Abstract
Background
Beyond known familial colorectal cancer (CRC) syndromes, the mechanisms underlying the elevated CRC risk associated with CRC family history remain largely unknown. A recent retrospective study suggests familial clustering of CRC with hypomethylation in long interspersed nucleotide element 1 (LINE-1). We tested the hypothesis that CRC family history might confer a higher risk of LINE-1 methylation-low CRC.
Methods
Using the Nurses’ Health Study and the Health Professionals Follow-up Study, we prospectively examined the association between CRC family history and the risk of rectal and colon cancer (N = 1224) according to tumor LINE-1 methylation level by duplication method Cox proportional hazards regression. We examined microsatellite instability (MSI) status to exclude the influence of Lynch syndrome. All statistical tests were two-sided.
Results
The association between CRC family history and non-MSI CRC risk differed statistically significantly by LINE-1 methylation level (P heterogeneity = .02). CRC family history was associated with a statistically significantly higher risk of LINE-1 methylation-low non-MSI cancer (multivariable hazard ratio [HR] = 1.68, 95% confidence interval [CI] = 1.19 to 2.38 for 1 vs 0 first-degree relatives with CRC; multivariable HR = 3.48, 95% CI = 1.59 to 7.6 for ≥2 vs 0 first-degree relatives with CRC; P trend < .001). In contrast, CRC family history was not statistically significantly associated with LINE-1 methylation-high non-MSI cancer (P trend = .35).
Conclusions
This molecular pathological epidemiology study shows that CRC family history is associated with a higher risk of LINE-1 methylation-low CRC, suggesting previously unrecognized heritable predisposition to epigenetic alterations. Additional studies are needed to evaluate tumor LINE-1 methylation as a molecular biomarker for familial cancer risk assessment.
Epidemiological evidence indicates that a family history of colorectal cancer (CRC) is associated with higher personal CRC risk (1–5). Beyond known familial CRC syndromes (including Lynch and polyposis syndromes), which together constitute less than 5% of CRC cases (6), the mechanisms underlying familial clustering of CRC remain largely unknown (7–9). Recent data by Goel et al. (10) suggest that one potential mechanism of familial clustering of CRC may be heritable predisposition to epigenomic instability and the development of tumors with hypomethylation in long interspersed nucleotide element 1 [LINE-1, which comprises approximately 17% of the human genome (11)]. Thus, we hypothesized that a family history of CRC might be associated with higher risk of CRC with low-level LINE-1 methylation.
Because LINE-1 methylation-low CRC has been associated with aggressive tumor behavior (12–14), it is imperative to develop effective prevention strategies tailored to those who are susceptible to the development of this unfavorable cancer subtype.
To test our hypothesis of possible familial clustering of LINE-1 methylation-low CRC, we utilized two US nationwide prospective cohort studies. We prospectively examined the relationship between a history of CRC in a first-degree relative and subsequent risk of developing CRC with varying degrees of LINE-1 methylation. We also examined the status of tumor microsatellite instability (MSI) and CpG island methylator phenotype (CIMP), both of which have been inversely associated with LINE-1 methylation level (15,16). In particular, we utilized tumor MSI status in our attempt to exclude the potential influence of Lynch syndrome (ie, heritable susceptibility to mismatch repair-deficient cancer, most likely to be MSI-high cancer). We aimed to support not only a possible link between the heritability of CRC and tumor epigenetic instability but also the possible presence of a previously unrecognized familial cancer trait. Akin to current MSI testing in CRC, tumor LINE-1 methylation level may potentially serve as a tumor biomarker for familial cancer risk assessment.
Methods
Study Population
Details on our study population are described in the Supplementary Materials (available online). We utilized the Nurses’ Health Study (NHS) and the Health Professionals Follow-up Study (HPFS) (5,17,18). Eligible participants included 86172 women (NHS) and 47907 men (HPFS). The Harvard School of Public Health and Brigham and Women’s Hospital Institutional Review Boards approved this study. All subjects provided informed consent.
Family History Data Collection
We utilized family history data prospectively collected from questionnaires (before a participant developed CRC if it occurred) to avoid recall bias, which is a major problem in assessing family history of cancer (19–22). A history of CRC in first-degree relatives was collected in 1982, 1988, 1992, 1996, 2000, and 2004 in the NHS and in 1986, 1990, 1992, and 1996 in the HPFS.
Assessment of Incident CRC Cases
On each biennial follow-up questionnaire, participants were asked whether they had a diagnosis of CRC. For nonresponders, we searched the National Death Index to discover deaths and ascertain any diagnoses of CRC. We collected paraffin-embedded CRC tissue blocks from hospitals where participants with CRC had undergone tumor resection (18). For rectal cancer, we collected diagnostic biopsy and resection specimens to avoid any effect of preoperative treatment on tumor tissue analyses. Based on the colorectal continuum model (23,24), we used both colon and rectal cancers as outcomes. Based on the availability of tumor tissue data, 1224 CRC cases diagnosed up to 2008 were included as outcome data. A pathologist (S. Ogino) reviewed the histopathology of all 1224 CRC cases. Distributions of age, sex, tumor subsite location, disease stage, and pathologic features of our CRC cases have been previously described (23) and are generally consistent with cancer registry data in the United States. Furthermore, patient characteristics did not appreciably differ between cases with and without available tissue (18), so no analyses by major racial/ethnic group were done.
Tumor LINE-1 Methylation Analysis
DNA was extracted from archival tumor tissue. We employed validated bisulfite DNA treatment (25), polymerase chain reaction, and pyrosequencing assay to measure LINE-1 methylation level (26). Precision of the LINE-1 methylation assay was high, with a coefficient of variation of approximately 3% to 4% (26). Moreover, DNA from a whole tissue tumor section yielded LINE-1 methylation values comparable with DNA from pure tumor cells collected by laser capture microdissection (26). We classified LINE-1 methylation level into low (<55%), intermediate (55%–64.9%), and high (≥65%) designations, which reflected methylation levels relative to the overall distribution of the 1224 tumors. The cut points of 55% and 65% LINE-1 methylation levels were chosen to subclassify tumors, as previously described (27), to keep consistency in classification. An alternative strategy would be tertile or quartile classification. Although distribution of LINE-1 methylation level in our dataset resembled a normal distribution, the lower tail had many more cases than expected by a normal distribution (28). Neither tertile nor quartile cut points worked well to capture the nature of methylation-low cases because their lowest cut points were greater than 55%. Quintile cut points would have made too many categories and yielded less robust effect estimates.
Analysis for CIMP
Using a validated real-time polymerase chain reaction assay (MethyLight) on bisulfite-treated DNA (25), we quantified DNA methylation in eight CIMP-specific promoters (CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) (29–31). CIMP-high was defined as the presence of six or more of eight methylated promoters, and CIMP-low/negative (CIMP-low/0) was defined as zero of eight to five of eight methylated promoters, as per established criteria (31,32).
MSI Analysis
MSI analysis was performed utilizing 10 microsatellite markers (31). MSI-high cancer was defined as instability in 30% or more of the markers, and microsatellite stable (MSS) cancer was defined as instability in 0% to 29% of the markers (31).
Statistical Analysis
Detailed statistical analysis methods are described in the Supplementary Materials (available online). All statistical analyses were performed using SAS version 9.2 (SAS Institute, Cary, NC). Our primary study hypothesis was that CRC family history was associated with an increased risk of LINE-1 methylation-low MSS cancer (after controlling for the effect of Lynch syndrome) but not with the risk of LINE-1 methylation-high MSS cancer. We used Cox proportional hazards regression model to estimate a hazard ratio (HR) with a 95% confidence interval (CI) of developing a specific CRC subtype by CRC family history, adjusted for multiple potential confounders. Nonetheless, we cautiously interpreted statistical significance in subset analyses that resulted from our molecular pathological epidemiology design (33,34). All statistical tests were two-sided. A P value less than .05 was considered statistically significant.
Results
Family History and CRC Risk According to Molecular Subtypes
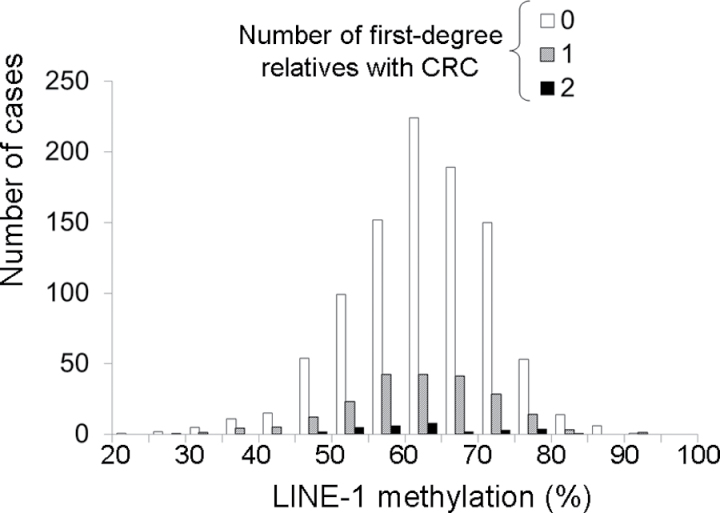
We followed a baseline population of 86172 women and 47907 men in the NHS and the HPFS, respectively. Characteristics of the population during the follow-up period are summarized in Table 1. There was no substantial difference in characteristics according to CRC family history status, except for age. During 3184415 person-years of follow-up in both cohorts, we documented 1224 incident CRC cases with available tumor molecular data. Distributions of LINE-1 methylation levels are shown in Figure 1 (overall CRCs) and Figure 2 (in relation to family history status).
Table 1.
Age-adjusted characteristics of participants (during follow-up), according to family history of colorectal cancer (CRC) in first-degree relatives*
| No. of first-degree relatives with CRC† | Women (Nurses’ Health Study) | Men (Health Professionals Follow-up Study) | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | ≥2 | 0 | 1 | ≥2 | 0 | 1 | ≥2 | |
| No. of study participants in each family history category as final status‡ | 70770 (82.1%) | 13831 (16.1%) | 1571 (1.8%) | 41336 (86.3%) | 6244 (13.0%) | 327 (0.7%) | 112106 (83.6%) | 20075 (15.0%) | 1898 (1.4%) |
| Total person-years | 1967983 | 262761 | 22250 | 819660 | 107327 | 4433 | 2787643 | 370088 | 26684 |
| Age, y | 58.6 (10.5) | 62.2 (10.1) | 66.5 (9.4) | 62.7 (10.9) | 65.2 (10.6) | 69.6 (9.9) | 59.8 (10.8) | 63.1 (10.3) | 67.0 (9.6) |
| Regular aspirin use, % | 42 | 42 | 42 | 51 | 50 | 51 | 44 | 44 | 44 |
| Multivitamin use, % | 49 | 49 | 48 | 54 | 55 | 55 | 51 | 51 | 49 |
| Physical activity, MET score/week | 16.7 (22.0) | 17.1 (21.9) | 17.0 (21.5) | 34.2 (40.0) | 34.7 (38.9) | 33.0 (38.3) | 21.8 (29.6) | 21.8 (28.5) | 19.2 (25.0) |
| Body mass index, BMI, kg/m2 | 24.3 (4.7) | 24.3 (4.8) | 24.1 (4.5) | 25.5 (3.3) | 25.5 (3.3) | 25.2 (3.5) | 24.6 (4.4) | 24.6 (4.5) | 24.3 (4.4) |
| Folate intake, µg/day § | 365 (276) | 366 (263) | 357 (245) | 481 (277) | 485 (278) | 504 (315) | 400 (282) | 399 (273) | 378 (262) |
| Methionine intake, g/day § | 1.9 (0.5) | 1.9 (0.5) | 1.9 (0.5) | 2.2 (0.5) | 2.2 (0.5) | 2.2 (0.4) | 2.0 (0.5) | 2.0 (0.5) | 1.9 (0.5) |
| Calcium intake, mg § | 908 (350) | 914 (343) | 905 (343) | 924 (375) | 921 (377) | 869 (346) | 912 (357) | 917 (353) | 905 (346) |
| Red meat intake, serving/week § | 2.1 (1.5) | 2.1 (1.4) | 2.0 (1.3) | 1.7 (1.4) | 1.7 (1.4) | 1.7 (1.4) | 2.0 (1.5) | 2.0 (1.4) | 2.0 (1.3) |
| Alcohol intake, g/day | 6.3 (10.3) | 6.4 (10.5) | 6.1 (10.2) | 11.1 (15.0) | 11.1 (14.8) | 11.9 (17.3) | 7.7 (12.1) | 7.7 (12.0) | 6.9 (11.7) |
| Smoking, current/former, % | 56 | 55 | 56 | 55 | 54 | 50 | 55 | 55 | 56 |
* Updated information of exposures from biennial questionnaires was averaged using person-years in each category of family history status up to censoring (including death from other causes) or immediately before personal CRC diagnosis if it occurred. In the rows from “Age” to “Smoking,” each value represents a mean (standard deviation) or percentage number and is standardized to the age distribution of the study population (except for age). BMI = body mass index; MET = metabolic equivalent task.
† Not including offspring.
‡ Final status was derived from updated family history status at censoring (including death from other causes) or immediately before personal CRC diagnosis if it occurred (to avoid recall bias).
§ Energy-adjusted intake.
Figure 1.
Distribution of long interspersed nucleotide element 1 (LINE-1) methylation levels in 1224 colorectal cancer cases in the Nurses’ Health Study and the Health Professionals Follow-up Study. LINE-1 methylation level was determined by polymerase chain reaction on bisulfite-modified genomic DNA from archival paraffin-embedded tumor tissue, followed by pyrosequencing, as described in the Methods.
Figure 2.
Distribution of long interspersed nucleotide element 1 (LINE-1) methylation levels in 1224 colorectal cancer cases according to the number of first-degree relatives with colorectal cancer (CRC) in the Nurses’ Health Study and the Health Professionals Follow-up Study. Information on the number of first-degree relatives with CRC was obtained from cohort participants before CRC diagnosis. LINE-1 methylation level was determined by polymerase chain reaction on bisulfite-modified genomic DNA, followed by pyrosequencing, as described in the Methods.
In the NHS (women), compared with individuals without CRC family history, those with CRC family history experienced a statistically significantly higher overall CRC risk and higher risks for all tumor subtypes examined (Table 2). The cancer risk associated with family history appeared to be higher for the LINE-1 methylation-low tumor subtype than the LINE-1 methylation-high subtype and higher for the MSI-high subtype than the MSS subtype, although the differences were not statistically significant (P heterogeneity > .10).
Table 2.
Family history of colorectal cancer (CRC) and subsequent risk of developing CRC according to molecular subtypes in the Nurses’ Health Study*
| No. of first-degree relatives with CRC† | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC molecular subtype | 0 | 1 | ≥2 | P trend | P heterogeneity‡ | ||||||
| Person-years | 1967983 | 262761 | 22250 | ||||||||
| All CRCs | |||||||||||
| No. of cancers | 517 | 119 | 23 | ||||||||
| Age-adjusted incidence rate§ | 28.3 | 43.6 | 96.3 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.41 (1.16 to 1.73) | 2.66 (1.74 to 4.06) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.39 (1.13 to 1.70) | 2.60 (1.70 to 3.97) | <.001 | |||||||
| LINE-1 methylation | .25 | ||||||||||
| Methylation-low, <55% (n = 115, 17%) | |||||||||||
| No. of cancers | 86 | 24 | 5 | ||||||||
| Age-adjusted incidence rate§ | 4.6 | 9.9 | 28.9 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.79 (1.12 to 2.86) | 3.98 (1.57 to 10.1) | .001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.77 (1.11 to 2.82) | 3.96 (1.58 to 9.94) | .001 | |||||||
| Methylation-intermediate 55%–64.9% (n = 270, 41%) | |||||||||||
| No. of cancers | 216 | 44 | 10 | ||||||||
| Age-adjusted incidence rate§ | 12.4 | 16.2 | 38.7 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.28 (0.92 to 1.77) | 2.76 (1.45 to 5.25) | .006 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.25 (0.90 to 1.74) | 2.69 (1.42 to 5.12) | .009 | |||||||
| Methylation-high, ≥65% (n = 274, 42%) | |||||||||||
| No. of cancers | 215 | 51 | 8 | ||||||||
| Age-adjusted incidence rate§ | 11.4 | 17.6 | 28.4 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.40 (1.03 to 1.91) | 2.12 (1.04 to 4.32) | .005 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.37 (1.01 to 1.87) | 2.06 (1.01 to 4.20) | .008 | |||||||
| MSI status | .11 | ||||||||||
| MSS (n = 515, 80%) | |||||||||||
| No. of cancers | 411 | 93 | 11 | ||||||||
| Age-adjusted incidence rate§ | 23.0 | 36.3 | 47.4 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.41 (1.12 to 1.77) | 1.68 (0.92 to 3.07) | .001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.39 (1.10 to 1.75) | 1.66 (0.91 to 3.02) | .002 | |||||||
| MSI-high (n = 126, 20%) | |||||||||||
| No. of cancers | 93 | 23 | 10 | ||||||||
| Age-adjusted incidence rate‡ | 5.2 | 6.1 | 38.7 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.40 (0.89 to 2.18) | 5.23 (2.66 to 10.3) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.37 (0.87 to 2.14) | 4.98 (2.54 to 9.76) | <.001 | |||||||
| CIMP status | .87 | ||||||||||
| CIMP-low/negative (n = 516, 79%) | |||||||||||
| No. of cancers | 409 | 92 | 15 | ||||||||
| Age-adjusted incidence rate§ | 22.9 | 36.0 | 63.7 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.42 (1.13 to 1.79) | 2.37 (1.41 to 4.00) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.40 (1.11 to 1.76) | 2.34 (1.40 to 3.94) | <.001 | |||||||
| CIMP-high (n = 140, 21%) | |||||||||||
| No. of cancers | 107 | 26 | 7 | ||||||||
| Age-adjusted incidence rate§ | 5.4 | 7.3 | 23.6 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.34 (0.88 to 2.04) | 3.01 (1.37 to 6.57) | .01 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.31 (0.86 to 2.00) | 2.87 (1.31 to 6.27) | .02 | |||||||
* CI = confidence interval; CIMP = CpG island methylator phenotype; HR = hazard ratio; LINE-1 = long interspersed nucleotide element 1; MSI = microsatellite instability; MSS = microsatellite stable.
† Not including offspring.
‡ P for heterogeneity for trends (0 vs 1 vs ≥2 affected first-degree relatives) between tumor molecular subtypes. A test for LINE-1 methylation subtypes assessed an ordinal linear trend for exposure (0 vs 1 vs ≥2 affected first-degree relatives) and for LINE-1 methylation-low to methylation-intermediate to methylation-high subtype.
§ Age-adjusted incidence rates (per 100000) were standardized to the age distribution of the population.
|| Adjusted for body mass index, cumulative mean physical activity, alcohol, folate, methionine, calcium, red meat intake, current smoking status, current multivitamin use, and regular aspirin use.
In the HPFS (men), compared with individuals without CRC family history, those with CRC family history experienced a statistically significantly higher overall CRC risk and higher risks for all tumor subtypes examined except for the LINE-1 methylation-high subtype (Table 3). The cancer risk associated with family history appeared to be higher for the LINE-1 methylation-low subtype than the LINE-1 methylation-high subtype and higher for the MSI-high subtype than the MSS subtype, although the differences were not statistically significant (P heterogeneity > .09).
Table 3.
Family history of colorectal cancer (CRC) and subsequent risk of developing CRC according to molecular subtypes in Health Professionals Follow-up Study*
| No. of first-degree relatives with CRC† | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC molecular subtype | 0 | 1 | ≥2 | P trend | P heterogeneity‡ | ||||||
| Person-years | 819660 | 107327 | 4433 | ||||||||
| All CRCs | |||||||||||
| No. of cancers | 459 | 97 | 9 | ||||||||
| Age-adjusted incidence rate§ | 59.4 | 85.1 | 123.3 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.41 (1.13 to 1.76) | 2.80 (1.43 to 5.50) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.41 (1.12 to 1.75) | 2.88 (1.46 to 5.67) | <.001 | |||||||
| LINE-1 methylation | .12 | ||||||||||
| Methylation-low, <55% (n = 125, 22%) | |||||||||||
| No. of cancers | 101 | 21 | 3 | ||||||||
| Age-adjusted incidence rate§ | 13.5 | 19.3 | 38.1 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.54 (0.95 to 2.49) | 4.75 (1.61 to 14.0) | .009 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.50 (0.93 to 2.44) | 5.18 (1.81 to 14.8) | .01 | |||||||
| Methylation-intermediate, 55%–64.9% (n = 204, 36%) | |||||||||||
| No. of cancers | 160 | 40 | 4 | ||||||||
| Age-adjusted incidence rate§ | 12.4 | 16.2 | 38.7 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.63 (1.15 to 2.31) | 3.22 (1.23 to 8.43) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.66 (1.17 to 2.35) | 3.22 (1.22 to 8.54) | <.001 | |||||||
| Methylation-high, ≥65% (n = 236, 42%) | |||||||||||
| No. of cancers | 198 | 36 | 2 | ||||||||
| Age-adjusted incidence rate§ | 25.6 | 32.1 | 25.5 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.17 (0.82 to 1.68) | 1.51 (0.37 to 6.09) | .31 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.16 (0.81 to 1.66) | 1.55 (0.38 to 6.36) | .33 | |||||||
| MSI status | .10 | ||||||||||
| MSS (n = 487, 89%) | |||||||||||
| No. of cancers | 399 | 82 | 6 | ||||||||
| Age-adjusted incidence rate§ | 51.2 | 71.0 | 76.2 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.38 (1.08 to 1.76) | 2.05 (0.95 to 4.45) | .002 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.38 (1.08 to 1.75) | 2.13 (0.99 to 4.59) | .002 | |||||||
| MSI-high (n = 62, 11%) | |||||||||||
| No. of cancers | 46 | 13 | 3 | ||||||||
| Age-adjusted incidence rate§ | 6.3 | 12.4 | 47.2 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.72 (0.93 to 3.18) | 13.8 (4.18 to 45.7) | .002 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.69 (0.92 to 3.10) | 13.2 (3.87 to 44.8) | .003 | |||||||
| CIMP status | .17 | ||||||||||
| CIMP-low/negative (n = 447, 88%) | |||||||||||
| No. of cancers | 367 | 75 | 5 | ||||||||
| Age-adjusted incidence rate§ | 47.3 | 66.5 | 63.6 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.37 (1.07 to 1.76) | 1.99 (0.86 to 4.59) | .004 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.37 (1.07 to 1.76) | 2.07 (0.90 to 4.77) | .004 | |||||||
| CIMP-high (n = 61, 12%) | |||||||||||
| No. of cancers | 45 | 14 | 2 | ||||||||
| Age-adjusted incidence rate§ | 5.5 | 12.0 | 34.3 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.95 (1.05 to 3.60) | 6.22 (1.33 to 29.2) | .005 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.91 (1.03 to 3.54) | 5.89 (1.22 to 28.5) | .007 | |||||||
* CI = confidence interval; CIMP = CpG island methylator phenotype; HR = hazard ratio; LINE-1 = long interspersed nucleotide element 1; MSI = microsatellite instability; MSS = microsatellite stable.
† Not including offspring.
‡ P for heterogeneity for trends (0 vs 1 vs ≥2 affected first-degree relatives) between tumor molecular subtypes. A test for LINE-1 methylation subtypes assessed an ordinal linear trend for exposure (0 vs 1 vs ≥2 affected first-degree relatives) and for LINE-1 methylation-low to methylation-intermediate to methylation-high subtype.
§ Age-adjusted incidence rates (per 100000) were standardized to the age distribution of the population.
|| Adjusted for body mass index, cumulative mean physical activity, alcohol, folate, methionine, calcium, red meat intake, current smoking status, current multivitamin use, and regular aspirin use.
There was no statistically significant heterogeneity between the two cohorts (ie, women and men) in the relationships of CRC family history with overall CRC risk or with the risk of each tumor subtype (Q statistics P ≥ .30). To increase statistical power, we combined the two cohorts for further analyses.
In the combined cohorts, the association between the number of first-degree relatives with colorectal cancer (0, 1, ≥2; an ordinal scale) and CRC risk appeared to differ by LINE-1 methylation status (an ordinal scale of 3 levels), although the difference was not statistically significant (P heterogeneity = .06) (Table 4). Compared with individuals without CRC family history, those with CRC family history experienced a substantially higher risk of LINE-1 methylation-low CRC (multivariable HR = 1.63, 95% CI = 1.16 to 2.28 for 1 vs 0 first-degree relatives with CRC; multivariable HR = 4.32, 95% CI = 2.15 to 8.67 for ≥2 vs 0 first-degree relatives with CRC; P trend < .001) (Table 4). To a lesser degree, CRC family history was associated with higher risks of LINE-1 methylation-intermediate CRC (P trend < .001) and LINE-1 methylation-high CRC (P trend = .007) (Table 4).
Table 4.
Family history of colorectal cancer (CRC) and subsequent risk of developing CRC according to molecular subtypes in the combined cohorts*
| No. of first-degree relatives with CRC† | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC molecular subtype | 0 | 1 | ≥2 | P trend | P heterogeneity‡ | ||||||
| Person-years | 2787643 | 370088 | 26684 | ||||||||
| All CRCs | |||||||||||
| No. of cancers | 976 | 216 | 32 | ||||||||
| Age-adjusted incidence rate§ | 37.2 | 54.6 | 100.4 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.41 (1.22 to 1.64) | 2.70 (1.88 to 3.86) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.40 (1.20 to 1.62) | 2.67 (1.86 to 3.82) | <.001 | |||||||
| LINE-1 methylation | .06 | ||||||||||
| Methylation-low, <55% (n = 240, 20%) | |||||||||||
| No. of cancers | 187 | 45 | 8 | ||||||||
| Age-adjusted incidence rate§ | 7.0 | 12.4 | 29.1 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.66 (1.19 to 2.32) | 4.21 (2.08 to 8.56) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.63 (1.16 to 2.28) | 4.32 (2.15 to 8.67) | <.001 | |||||||
| Methylation-intermediate, 55%–64.9% (n = 474, 39%) | |||||||||||
| No. of cancers | 376 | 84 | 14 | ||||||||
| Age-adjusted incidence rate§ | 14.7 | 21.3 | 42.6 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.42 (1.12 to 1.81) | 2.90 (1.70 to 4.95) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.42 (1.12 to 1.80) | 2.83 (1.66 to 4.83) | <.001 | |||||||
| Methylation-high, ≥65% (n = 510, 42%) | |||||||||||
| No. of cancers | 413 | 87 | 10 | ||||||||
| Age-adjusted incidence rate§ | 15.7 | 21.4 | 28.3 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.30 (1.03 to 1.64) | 1.94 (1.03 to 3.66) | .005 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.28 (1.01 to 1.61) | 1.92 (1.02 to 3.63) | .007 | |||||||
| MSI status | .03 | ||||||||||
| MSS (n = 1002, 84%) | |||||||||||
| No. of cancers | 810 | 175 | 17 | ||||||||
| Age-adjusted incidence rate§ | 30.9 | 45.0 | 49.7 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.40 (1.18 to 1.65) | 1.79 (1.11 to 2.89) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.38 (1.17 to 1.63) | 1.79 (1.12 to 2.89) | <.001 | |||||||
| MSI-high (n = 188, 16%) | |||||||||||
| No. of cancers | 139 | 36 | 13 | ||||||||
| Age-adjusted incidence rate§ | 5.6 | 8.1 | 41.1 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.50 (1.04 to 2.15) | 6.20 (3.43 to 11.2) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.47 (1.02 to 2.10) | 5.86 (3.25 to 10.6) | <.001 | |||||||
| CIMP status | .40 | ||||||||||
| CIMP-low/negative (n = 963, 83%) | |||||||||||
| No. of cancers | 776 | 167 | 20 | ||||||||
| Age-adjusted incidence rate§ | 30.1 | 44.1 | 62.6 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.40 (1.18 to 1.66) | 2.26 (1.45 to 3.52) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.39 (1.17 to 1.64) | 2.27 (1.46 to 3.52) | <.001 | |||||||
| CIMP-high (n = 201, 17%) | |||||||||||
| No. of cancers | 152 | 40 | 9 | ||||||||
| Age-adjusted incidence rate§ | 5.6 | 8.8 | 26.7 | ||||||||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.51 (1.07 to 2.13) | 3.45 (1.72 to 6.92) | <.001 | |||||||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.48 (1.04 to 2.09) | 3.29 (1.64 to 6.59) | <.001 | |||||||
* CI = confidence interval; CIMP = CpG island methylator phenotype; HR = hazard ratio; LINE-1 = long interspersed nucleotide element 1; MSI = microsatellite instability; MSS = microsatellite stable.
† Not including offspring.
‡ P for heterogeneity for trends (0 vs 1 vs ≥2 affected first-degree relatives) between tumor molecular subtypes. A test for LINE-1 methylation subtypes assessed an ordinal linear trend for exposure (0 vs 1 vs ≥2 affected first-degree relatives) and for LINE-1 methylation-low to methylation-intermediate to methylation-high subtype.
§ Age-adjusted incidence rates (per 100000) were standardized to the age distribution of the population.
|| Adjusted for body mass index, cumulative mean physical activity, alcohol, folate, methionine, calcium, red meat intake, current smoking status, current multivitamin use, and regular aspirin use.
In the combined cohorts, the association between the number of first-degree relatives with CRC (0, 1, ≥2; an ordinal scale) and CRC risk statistically significantly differed by MSI status (P heterogeneity = .03) (Table 4). In contrast, CRC risk associated with family history did not differ statistically significantly by CIMP status (P heterogeneity = .40).
CRC Family History and MSS Cancer Risk According to LINE-1 Methylation Level
We aimed to examine the relationship between CRC family history and CRC risk according to LINE-1 methylation level while attempting to exclude influence of Lynch syndrome (ie, hereditary susceptibility to mismatch repair-deficient cancer, most likely to be MSI-high cancer). For this purpose, we examined the risk of MSS (non-MSI-high) CRC according to LINE-1 methylation level (censoring MSI-high cancer incidence) (Table 5). The association between the number of first-degree relatives with CRC (0, 1, ≥2; an ordinal scale) and MSS CRC risk statistically significantly differed by LINE-1 methylation status (ordinal 3 levels) (P heterogeneity = .02). CRC family history was associated with a statistically significant increase in LINE-1 methylation-low MSS cancer (multivariable HR = 1.68, 95% CI = 1.19 to 2.38 for 1 vs 0 first-degree relatives with CRC; multivariable HR = 3.48, 95% CI = 1.59 to 7.63 for ≥2 vs 0 first-degree relatives with CRC; P trend < .001). To a lesser degree, CRC family history was associated with a higher risk of LINE-1 methylation-intermediate MSS cancer (multivariable HR = 1.50, 95% CI = 1.17 to 1.93 for 1 vs 0 first-degree relatives with CRC; multivariable HR = 1.42, 95% CI = 0.64 to 3.16 for ≥2 vs 0 first-degree relatives with CRC; P trend = .002) (Table 5). In contrast, CRC family history was not statistically significantly associated with LINE-1 methylation-high MSS cancer (P trend = .35). Our data implied that the association between CRC family history and MSS cancer risk was strongest for the LINE-1 methylation-low tumor subtype, followed by the LINE-1 methylation-intermediate subtype, and weakest or null for the LINE-1 methylation-high tumor subtype.
Table 5.
Family history of colorectal cancer (CRC) and subsequent risk of developing microsatellite stable (MSS) CRC by long interspersed nucleotide element 1 (LINE-1) methylation level (censoring microsatellite instability-high cancers) in the combined cohorts*
| No. of first-degree relatives with CRC† | ||||||
|---|---|---|---|---|---|---|
| MSS CRC molecular subtype | 0 | 1 | ≥2 | P trend | P heterogeneity‡ | |
| Person-years | 2787643 | 370088 | 26684 | |||
| LINE-1 subtyping in MSS tumors | .02 | |||||
| Methylation-low, <55% | ||||||
| No. of cancers | 172 | 43 | 6 | |||
| Age-adjusted incidence rate§ | 6.8 | 11.8 | 17.4 | |||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.71 (1.21 to 2.42) | 3.37 (1.52 to 7.50) | <.001 | ||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.68 (1.19 to 2.38) | 3.48 (1.59 to 7.63) | <.001 | ||
| Methylation-intermediate, 55%–64.9% | ||||||
| No. of cancers | 330 | 77 | 6 | |||
| Age-adjusted incidence rate§ | 12.5 | 19.7 | 20.0 | |||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.50 (1.17 to 1.93) | 1.42 (0.64 to 3.16) | .002 | ||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.50 (1.17 to 1.93) | 1.42 (0.64 to 3.14) | .002 | ||
| Methylation-high, ≥65% | ||||||
| No. of cancers | 308 | 55 | 5 | |||
| Age-adjusted incidence rate§ | 11.6 | 14.1 | 12.4 | |||
| Age-adjusted HR (95% CI) | 1 (referent) | 1.12 (0.84 to 1.50) | 1.43 (0.59 to 3.48) | .31 | ||
| Multivariable HR (95% CI)|| | 1 (referent) | 1.11 (0.83 to 1.48) | 1.43 (0.59 to 3.49) | .35 | ||
* CI = confidence interval; HR = hazard ratio.
† Not including offspring.
‡ A heterogeneity test for LINE-1 methylation subtypes assessed an ordinal linear trend for exposure (0 vs 1 vs ≥2 affected first-degree relatives) as well as for LINE-1 methylation-high to methylation-intermediate to methylation-low subtype.
§ Age-adjusted incidence rates (per 100000) were standardized to the age distribution of the population.
|| Adjusted for body mass index, cumulative mean physical activity, alcohol, folate, methionine, calcium, and red meat intake, current smoking status, current multivitamin use, and regular aspirin use.
Discussion
In this large, prospective study, we showed that a family history of CRC was associated with a high risk of CRC with low-level LINE-1 methylation. After attempting to exclude the potential influence of Lynch syndrome (ie, familial predisposition to MSI-positive cancer), the relationship between CRC family history and the risk of LINE-1 methylation-low MSS cancer persisted. The relationship between CRC family history and subsequent MSS cancer risk differed statistically significantly by tumor LINE-1 methylation level. Our data suggest a possible link between the heritability of CRC and tumoral epigenetic changes. It is possible that at least some LINE-1 methylation-low MSS cancer may represent a previously unrecognized familial CRC trait.
Genetic predisposition to CRC may underlie specific molecular alterations in neoplastic cells (35–39). Familial clustering of CRCs has been commonly associated with a specific molecular subtype, namely MSI-high tumors in the setting of Lynch syndrome (36,40,41). Nonetheless, it has also been shown that familial clustering suggestive of Lynch syndrome is not always associated with hereditary mismatch repair defects or MSI-high tumors (10,40,42). In a recent study by Goel et al. (10), non-MSI-high tumors occurring in a familial pattern suggestive of Lynch syndrome frequently exhibited LINE-1 hypomethylation, which is consistent with our current data. Besides intense familial cancer clustering, a common family history of colorectal cancer in a first-degree relative (observed in approximately 20% of all colorectal cancer cases) may help to identify novel risk alleles by genome-wide linkage analysis (7). A recent study highlights the novel opportunities of using family history data in genome-wide association study datasets to gain new evidence for disease risk alleles (43). Family history of cancer remains pivotal in our attempts to decipher genetic etiologies (beyond genome-wide association studies) and their interactions with environment.
LINE-1 represents a major repetitive element and occupies approximately 17% of the human genome (11). Thus, methylation level in LINE-1 has been shown to correlate with global DNA methylation level in tumor cells (44). Genomic hypomethylation has been linked to genomic and chromosomal instability leading to carcinogenesis (45–49). In addition to its role as a surrogate of genomic hypomethylation, LINE-1 hypomethylation may, in itself, have carcinogenic effects through deregulation of gene transcription and activation of retrotransposons (50–52). Tumor LINE-1 hypomethylation has been associated with inferior prognosis in not only colon cancer (12–14) but also many different human cancer types (53). We previously found that the degree of colon cancer LINE-1 hypomethylation was linearly associated with aggressive tumor behavior and observed an approximately fivefold increase in cancer-specific mortality associated with tumors at the low end of the methylation spectrum, compared with those at the high end (12). In addition, LINE-1 hypomethylated CRC has been associated with young age of onset (28) and CRC familial clustering (10), but not with polymorphisms in one-carbon metabolizing enzyme genes (54). Taking these findings together with our current data, LINE-1 methylation level may serve as a potential tumor biomarker for prognostication as well as familial cancer risk assessment.
Interestingly, we showed that CRC family history was associated with elevated risk of almost all molecular subtypes that we examined. One reason for this phenomenon may be that many common CRC susceptibility variants may increase the risks of many different molecular subtypes. As indicated by the unique tumor principle (55,56), each tumor confers its own differential familial risk given the uniqueness of each human genome. Nonetheless, we use molecular classification (such as MSI-high vs MSS) to better predict familial risk, natural history, and response to treatment (55,56). A second reason is mutual confounding of molecular subtypes (55). For example, MSI-high cancers are strongly associated with CRC family history, and are present in multiple molecular subtypes, including LINE-1 methylation-high, CIMP-low, and CIMP-negative subtypes, which is why we needed to perform our primary hypothesis testing within MSS cancers.
In CRC, CIMP has been associated with sporadic MSI-high cancers due to epigenetic silencing of MLH1 (57,58). One retrospective case–case study (N = 47) (59) reported a statistically significant association between CRC family history and CIMP, whereas our much larger current study and retrospective case–case study (N = 547) (60) failed to confirm this association, indicating overall lack of evidence for a strong link between CRC family history and CIMP.
Our study possesses several key strengths. First, collection of family history information and data on numerous dietary and lifestyle covariables relevant to cancer risk was performed repeatedly over decades, with high rates of follow-up in both cohorts. Second, our study participants were distributed throughout the United States, and thus our CRC cases were more representative of cases in the US white population than CRC cases from several referral hospitals. Third, data collection was prospective, eliminating differential recall bias, which is a major concern in analyses involving family history of cancer (19,20). In addition, our robust and extensive molecular characterization of CRC enabled a molecular pathological epidemiology study design (33,34). Molecular pathological epidemiology studies have yielded several interesting observations relevant to carcinogenesis and personalized patient management strategies (18,33,34,61–66).
One limitation of our study is the use of questionnaire-based data collection for family history, which relies on knowledge and compliance of study participants. Nonetheless, a recent study (67) has validated the predictive accuracy of self-reporting of CRC in first-degree relatives (positive predictive value = 0.86, 95% CI = 0.64 to 0.95; negative predictive value = 0.98, 95% CI = 0.94 to 0.99), in keeping with an existing report (68). Furthermore, all of our study participants were health professionals, which increased accuracy of family history information. In addition, we ascertained family history at multiple time points, further increasing accuracy. A second limitation is that we could not obtain tumor tissue specimens from all incident CRCs. Nonetheless, the demographic and clinical characteristics did not statistically significantly differ between cases with and without available tumor tissue, and the pathologic and molecular features of our CRC cases were generally compatible with data from the US cancer registry and published literature (23). Additionally, any biomarker research should be interpreted with caution, and study findings must be carefully evaluated (69).
Clinical implications of our findings warrant further discussion. As shown in Table 4, the multivariable hazard ratio for MSI-high cancer by the presence of two or more affected first-degree relatives was 5.86 (95% CI = 3.25 to 10.6), and the multivariable hazard ratio for LINE-1 methylation-low cancer was 4.32 (95% CI = 2.15 to 8.67). Thus, the presence of these tumor molecular features imply further elevated familial CRC risk, compared with the approximately twofold increase in CRC risk associated with CRC family history (without considering tumor molecular features). As the effect estimates imply, the ability of MSI testing to predict familial risk is higher than that of LINE-1 testing. Nonetheless, considering the importance of MSI testing in clinical and pathology practice, additional studies are necessary to assess implications of LINE-1 methylation status in the estimation of familial CRC risk.
In summary, our current study suggests that a family history of CRC is associated with a high risk of CRC with low-level LINE-1 methylation and that genetic predisposition likely underlies somatic epigenetic changes. It is possible that at least a subset of LINE-1 methylation-low CRC may constitute a previously unrecognized familial cancer trait. Previous studies have shown that LINE-1 methylation-low colon cancer is an aggressive cancer subtype (12–14), which demands effective prevention and surveillance strategies. Therefore, our findings may have considerable clinical and public health implications. Additional studies are needed to assess utility of tumor LINE-1 methylation testing in clinical settings before it can be implemented as a tumor molecular biomarker for prognostication in the proband and for cancer risk assessment in family members, akin to current MSI testing in CRC. We also anticipate that future studies will further elucidate the mechanisms that underlie the association between the heritability of CRC and somatic epigenetic alterations.
Funding
This work was supported by the National Institute of Health (P01 CA87969 [to S. E. Hankinson], P01 CA55075 [to W. C. Willett], P50 CA127003 [to CSF], R01 CA151993 [to SO], and R01 CA137178 [to ATC]; the Bennett Family Fund for Targeted Therapies Research; and the National Colorectal Cancer Research Alliance. PL is a Scottish Government Clinical Academic Fellow and is supported by a Harvard University Knox Memorial Fellowship. ATC is a Damon Runyon Clinical Investigator.
Supplementary Material
S. Ogino, R. Nishihara, and P. Lochhead contributed equally to this study. A. T. Chan, E. Giovannucci, and C. S. Fuchs contributed equally to this study.
A.T. Chan was a consultant of Bayer Healthcare, Millennium Pharmaceuticals, and Pfizer Inc. This study was not funded by Bayer Healthcare, Millennium Pharmaceuticals, or Pfizer Inc. No other authors report any conflicts of interest.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
Recently, Antelo et al. (70) reported that LINE-1 hypomethylated colorectal cancer was associated with early onset disease, consistent with our previous data (28), and possible genetic susceptibility that may also lead to synchronous LINE-1 hypomethylated colorectal cancers (71).
We deeply thank the Nurses’ Health Study and Health Professionals Follow-up Study cohort participants who have agreed to provide us with information through questionnaires and biological specimens and hospitals and pathology departments throughout the United States for generously providing us with tissue specimens. In addition, we thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study for their valuable contributions and the US state cancer registries for their help.
References
- 1. Le Marchand L, Zhao LP, Quiaoit F, Wilkens LR, Kolonel LN. Family history and risk of colorectal cancer in the multiethnic population of Hawaii. Am J Epidemiol. 1996; 144(12):1122–1128 [DOI] [PubMed] [Google Scholar]
- 2. Slattery ML, Kerber RA. Family history of cancer and colon cancer risk: the Utah Population Database. J Natl Cancer Inst. 1994; 86(21):1618–1626 [DOI] [PubMed] [Google Scholar]
- 3. Butterworth AS, Higgins JP, Pharoah P. Relative and absolute risk of colorectal cancer for individuals with a family history: a meta-analysis. Eur J Cancer. 2006; 42(2):216–227 [DOI] [PubMed] [Google Scholar]
- 4. Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol. 2001; 96(10):2992–3003 [DOI] [PubMed] [Google Scholar]
- 5. Fuchs CS, Giovannucci EL, Colditz GA, Hunter DJ, Speizer FE, Willett WC. A prospective study of family history and the risk of colorectal cancer. N Engl J Med. 1994; 331(25):1669–1674 [DOI] [PubMed] [Google Scholar]
- 6. Scheuner MT, McNeel TS, Freedman AN. Population prevalence of familial cancer and common hereditary cancer syndromes. The 2005 California Health Interview Survey. Genet Med. 2010; 12(11):726–735 [DOI] [PubMed] [Google Scholar]
- 7. Cicek MS, Cunningham JM, Fridley BL, et al. Colorectal cancer linkage on chromosomes 4q21, 8q13, 12q24, and 15q22. PLoS One. 2012; 7(5):e38175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ku CS, Cooper DN, Wu M, et al. Gene discovery in familial cancer syndromes by exome sequencing: prospects for the elucidation of familial colorectal cancer type X. Mod Pathol. 2012; 25(8):1055–1068 [DOI] [PubMed] [Google Scholar]
- 9. Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, et al. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004; 1(3):–e65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goel A, Xicola RM, Nguyen TP, et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology. 2010; 138(5):1854–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009; 10(10):691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008; 100: 1734–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011; 117(9):1847–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rhee YY, Kim MJ, Bae JM, et al. Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level. Ann Surg Oncol. 2012; 19(11):3441–3448 [DOI] [PubMed] [Google Scholar]
- 15. Estecio MR, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007; 2(5):e399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG methylator phenotype in colorectal cancer. Int J Cancer. 2008; 122: 2767–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. New Engl J Med. 2007; 356(21):2131–2142 [DOI] [PubMed] [Google Scholar]
- 18. Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011; 305(16):1685–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Floderus B, Barlow L, Mack TM. Recall bias in subjective reports of familial cancer. Epidemiology. 1990; 1(4):318–321 [DOI] [PubMed] [Google Scholar]
- 20. Chang ET, Smedby KE, Hjalgrim H, Glimelius B, Adami HO. Reliability of self-reported family history of cancer in a large case-control study of lymphoma. J Natl Cancer Inst. 2006; 98(1):61–68 [DOI] [PubMed] [Google Scholar]
- 21. Ziogas A, Horick NK, Kinney AY, et al. Clinically relevant changes in family history of cancer over time. JAMA. 2011; 306(2):172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Acheson LS. Recording, interpreting, and updating the family history of cancer: implications for cancer prevention. JAMA. 2011; 306(2):208–210 [DOI] [PubMed] [Google Scholar]
- 23. Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012; 61(6):847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum?. Gut. 2012; 61(6):794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006; 8(2):209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa and peripheral blood cells. J Mol Diagn. 2010; 12(2):177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schernhammer ES, Giovannucci E, Kawasaki T, Rosner B, Fuchs CS, Ogino S. Dietary folate, alcohol, and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut. 2010; 59(6):794–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baba Y, Huttenhower C, Nosho K, et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer. 2010; 9: 125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006; 55(7):1000–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006; 38(7):787–793 [DOI] [PubMed] [Google Scholar]
- 31. Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008; 3(11):e3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009; 58(1):90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: The evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010; 102(6):365–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011; 60(3):397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Slattery ML, Curtin K, Schaffer D, Anderson K, Samowitz W. Associations between family history of colorectal cancer and genetic alterations in tumors. Int J Cancer. 2002; 97(6):823–827 [DOI] [PubMed] [Google Scholar]
- 36. Bapat B, Lindor NM, Baron J, et al. The association of tumor microsatellite instability phenotype with family history of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009; 18(3):967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindor NM, Rabe K, Petersen GM, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA. 2005; 293(16):1979–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bonadona V, Bonaiti B, Olschwang S, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011; 305(22):2304–2310 [DOI] [PubMed] [Google Scholar]
- 39. Baglietto L, Lindor NM, Dowty JG, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010; 102(3):193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lubbe SJ, Webb EL, Chandler IP, Houlston RS. Implications of familial colorectal cancer risk profiles and microsatellite instability status. J Clin Oncol. 2009; 27(13):2238–2244 [DOI] [PubMed] [Google Scholar]
- 41. Funkhouser WK, Lubin IM, Monzon FA, et al. Relevance, pathogenesis, and testing algorithm for mismatch repair-defective colorectal carcinomas: a report of the Association for Molecular Pathology. J Mol Diagn. 2012; 14(2):91–103 [DOI] [PubMed] [Google Scholar]
- 42. Abdel-Rahman WM, Ollikainen M, Kariola R, et al. Comprehensive characterization of HNPCC-related colorectal cancers reveals striking molecular features in families with no germline mismatch repair gene mutations. Oncogene. 2005; 24(9):1542–1551 [DOI] [PubMed] [Google Scholar]
- 43. Ghosh A, Hartge P, Purdue MP, et al. Assessing disease risk in genome-wide association studies using family history. Epidemiology. 2012; 23(4):616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004; 32(3):e38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003; 300(5618):489–492 [DOI] [PubMed] [Google Scholar]
- 46. Yamada Y, Jackson-Grusby L, Linhart H, et al. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci U S A. 2005; 102(38):13580–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006; 66(17):8462–9468 [DOI] [PubMed] [Google Scholar]
- 48. Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011; 8(12):686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rawson JB, Bapat B. Epigenetic biomarkers in colorectal cancer diagnostics. Expert Rev Mol Diagn. 2012; 12(5):499–509 [DOI] [PubMed] [Google Scholar]
- 50. Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004; 429(6989):268–274 [DOI] [PubMed] [Google Scholar]
- 51. Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008; 27(3):404–408 [DOI] [PubMed] [Google Scholar]
- 52. Aporntewan C, Phokaew C, Piriyapongsa J, et al. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One. 2011; 6(3):e17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kitkumthorn N, Mutirangura A. Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenet. 2012; 2: 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hazra A, Fuchs CS, Kawasaki T, Kirkner GJ, Hunter DJ, Ogino S. Germline polymorphisms in the one-carbon metabolism pathway and DNA methylation in colorectal cancer. Cancer Causes Control. 2010; 21(3):331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ogino S, Giovannucci E. Lifestyle factors and colorectal cancer microsatellite instability: molecular pathological epidemiology science, based on unique tumour principle. In J Epidemiol. 2012;41(4):1072–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012; 12(6):621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathol Res Int. 2011; doi:10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hughes LA, Khalid-de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: progress and problems. Biochim Biophys Acta. 2012; 1825(1):77–85. [DOI] [PubMed] [Google Scholar]
- 59. Frazier ML, Xi L, Zong J, et al. Association of the CpG island methylator phenotype with family history of cancer in patients with colorectal cancer. Cancer Res. 2003; 63(16):4805–4808 [PubMed] [Google Scholar]
- 60. Ward RL, Williams R, Law M, Hawkins NJ. The CpG island methylator phenotype is not associated with a personal or family history of cancer. Cancer Res. 2004; 64(20):7618–7621 [DOI] [PubMed] [Google Scholar]
- 61. Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology-analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011; 8(12):711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Samowitz WS, Albertsen H, Sweeney C, et al. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst. 2006; 98(23):1731–1738. [DOI] [PubMed] [Google Scholar]
- 63. Ballard-Barbash R, Friedenreich CM, Courneya KS, Siddiqi SM, McTiernan A, Alfano CM. Physical activity, biomarkers, and disease outcomes in cancer survivors: a systematic review. J Natl Cancer Inst. 2012; 104(11):815–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Limsui D, Vierkant RA, Tillmans LS, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J Natl Cancer Inst. 2010; 102(14):1012–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Campbell PT, Jacobs ET, Ulrich CM, et al. Case-control study of overweight, obesity, and colorectal cancer risk, overall and by tumor microsatellite instability status. J Natl Cancer Inst. 2010; 102(6):391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kuchiba A, Morikawa T, Yamauchi M, et al. Body mass index and risk of colorectal cancer according to fatty acid synthase expression in the Nurses’ Health Study. J Natl Cancer Inst. 2012; 104(5):415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mai PL, Garceau AO, Graubard BI, et al. Confirmation of family cancer history reported in a population-based survey. J Natl Cancer Inst. 2011; 103(10):788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Murff HJ, Spigel DR, Syngal S. Does this patient have a family history of cancer? An evidence-based analysis of the accuracy of family cancer history. JAMA. 2004; 292(12):1480–1489 [DOI] [PubMed] [Google Scholar]
- 69. Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011; 9(Suppl 5):S1–32; quiz S33 [DOI] [PubMed] [Google Scholar]
- 70. Antelo M, Balaguer F, Shia J, et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS One. 2012; 7(9):e45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nosho K, Kure S, Irahara N, et al. A prospective cohort study shows unique epigenetic, genetic, and prognostic features of synchronous colorectal cancers. Gastroenterology. 2009; 137(5):1609–1620.e1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.