was discovered that HeLa cells had probably cross-contaminated many other supposedly unique human cancer cell lines (5). Despite a constant stream of reports demonstrating evidence of inter- and intraspecies cross-contamination (6–10), cross-contamination continues to occur at an intolerably high rate (11–13). In a recent paper describing new cell lines deposited at the German Cell Line Bank, 18% of the 252 “new” cell lines were cross-contaminants (14).
There are many methods of detecting cell line cross-contamination, including enzyme polymorphisms (15, 17), HLA typing (8), karyotyping (18), and DNA polymorphisms (16). The description of hypervariable regions within DNA led to the concept of DNA fingerprinting (19), which was applied to the authentication of human cell lines (20, 21). Locus-specific probes were also used for the same purpose (22, 23). Although highly informative, neither DNA fingerprinting nor locus-specific probes provided data that could be exactly described or reproduced in a format suitable for a database of reference standards. Until recently, there has been no standard inexpensive method that could be universally applied to give a simple numerical code that is reproducible in different laboratories.
Authentication and standardization are now possible by using the short tandem repeat (STR) profiling techniques developed for forensic applications (24). By using this approach, a number of polymorphic STR loci are amplified by using commercially available sets of primers. The PCR products are analyzed simultaneously with size standards by using automated fluorescent detection techniques. The result is a simple numerical code corresponding to the lengths of the PCR products amplified at each locus, accurate to less than one base pair. By applying this method to cell lines, every laboratory could either check the authenticity of its cell lines or have them checked commercially at a minimal cost, less than $200 each.
The aim of this study was to evaluate STR profiling for detecting cross-contamination in samples obtained worldwide and including likely cross-contaminating cell lines. The leading cell banks from the U.S., Europe, Asia, and five large cancer research institutes contributed samples. It was demonstrated that STR profiling is an efficient and reliable means of detecting cross-contaminated cell lines. These data could provide the basis for an international reference standard for human cell lines. It is to the benefit of all the scientific community that all cell lines included in publications be authenticated by DNA profiling at the time they are being used.
Materials and Methods
In a pilot study, DNA derived from 33 human cancer cell lines was compared by using two STR profile multiplex systems: Second Generation Multiplex (United Kingdom Forensic Science Service) (25) and Powerplex 1 (Promega) (26). The information on cell line identification and assortment obtained was identical in the two systems, with the exception of a single pair of samples that showed identical SGM profiles but an additional allele in one sample's Powerplex 1 profile (at the D13S317 locus). On the basis of the pilot study, the SGM STR multiplex was chosen for an extended study.
For the extended study, 20 samples of DNA (1 μg/ml) or cells were requested from each of the major cell banks in the U.S. (American Type Culture Collection, Manassas, VA; Coriell Cell Repositories, Camden, NJ), Europe (European Collection of Animal Cell Cultures, Salisbury, U.K.; Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) and Asia [The Institute of Physical and Chemical Research (Japan) RIKEN, Tsukuba, Japan; Japanese Collection of Research Bioresources, Tokyo] and from five cancer research institutes carrying large stocks of human cancer cell lines. The purpose of the project was explained to the 11 contributors, and each was asked to provide 20 DNA or cell samples of their own choosing. The analysis was blind and confidential for both the contributing centers and the analytical center (LGC, Teddington, U.K.). No data on the claimed origin or the STR profile of any cell line were released before the study was complete. A small number of unexpected findings were made, and requests were made for additional samples to be analyzed, in every case with an identical result.
DNA was prepared from the cells, where necessary, by using a commercial silica-gel-based purification kit (Qiagen, Crawley, U.K.). The SGM kit comprises the six STR loci: tyrosine hydroxylase, HUMTH01, 11p15.5; von Willebrand factor (vWF), HUMVWFA31/A, 12p-12pter; D8S1179, chromosome 8; D21S11, 21q11.2–21q21; α Fibrinogen (FGA), HUMFIBRA, 4q28 and D18S51, 18q21.3 plus the sex chromosome marker amelogenin, HUMAMGX/Y, Xp22.1–22.3 and Yp11.2. SGM amplifications contained 2 ng of target DNA in a 50-μl reaction volume containing 1× PARR buffer (Cambio, Cambridge, U.K.), 1.25 units AmpliTaq Gold DNA Polymerase (Perkin–Elmer) and 200 μM of each dNTP (Amersham Pharmacia). Samples were amplified by using the following conditions: 95°C for 18 min, 30 cycles of 95°C for 30 seconds, 58°C for 75 seconds, 72°C for 15 seconds, and 72°C for 25 min. Primer concentrations were 45 nM AMG 1/2, 87.5 nM TH01 1/2, 125 nM vWF 1/2, 560 nM D8S1179 1/2, 210 nM D21S11 1/2, 100 nM FGA 1/2, and 100 nM D18S51 1/2. Samples that failed to give measurable peaks at all loci were reanalyzed by using a different concentration of DNA. A small proportion of samples failed twice but were not processed further, as the aim of this study was to determine the utility of the method under routine conditions.
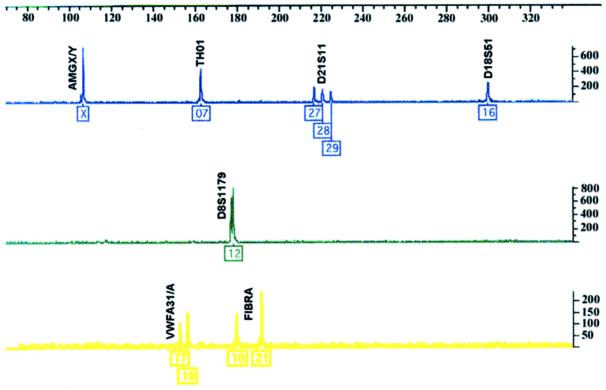
All of the STRs used in this study have a tetranucleotide repeat sequence, although intermediate sized alleles have been observed for TH01, FGA, and D21S11. Multiplex PCR reactions were carried out by using fluorescent dye-linked primers. Labeled products were detected by electrophoretic size fractionation on a Perkin–Elmer–ABI Prism 377 Genetic Analyser and analyzed by using genescan and genotyper analysis software (Perkin–Elmer). The end result for each cell line was an electropherogram, with each STR allele represented as one or more peaks of the appropriate color (see Fig. 1).
Figure 1.
Fully analyzed data derived from the DNA of cell line HeLaS3, showing the blue, green, and yellow amplified STR peaks and allele classifications. These data are derived from a single lane containing all the PCR products and the red size standards. The PCR products and size standards are detected by laser excitation of the four fluorescent dye labels by using an Applied Biosystems Prism 377 instrument. Profile quality is assessed by two independent analysts, and the peaks are then assigned allele values corresponding to the number of repeat units by using genescan and genotyper software. Allele designation is based on fixed size windows (±0.5 bp) derived from multiple analyses of allelic ladder samples containing all the commonly occurring alleles (data not shown). The loci are as follows: blue (Left to Right): HUMAMGX/Y, HUMTH01, D21S11, D18S51; green: D8S1179; yellow: HUMVWFA31/A, HUMFIBRA.
The data were further analyzed to categorize peaks according to their size in relation to an internal standard run (GS500, Perkin–Elmer) in every lane in the gel. This analysis enabled every peak to be allocated a size corresponding to the number of repeat units present (e.g., TH01 9 has 9 AATG repeats and results in a peak at 170.5 ± 0.5 bp in SGM analysis). An algorithm was developed to compare the allelic profiles, with each profile (questioned profile) being checked against every other profile (reference profiles) in the database. For each comparison, the number of alleles present in both reference and questioned profiles was scored and expressed as a percentage of the total number of alleles in the questioned profile. The principle can be seen operating in Table 1, where a consensus profile is taken as the reference profile, and 16 examples of HeLa cross-contaminants taken as questioned profiles.
Table 1.
STR profiles of HeLa lines
| Trial identification | Reported name | Comparison with consensus | Percent match | SGM profile results
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| amg | D18S1179 | D21S11 | D8S1179 | FGA | TH01 | vWF | |||||
| Consensus profile |
X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 | ||||
| 1 | 43 | HeLa | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 2 | 229 | HeLa | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 3 | 243 | HeLa | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 4 | 162 | HeLa S3 | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 5 | 181 | HeLa S3 | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 6 | 169 | Hep II | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 7 | 254 | Intestine 407 | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 8 | 14H | HPC36M | Identical | 100 | X | 16 | 27, 28 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 9 | 127 | KB | FGA (18) loss | 100 | X | 16 | 27, 28 | 12, 13 | 21 | 7 | 16, 18 |
| 10 | 183 | KB | FGA (18) loss | 100 | X | 16 | 27, 28 | 12, 13 | 21 | 7 | 16, 18 |
| 11 | 250 | Chang Liver | FGA (18) loss | 100 | X | 16 | 27, 28 | 12, 13 | 21 | 7 | 16, 18 |
| 12 | 233 | J-111 | D8 (13) loss | 100 | X | 16 | 27, 28 | 12 | 18, 21 | 7 | 16, 18 |
| 13 | 236 | HeLa.P3 | D21 (28) loss | 100 | X | 16 | 27 | 12, 13 | 18, 21 | 7 | 16, 18 |
| 14 | 242 | HMV-1 | D8 (13) loss vWF (19) gain | 91 | X | 16 | 27, 28 | 12 | 18, 21 | 7 | 16, 18, 19 |
| 15 | 256 | IMC-3 | D8 (13) loss vWF (18 to 19) change | 90 | X | 16 | 27, 28 | 12 | 18, 21 | 7 | 16, 19 |
| 16 | 223 | HeLa S3 | D21 (29) gain | 82 | X | 16 | 27, 28, 29 | 12 | 18, 21 | 7 | 17, 18 |
| D8 (13) loss | |||||||||||
| vWF (16 to 17) change | |||||||||||
amg, amelogenin.
The term cross-contamination is used in this study to indicate misidentification of one cell line by another, rather than contamination by a microbiological organism. Other tissue culture terminology follows the internationally agreed nomenclature (27), as used in the recently published United Kingdom Coordinating Committee on Cancer Research guidelines for the use of cell lines in cancer research (28).
Results
DNA from 253 human cell lines, including 33 cell lines in a pilot study, was analyzed by STR profiling. Two further samples of Vero cells, which are of monkey origin, were sent by different centers and failed to amplify, as would be expected. One center was excluded, because the DNA was too dilute for routine analysis. Of the remaining 10 centers, 173/198 (87.4%) of samples gave complete results within two runs. Including the samples where partial data were obtained (up to two loci missing), the success rate increased to 188/198 (94.9%). Three centers sent cells rather than DNA, and the success rate for complete analysis was 46/59 (78.0%) and for partial analysis was 55/59 (93.2%).
Matching Profiles.
Among the 221 cell lines successfully analyzed, 139 different profiles were obtained (see Table 5, which is published as supplemental data on the PNAS web site, www.pnas.org). In some cases, the profiles were closely related, differing by only one or two alleles. Seven groups, each containing four or more identical or closely related profiles, were identified blind, solely on the basis of the criterion that profiles within a group should match at 80% or more of alleles, according to the matching algorithm described in Materials and Methods. Subsequent identification of cell lines within these groups confirmed that they represent HeLa (16 samples), T24 (9 samples), K562 (8 samples), MRC5 (5 samples), HL60 (5 samples), 293 (7 samples), and MCF7 (4 samples). All examples of these lines submitted in the study were identified by the blind analysis.
Sixteen examples of HeLa cells or its subline HeLaS3 or its cross-contaminants (Hep2, Intestine 407, KB, J-111, HMV-1, Chang Liver, and IMC-3) were included among the samples submitted for analysis. Although these samples were shown to be closely related by STR profiling, some were not identical. With respect to a consensus profile, two contained an additional allele at one locus, two showed a change of an allele, and seven showed loss of an allele (Table 1). There were nine samples of another common cross-contaminant, T24, and its cross-contaminants EJ-1 and ECV304, MGH-U1, MGH-U2, HU456, and HU961T. Again, the nine profiles were very similar, but five showed minor changes with respect to the consensus sequence (Table 2). Although some stocks of the J82 bladder cancer cell line have been shown to be cross-contaminated with T24, the stocks tested here and obtained from the originator were distinct. However, it was discovered that another human bladder cell line, HU609, was over 90% identical to J82.
Table 2.
STR profiles of T24 cell lines
| Trial identification | Reported name | Comparison with consensus | Percent match | SGM profile results
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| amg | D18S1179 | D21S11 | D8S1179 | FGA | TH01 | vWF | |||||
| Consensus profile |
X | 16, 18 | 29 | 14 | 17, 22 | 6 | 17 | ||||
| 1 | 228 | ECV304 | Identical | 100 | X | 16, 18 | 29 | 14 | 17, 22 | 6 | 17 |
| 2 | 222 | EJ-1 (T24) | Identical | 100 | X | 16, 18 | 29 | 14 | 17, 22 | 6 | 17 |
| 3 | 9M | MGH-U2 | Identical | 100 | X | 16, 18 | 29 | 14 | 17, 22 | 6 | 17 |
| 4 | 234 | T24 | Identical | 100 | X | 16, 18 | 29 | 14 | 17, 22 | 6 | 17 |
| 5 | 5H | HU456 | FGA (17) loss | 100 | X | 16, 18 | 29 | 14 | 22 | 6 | 17 |
| 6 | 10H | HU961T | FGA (17) loss | 100 | X | 16, 18 | 29 | 14 | 22 | 6 | 17 |
| 7 | 18M | MGH-U2 | FGA (17) loss | 100 | X | 16, 18 | 29 | 14 | 22 | 6 | 17 |
| 8 | 41 | T24 | vWF (19) gain | 90 | X | 16, 18 | 29 | 14 | 17, 22 | 6 | 17, 19 |
| 9 | 7T | T24 | D8 (9) gain vWF (19) gain | 82 | X | 16, 18 | 29 | 9, 14 | 17, 22 | 6 | 17, 19 |
amg, amelogenin.
To investigate the discriminatory power of the similarity measure with an 80% threshold, the data were divided into two groups, one containing all cell lines thought to be unrelated, and the other containing all of the cell lines known to be of the same origin. In this analysis, known kindreds were excluded by selecting parental profiles only, and from each subgroup of lines with the same origin, one example was selected randomly for inclusion in the unrelated cell lines. In the group of lines with the same origin (total of 131), there were 41 subgroups, each containing between 2 and 16 cell lines. The unrelated group contained 127 lines, of which 9 were partial profiles.
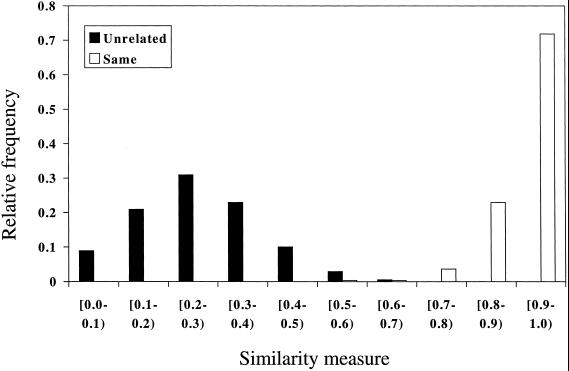
The algorithm was applied to every possible pair of lines within the unrelated group and to every known pair of cell lines within the related group. The algorithm was applied twice for each pair, with each line taken as the reference. This resulted in two similarity measures for each pair, which were then averaged to obtain a percentage similarity. The results are shown in Fig. 2 for the complete profiles. These results are based on 264 pairs of profiles for the same origin lines, of which 12 pairs resulted in a similarity statistic of less than 80%. Nine of these were comparisons of HeLa sublines, all of which were greater than 70% similar. Two were drug-resistant sublines of A2780. It has been reported that drug-resistant sublines of A2780, in contrast to the parental cell line, show strand-specific mismatch repair, which may be the cause of the lack of similarity. One was a pair of cell lines derived from normal and cancer tissue in the same individual, which was only 78% similar because of three losses of heterozygosity in the cancer cell line. Of the 6,903 pairs of lines of supposedly different origin, only one resulted in a similarity measure greater than 70%, the J82 and HU609 cell lines.
Figure 2.
Histogram showing relative frequency of the similarity measure at 10% intervals for the 6,903 unrelated pairs of cell lines (black bars) and 264 pairs of cell lines of the same origin (open bars).
Viral Transformation.
In this series, it was possible to make a number of comparisons between cell lines before and after transformation with viral genes. A MRC5 human fibroblast cell line transformed with simian virus (SV) 40 had lost the Y allele and one further allele but was otherwise identical to the untransformed cells (Table 3). In another pair of cell lines (trial number 69, AG/NA0090, and trial number 70, AG/NA02804), the SV40-transformed cells had gained one allele but were otherwise identical. A pSV3neo-transformed subline (trial number 121) of the IBR.3 cell line (trial number 125) had lost an allele. TTD1BR (trial number 124) has been transformed by using different agents. A pSV3neotransformed subline (trial number 138) had lost one allele and a heavily mutagenized (with ultraviolet-C irradiation) subline had lost a different allele, whereas an Epstein–Barr virus-transformed lymphoblastoid cell line (trial number 133) from the same individual was identical to TTD1BR.
Table 3.
MRC5 cell lines transformed with viral genes
| Trial identification | Reported name | Comparison with consensus | Percent match | SGM profile results
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| amg | D18S1179 | D21S11 | D8S1179 | FGA | TH01 | vWF | ||||
| Consensus profile |
X, Y | 15, 21 | 31.2 | 13 | 21, 23 | 8 | 15 | |||
| 132 | MRC5/28 Primary untransformed cell line | Identical | 100 | X, Y | 15, 21 | 31.2 | 13 | 21, 23 | 8 | 15 |
| 139 | MRC5neoA/48 PSV3 neo-transformed | Identical | 100 | X, Y | 15, 21 | 31.2 | 13 | 21, 23 | 8 | 15 |
| 180 | MRC5 | Identical | 100 | X, Y | 15, 21 | 31.2 | 13 | 21, 23 | 8 | 15 |
| 160 | MRC5 | Identical | 100 | X, Y | 15, 21 | 31.2 | 13 | 21, 23 | 8 | 15 |
| 200 | MRC-5 | FGA (21) loss | 90 | X, Y | 15, 21 | 31.2 | 13 | 23 | 8 | 15 |
| 135 | MRC SV1 SV40-transformed | FGA (21) loss amg (Y) loss | 80 | X | 15, 21 | 31.2 | 13 | 23 | 8 | 15 |
Drug-Resistant Lines.
Sublines with high levels of drug resistance can be obtained by growing the cells over long periods (usually 12 months or more) in the presence of increasing concentrations of the drug of interest. This treatment can lead to major changes in karyotype. The drug-resistant sublines were shown to be related to the parental cells within the limits of the algorithm, with the exception of the A2780 ovarian cancer cell lines.
Familial Studies.
STR profiling is now used routinely to determine family relationships. Some DNA samples from cell lines derived from related individuals were included. For example, samples 62, 63, and 66 are father, mother, and daughter, respectively, and the observed inheritance of alleles within the STR profile is consistent with this claim. Samples 126 and 137 are the father and mother of siblings 122, 129, 131, and 140, and again the data are consistent.
Peak Height Imbalance.
For normal diploid human DNA, STR profiling will usually produce profiles with approximately equal peak heights for both alleles at heterozygous loci, although more variation may be seen between loci. Many samples in this study showed large height differences between peaks at heterozygous loci. This preferential amplification of one allele over the other may indicate gene duplication events, aneuploidy, or a chimeric cell population. Data on relative peak heights, evident from the electropherograms (e.g., see Fig. 1), could provide useful additional information in identifying related cell lines.
Loss of Heterozygosity.
The STR loci used were chosen on the basis of their high degree of heterozygosity. In STR profiles from 200 Caucasian adults randomly selected from samples submitted to LGC for forensic analysis, heterozygosity at the six STR loci ranged from 79.1 to 87.8%. In this series of transformed and cancer cell lines, the degree of heterozygosity was far lower, ranging from 48.2 to 70.3% (see Table 4). Although these figures are biased by multiple representation of the same cell lines, they still indicate additional loss of heterozygosity. This finding is consistent with the neoplastic origin of most of the cell lines included.
Table 4.
Frequency of genetic changes in the general population of Caucasian adults and in the cell lines in this series
| D18S1179 | D21S11 | D8S1179 | FGA | TH01 | vWF | |
|---|---|---|---|---|---|---|
| Heterozygosity in this trial | 59.9% | 69.8% | 70.3% | 69.9% | 48.2% | 61.5% |
| Heterozygosity in Caucasian adults | 85.7% | 82.7% | 82.7% | 87.8% | 79.1% | 82.7% |
| Occurrence of three alleles in this trial | 2/177 | 6/192 | 4/192 | 3/183 | 0/193 | 5/192 |
| Occurrence of four alleles in this trial | 0 | 0 | 0 | 0 | 0 | 1/192 |
Three or More Alleles at a Locus.
The occurrence of three detectable alleles at one locus is a rarity among normal human SGM profiles [≈1 in 1,000 individuals (J.A.T., unpublished work)]. There were 18 examples of triple peaks in this series, with examples at every locus except TH01 (see Table 4). A leukemia cell line, MOLT-4, showed three alleles at two loci and an ovarian cancer cell line, SKOV-3, showed three alleles at three loci. Two samples of the lymphoma cell line, U937, were identical, except one had two and the other four alleles at the vWF locus. As with peak height imbalance, multiple alleles may be a result of genuine trisomy or may represent other gene duplication events or cell mixtures.
Cell Mixtures and Somatic Cell Hybrids.
Three parental cell lines and eight distinct somatic cell hybrids were analyzed blind. The results were entirely consistent, although some alleles were lost in the somatic cell hybrids, as might be expected. Knowing the origin of the parent cells, STR profiling could be used to confirm the derivation of the somatic cell hybrids (data not shown). Two cancer cell lines and mixtures ranging from 1 to 99% of each cell line were tested blind. In this limited study, objective evidence for two profiles was not observed in mixtures containing less than 10% of the smaller fraction (data not shown).
Discussion
STR profiling was evaluated as a simple method for cell line identification suitable for the establishment of reference genotypes. Two hundred and fifty-three cell lines from international cell banks and cancer research institutes worldwide were analyzed. It was demonstrated that this method can provide a universal reference standard for human cell lines. If this approach is applied internationally by using a common set of STR primers, cross-contamination of human cell lines will be readily detectable and such misrepresentation almost entirely eliminated, except as a result of deliberate fraud.
The methodology used deliberately did not set out to customize analysis for different sample or cell types, and so the results illustrate the success rate for obtaining complete profiles under routine circumstances at minimal cost. Samples with low DNA concentrations could have been processed further, as is standard for certain forensic samples, or another sample obtained. STR profiles should be readily obtainable for all human cell lines.
Where appropriate primers are available, DNA profiling can be applied to DNA samples from all species. In addition, human specific STR primers, such as those used in this study, could provide rapid confirmation that particular human chromosomes are present in somatic cell hybrids between human cells and cells of other species. Within an established cell line designation, analysis of additional STR loci can be used to characterize subline specific markers if required, as shown in the pilot study.
The STR profiles indicate that the HeLa sublines are not identical, and that there are both gains and losses of alleles, which may reflect the phenotypic differences observed in different laboratories. For the same strain of each cell line to be used worldwide, the international cell banks will need to agree to have a common stock.
Apart from simple cross-contamination, we checked whether viral transformation or long-term exposure to chemotherapeutic drugs would produce sublines that appeared to be distinct by STR profiling. However, the differences between the parental and derived cell lines were no greater than those between the HeLa sublines, and in all cases the sublines were identified as being closely related to the parental line.
Electropherograms provide information about the relative heights of peaks at heterozygous loci, which may reflect the number of copies of that allele present (Fig. 1). This information is lost in the simple numerical code. The code could be modified to indicate the relative peak height at each locus, for example by indicating the ratio of the two peaks as a bracketed suffix, e.g., TH01 7, 9 (1:1.8). Large differences in the height of two peaks at the same locus can be caused by a variety of reasons, including cell mixtures, differential amplification efficiency between heterozygous loci (for example because of a mutation in the primer binding site), or the presence of additional copies of one allele. Many of the samples in this study showed large differences in peak height at one or more loci. This finding is typical of cancer cells and reflects their relatively high genetic instability compared with normal cells.
For normal human DNA, STR profiling will show two alleles at most loci, as expected for highly polymorphic loci in a diploid genotype. Additional alleles at a locus are rarely seen in normal profiles but theoretically can occur by trisomy, gene duplication, or mixed populations of cells or hybrids. If most of the loci show more than two peaks, this is an indication of an hybrid or mixture of cells, whereas single loci with more than two peaks are more likely explained by trisomy or gene duplication events.
Because the loci were chosen on the basis of their high degree of heterozygosity in human populations, relatively few homozygous loci with single alleles would be expected. The significantly elevated levels of homozygosity observed in this study are consistent with the characteristic loss of heterozygosity, common in many cancer cells, particularly in the later stages of cancer progression from which almost all cancer cell lines are derived. At the TH01 locus, the incidence of homozygosity increased from about 20 to over 50%.
STRs are subject to mutation at a relatively high rate: in 10,844 parent/child allelic transfers at 9 loci, 23 mismatches were observed, 22 of which were because of a single-step mutation (29). It is generally accepted that replication slippage (30) is the major mechanism causing new mutations in microsatellites (29). STR loci have a higher frequency of polymorphism than single nucleotide polymorphisms and are the preferred option for forensic applications, but technological developments in the near future will provide rapid automated DNA profiling on arrays.
If DNA profiling of cell lines becomes accepted as normal practice, then editors of scientific journals could require authors to confirm that all cell lines used have been checked for authenticity during the period of the study. This analysis could become a prerequisite for publication, so that the problem of cell line cross-contamination can be reduced to a minimum in the future. This advance will be of great benefit to the scientific community, which can in future confidently develop or extend cell culture-based studies from other researchers without fear of false premises and without their own results being attributable to contamination.
Supplementary Material
Abbreviations
- STR
short tandem repeat
- vWF
von Willebrand factor
- FGA
α fibrinogen
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 7656.
References
- 1.Stacey G N, Masters J R W, Hay R J, Drexler H G, MacLeod R A F, Freshney R I. Nature (London) 2000;403:356. [Google Scholar]
- 2.Gey G O, Coffman W D, Kubicek M T. Cancer Res. 1952;12:264–265. [Google Scholar]
- 3.Defendi V, Billingham R E, Silvers W K, Moorhead P. J Natl Cancer Inst. 1960;25:359–385. [PubMed] [Google Scholar]
- 4.Brand K G, Syverton J T. J Natl Cancer Inst. 1962;28:147–157. [PubMed] [Google Scholar]
- 5.Gartler S M. Natl Cancer Inst Monogr. 1967;26:167–195. [PubMed] [Google Scholar]
- 6.Povey S, Hopkinson D A, Harris H, Franks L M. Nature (London) 1976;264:60–63. doi: 10.1038/264060b0. [DOI] [PubMed] [Google Scholar]
- 7.Lavappa K S. In Vitro. 1978;14:469–475. doi: 10.1007/BF02616110. [DOI] [PubMed] [Google Scholar]
- 8.O'Toole C M, Povey S, Hepburn P, Franks L M. Nature (London) 1981;301:429–430. doi: 10.1038/301429a0. [DOI] [PubMed] [Google Scholar]
- 9.Harris N L, Gang D L, Quay S C, Poppema S, Zamecnik P C, Nelson-Rees W A, O'Brien S J. Nature (London) 1981;289:228–230. doi: 10.1038/289228a0. [DOI] [PubMed] [Google Scholar]
- 10.Nelson-Rees W A, Daniels D W, Flandermeyer R R. Science. 1981;212:446–452. doi: 10.1126/science.6451928. [DOI] [PubMed] [Google Scholar]
- 11.Markovic O, Markovic N. In Vitro Cell Dev Biol. 1998;34:1–8. doi: 10.1007/s11626-998-0040-y. [DOI] [PubMed] [Google Scholar]
- 12.Scudiero D A, Monks A, Sausville E A. J Natl Cancer Inst. 1998;90:862. doi: 10.1093/jnci/90.11.862. [DOI] [PubMed] [Google Scholar]
- 13.Dirks W G, Drexler H G, MacLeod R A F. In Vitro Cell Dev Biol. 1999;35:558–559. doi: 10.1007/s11626-999-0091-8. [DOI] [PubMed] [Google Scholar]
- 14.MacLeod R A F, Dirks W G, Matsuo Y, Kaufmann M, Milch H, Drexler H G. Int J Cancer. 1999;83:555–563. doi: 10.1002/(sici)1097-0215(19991112)83:4<555::aid-ijc19>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien S J, Shannon J E, Gail M H. In Vitro. 1980;16:119–135. doi: 10.1007/BF02831503. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert D A, Reid Y A, Gail M H, Pee D, White C, Hay R J, O'Brien S J. Am J Hum Genet. 1990;47:499–514. [PMC free article] [PubMed] [Google Scholar]
- 17.Nims R W, Shoemaker A P, Bauernschub M A, Rec L J, Harbell J W. In Vitro Cell Dev Biol. 1998;34:35–39. doi: 10.1007/s11626-998-0050-9. [DOI] [PubMed] [Google Scholar]
- 18.MacLeod R A F, Drexler H G. In: Human Cell Culture. Masters J R W, Palsson B O, editors. III. Dordrecht, The Netherlands: Kluwer; 2000. pp. 371–397. [Google Scholar]
- 19.Jeffreys A J, Wilson V, Thein S L. Nature (London) 1985;316:76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- 20.Thacker J, Webb M B, Debenham P G. Somat Cell Mol Genet. 1988;14:519–525. doi: 10.1007/BF01535307. [DOI] [PubMed] [Google Scholar]
- 21.Stacey G N, Bolton B J, Doyle A. Nature (London) 1992;357:261–262. doi: 10.1038/357261a0. [DOI] [PubMed] [Google Scholar]
- 22.Masters J R, Bedford P, Kearney A, Povey S, Franks L M. Br J Cancer. 1988;57:284–286. doi: 10.1038/bjc.1988.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honma M, Kataoka E, Ohnishi K, Ohno T, Takeuchi M, Nomura N, Mizusawa H. In Vitro Cell Dev Biol. 1992;28A:24–28. doi: 10.1007/BF02631076. [DOI] [PubMed] [Google Scholar]
- 24.Oldroyd N J, Urquhart A, Kimpton C P, Downes T J, Millican E S, Watson S K, Gill P. Electrophoresis. 1995;16:334–337. doi: 10.1002/elps.1150160155. [DOI] [PubMed] [Google Scholar]
- 25.Thomson J A, Pilotti V, Stevens P, Ayres K L, Debenham P G. Forensic Sci Int. 1999;100:1–16. doi: 10.1016/s0379-0738(98)00199-6. [DOI] [PubMed] [Google Scholar]
- 26.Lins A B, Micka K A, Sprecher C J, Taylor J A, Bacher J W, Rabbach D R, Bever R A, Creacy S D, Schumm J W. J Forensic Sci. 1998;43:1168–1180. [PubMed] [Google Scholar]
- 27.Schaeffer W I. In Vitro Cell Dev Biol. 1990;26:97–101. doi: 10.1007/BF02624162. [DOI] [PubMed] [Google Scholar]
- 28.United Kingdom Coordinating Committee on Cancer Research. Br J Cancer. 2000;82:1495–1509. doi: 10.1054/bjoc.2000.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B. Am J Hum Genet. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levinson G, Gutman G A. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.