Abstract
Understanding the genetic and ecological factors which support the emergence of new clones of pathogenic bacteria is vital to develop preventive measures. Vibrio cholerae the causative agent of cholera epidemics represents a paradigm for this process in that this organism evolved from environmental non-pathogenic strains by acquisition of virulence genes. The major virulence factors of V. cholerae, cholera toxin (CT) and toxin coregulated pilus (TCP) are encoded by a lysogenic bacteriophage (CTXφ) and a pathogenicity island, respectively. Additional phages which cooperate with the CTXφ in horizontal transfer of genes in V. cholerae have been characterized, and the potential exists for discovering yet new phages or genetic elements which support the transfer of genes for environmental fitness and virulence leading to the emergence of new epidemic strains. Phages have also been shown to play a crucial role in modulating seasonal cholera epidemics. Thus, the complex array of natural phenomena driving the evolution of pathogenic V. cholerae includes, among other factors, phages that either participate in horizontal gene transfer or in a bactericidal selection process favoring the emergence of new clones of V. cholerae.
Key words: Vibrio cholerae, bacteriophage, bacterial evolution, filamentous phage, CTX phage, lytic vibriophage, virulence genes
Introduction
Bacterial evolution is believed to have been significantly driven by their interactions with bacteriophages. Phages contribute to the evolution of bacteria by mediating horizontal transfer of clusters of genes and genomic rearrangements, as well as by bactericidal selection.1-3 In this latter process, bacterial strains that are able to resist phage predation have an advantage over the susceptible strains in their competition for survival. Toxigenic Vibrio cholerae, the causative agent of the epidemic diarrheal disease cholera, interacts with diverse phages, and the interaction can promote genetic diversity and/or cause selective enrichment of particular bacterial clones.3 V. cholerae represents a group of bacteria which is autochthonous to riverine, coastal and estuarine ecosystems, but at the same time, pathogenic for humans.4,5 In recent times, dramatic progress has been made in understanding the genetic determinants that account for virulence of this organism and how virulent strains emerge in nature.
Cholera is an ancient disease, and there have been seven recorded pandemics of cholera since the first pandemic began in 1817.4,5 However, the disease still affects millions of people worldwide, and causes an estimated 120,000 deaths and manifold more cases each year. The current seventh pandemic of cholera, which originated in Indonesia in 1961, is the most extensive in geographic spread and duration, and the causative agent is V. cholerae O1 of the El Tor biotype. The sixth pandemic and presumably the earlier pandemics were caused by the classical biotype, which is now extinct.6 The two biotypes of V. cholerae differ in certain phenotypic and genetic characteristics.4 The factors associated with the replacement of the classical biotype as the predominant epidemic strain by the El Tor biotype, and eventual disappearance of the classical strains have not been adequately explained. The displacement of the classical strains by the El Tor might have been driven by undefined environmental forces to which the El Tor strains adapted more efficiently. However, possible contribution of particular groups of phages have not been ruled out. Presumably, these phages selectively eliminated the classical biotype strains, while enriching the El Tor biotype.7
The best known example of a phage contributing to the emergence of toxigenic strains of V. cholerae is the CTXφ which carries the genes encoding cholera toxin in its genome.2 There is also a high probability for discovering new phages which are able to transfer other virulence associated genes, pathogenicity islands or genes that promote environmental fitness. This is because the pandemic V. cholerae strains carry a number of gene clusters which have structural features consistent with horizontally transferable elements,8 and evolutionary intermediate strains carrying incomplete sets of these gene clusters, and thus with a lower virulence potential than the pandemic strains have been shown to exist in nature.
The recent recognition that phage predation may play a role in the natural control of cholera epidemics3,9,10 reinforces prediction that changes in this pathogen may have been to a large extent driven by phages. The emergence of certain strains is likely to be enhanced by phages through the bactericidal mechanism in which phage sensitive strains are killed while providing a selective advantage to phage resistant strains. Therefore, the ability to evade phage predation constitutes important development in attaining evolutionary fitness. On the other hand, to achieve phage resistance there may be a fitness cost for the pathogen, due to altered expression of surface appendages such as flagella or pili or O-antigens which may often act as receptors for different phages. Therefore the evolutionary success of V. cholerae has presumably been dependent on a balance between these various factors. In this review we have summarized available information on the interactions of phage and V. cholerae, and the effect of these interactions on the genetics, epidemiology, and evolution of the pathogen.
Bacteriophages Acting on V. cholerae (Vibriophages)
As far back as the 19th century it was recognized that certain elements (most likely phages) in the waters of the Ganges and Jumna rivers in India had marked antibacterial activity against V. cholerae, and could protect against cholera.11-13 The role of phages in combating cholera, as well as other bacterial infections were eventually ignored due mainly to the discovery and availability of antibiotics. Two remarkable discoveries in recent times have renewed our interest in phages, in particular, phages that grow on V. cholerae, and generally known as vibriophages. The discovery that cholera toxin (CT) the major virulence factor of toxigenic V. cholerae, is encoded by a lysogenic filamentous phage (CTXφ), led to the exploration of other filamentous vibriophages. Accordingly, a number of other phages and satellite phages have been identified in V. cholerae O1 and O139 serogroup strains and the possible roles of these phages in horizontal gene transfer have been studied.11,14
In contrast to filamentous phages, which do not usually kill the host bacterial cells, the lytic phages can kill the host cells and thus contribute a strong selective force in nature, for the emergence of bacterial clones which are resistant to certain phages. The recent recognition that phage blooms coincide with the decline of V. cholerae in water in cholera-endemic areas3,9,10 has also renewed interest in lytic phages, due to their potential application in developing control measures against cholera epidemics. A considerable number of lytic and temperate vibriophages have now been found, of which the JSF series of environmental phages isolated in Bangladesh are of particular interest because of their role in epidemiology and ecology of V. cholerae. At least 16 distinct phages (JSF1 through JSF 16) have been isolated in Bangladesh,7,15 and efforts are under way to isolate and characterize more cholera phages.
The Cholera Toxin Converting Phage CTXφ
Bacteriophages can convert their bacterial host from a nonpathogenic strain to a pathogenic strain through a process called phage conversion, by providing the host with phage-encoded virulence genes. Toxigenic V. cholerae isolates carry the ctxAB genes encoded by a lysogenic filamentous phage CTXφ.2 Only those strains that carry CTXφ cause epidemic and pandemic cholera. The phage is similar in size, structure, and gene order to filamentous coliphages such as M13 and f1 from E. coli.11 The ctxAB genes which encode the A-B type enterotoxin CT reside in the CTXφ genome, which exists as a prophage, the integrated form of the phage genome in the host bacterial chromosome.
CTXφ has a 6.9-kb genome organized into two functionally distinct modules (Fig. 1). These are RS2 (repeat sequence 2), which comprises three genes, rstR, rstA and rstB, and a core region encoding psh, cep, orfU (gIII), ace, zot and ctxAB. The RstA protein is required for CTXφ DNA replication, which in E. coli Ff filamentous coliphage is performed by pII, the gene for which is located at the same genomic region as the rstA gene in CTXφ.16 The rstR gene encodes a repressor protein whereas the rstB gene encodes a protein required for integration of the CTXφ genome into the bacterial chromosome. The RstB protein contains a region similar to single-stranded (ss) DNA-binding proteins, which in other filamentous phages stabilize phage ssDNA.17 The Psh, Cep, OrfU and Ace proteins are phage structural proteins whereas Zot is a protein required for phage assembly.2 The genes cep, orfU, ace and zot correspond to genes gVIII, gIII, gVI and gI respectively of Ff coliphage. The ctxAB genes are unique to CTXφ, but are not essential for phage production, and are located at the same site as gIV in E. coli Ff phage. CTXφ does not encode a pIV protein homolog, which in Ff phage is required for secretion from the bacterial cell. Another important difference between CTXφ and typical E. coli Ff filamentous coliphages is that CTXφ integrates into the V. cholerae genome to form stable lysogens. However, the CTXφ genome does not encode an integrase gene which should perform this function in lysogenic phages.2 Instead, CTXφ uses host encoded integrases XerC and XerD for its integration into the bacterial chromosome. The XerCD proteins are conserved among eubacteria, as they serve to resolve chromosome dimers during cell division.18 Besides CT, a type IV pilus, the toxin co-regulated pilus (TCP) is the most important virulence factor of V. cholerae, and is essential for colonizing the intestine of the host. TCP consists of TcpA subunits encoded by the tcpA gene located in the TCP pathogenicity island. The TCP island is a 40 kb chromosomal region flanked on both sides by a 20-bp att-like attachment sequence, and comprised of tcp and acf gene clusters as well as genes encoding a putative integrase and a transposase.8
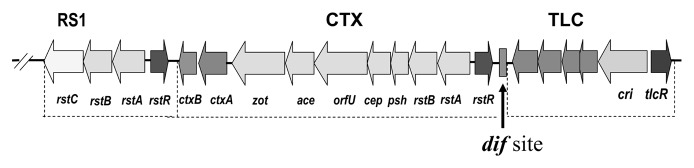
Figure 1. Chromosomal arrangement of RS1, CTX prophage and TLC gene cluster in V. cholerae. Open reading frames are shown as arrows, and the rectangle between CTX and TLC represents the dif site.
Interestingly TCP is also the receptor for CTXφ on the V. cholerae cell surface, whereas the periplasmic and inner membrane proteins TolQRA facilitate infection of the bacteria by CTXφ.2,19 The OrfU protein of CTXφ, which has been renamed pIIIctx, has been hypothesized to bind to the major pilin protein TcpA of TCP, similar to the binding of Ff coliphages to the F pilus mediated by the minor coat protein pIII. After CTXφ is adsorbed to the V. cholerae cell wall, the ssDNA is injected to the cell cytoplasm. Upon entry into the cell cytoplasm, the linear ssDNA circularizes, forming pCTX, which can remain as an extra chromosomal element or integrate into the V. cholerae genome at specific attachment sites.2,18 However, in contrast to the classical model of phage integration in which phage ssDNA is converted to dsDNA prior to integration, Val and colleagues20 proposed that Xer recombinases promote the direct insertion of the ssDNA genome into the dif1 site of V. cholerae. The wide distribution of XerCD recombinase system among bacteria and the prevalence of dif sites among filamentous phages suggest that this system may be widely involved in filamentous phage integration into the bacterial genome.
Similar to other filamentous phages, CTXφ is secreted from the bacterial cell without lysis of the cell. However, the genome of CTXφ differs from that of filamentous phages from E. coli in that it lacks the gene for a putative bacteriophage export or secretin protein (a homolog of Ff gIV). Instead, CTXφ uses the EspD secretin, part of a type II extracellular protein secretion system. The EspD protein, an outer membrane component, appears to be the only component of the type II secretion system required for secretion of CTXφ.21 The type II secretion system is the pathway used for export of CT, hemagglutinin-protease, chitinase and other proteins out of the V. cholerae cell.21,22
Origin of CTXφ
A number of features of the CTXφ genome suggest that the ctxAB genes were acquired by a CTXφ progenitor (preCTXφ) from an unknown source.2,23 The percent GC content of the ctxAB genes (38%), is different from that of the rest of the CTXφ genome (40%). The production of CT is promoted by two promoters: the PrstA promoter, which is ∼5 kb upstream of ctxA is required for CTXφ virion synthesis but can extend into ctxAB, and PctxAB, the promoter found immediately upstream of ctxAB. Furthermore, V. cholerae strains carrying CTX prophage sequences that do not encode the ctxAB genes have been identified.23 These precursor CTX (preCTX) prophages also produce the replicative form (RF) of their genomes, which are very similar to the RF of CTXφ genome except that they do not carry the ctxAB genes. Molecular analysis of these preCTX plasmids revealed that they also lacked the upstream control region normally found 5′ of ctxA, as well as the ctxAB promoter region and coding sequences. These observations suggest that a preCTX simultaneously acquired the ctxAB genes and their regulatory sequences. It has been proposed that in the event that led to the acquisition of ctxAB genes by a preCTXφ, integration of the preCTXφ genome occurred next to the ctxAB genes carried by an unknown organism, and because CTX virion production occurs by rolling circle replication from the chromosome, ctxAB genes and the regulatory sequences were acquired from the unknown source by imprecise prophage replication.11,23
The acquisition of ctxAB genes by the pre-CTXφ has increased its fitness since CTXφ confers a selective advantage to its V. cholerae host by producing CT; the action of the enterotoxin results in profuse diarrhea, promoting dissemination of the bacterium and hence the phage. CTXφ forms stable lysogens in V. cholerae, and replication of the phage does not cause V. cholerae cell lysis; therefore, CTXφ genome can be both vertically and horizontally transferred.2,11
Diversity of CTXφ
The sequence of rstR gene of CTXφ can vary considerably among El Tor, classical and O139 isolates, although rstA and rstB show a high level of sequence conservation among different V. cholerae isolates. The rstR sequence variation has been used to classify CTXφ. For example, CTXφ from classical isolates carry rstRclass and are designated CTXclassφ; whereas that from El Tor isolates usually carry rstRET, and are designated CTXETφ. V. cholerae O139 carry either CTXETφ or a CTXφ with a distinct rstR designated rstRCalc and the phage is designated CTXCalcφ.24,25 V. cholerae O139 strains may also carry both CTXETφ or CTXCalcφ prophages simulataneously.26 In addition several novel rstR sequences have been identified in V. cholerae non-O1/non-O139 isolates, and hence there may be diverse CTXφ carried by these environmental V. cholerae strains.11 Molecular evolutionary analysis of CTXφs derived from a variety of V. cholerae natural isolates have been done based on sequence analysis of the rstR and the gIIICTX. These data taken together indicate distinct lineages among CTXφ, independent acquisition of CTXφ by different V. cholerae isolates, and acquisition of novel CTXφ23,27 by classical, El Tor and O139 strains.
Chromosomal organization of CTX prophages
The arrangement of CTX prophage on the genome of V. cholerae varies depending on (1) whether it is integrated at a single site or two sites, and (2) the biotype or serogroup of the strain. Among El Tor strains, the CTX prophage is located on the large chromosome (chromosome I) ∼842 bp downstream from a tandemly duplicated DNA sequence referred to as toxin linked cryptic (TLC) (Fig. 1), which was later shown to be a satellite phage genome.14,28 CTX prophage in the chromosome of El Tor strains is also flanked by a 2.7 kb repetitive sequence originally referred to as RS1 element, which was also later found to be the genome of a satellite phage, RS1φ.29,30 Many different CTX and RS1 arrangements have been observed among V. cholerae O1 El Tor biotype strains.28 For example, El Tor strain E7946 isolated in Bahrain in 1978 was shown to contain an RS1-CTXET-RS1-CTXET-RS1 arrangement, whereas strain C6709 isolated in Peru in 1991, contained a CTXET-RS1 arrangement.28,31 The production of infectious CTXφ particles requires the presence of either a CTX-CTX array comprising two CTX prophages or a CTX-RS1 array, and the DNA in CTXφ is a hybrid sequence that is derived from these elements.32 In V. cholerae classical biotype strains, CTX prophage is located on both chromosomes and without an RS1 element.33 On chromosome I, CTX is present in the intergenic region between TLC and repeat in toxin (RTX) gene clusters, often designated as the El Tor site, and on chromosome II in the intergenic region between the genes designated VCA0569 and VCA0570, defined as the classical site.33 Within certain classical strains (O395, GP12 and C33), CTXclass is present as an array of truncated, fused prophages at the El Tor integration site on chromosome I whereas at the classical integration site on chromosome II, a solitary CTXclass prophage is present.33 In several other classical strains (CA401, C1, C14, C21 and C34), a single CTXclass prophage is present in the El Tor site whereas at the classical site on Chromosome II, a truncated CTXclass array is found. The classical prophage arrangements cannot generate pCTXclass and CTXclassφ.33
A number of CTX prophage arrangements have been described within V. cholerae O139 serogroup strains.24,25,32-35 Whereas strain SG24 contains a CTXET-RS1-CTXET array, strains AS197 and AS209 contain a RS1-CTXET-CTXCalc-CTXCalc array. CTX prophage arrangement appears to be dynamic and continually giving rise to new variant strains. For example, genomic analysis of a V. cholerae strain B-33 isolated in Mozambique in 2004 revealed the presence of CTXclass with a CTXclass-CTXclass array at the classical insertion site on chromosome II.36
Distribution of CTXφ
Although most toxigenic V. cholerae strains belonging to the O1 or O139 serogroups carry CTX prophages, the distribution of CTX is sporadic among non-O1/non-O139 serogroup strains.5,37-39 The limited distribution of CTXφ among non-O1/non-O139 strains is possibly due to the limited distribution of TCP, which is the host receptor for CTXφ. TCP is encoded on the TCP pathogenicity island, and the bacterium must acquire this island before it can uptake CTXφ.2 A range of V. mimicus isolates are also known to carry CTXφ and most of these isolates encode TCP, indicating a similar method of phage infection. There have been preliminary reports on the presence of homologs of CTX prophage as well as the CTXφ receptor TCP in the genomic sequence of V. fischeri. However, it is not known whether the TCP sequence is functional in this species. The CTX prophage from V. fischeri has been found to lack the ctxAB genes.11 Nevertheless, the presence of CTXφ and TCP island or their homologs in Vibrio species other than V. cholerae may represent additional reservoirs of potential V. cholerae virulence genes.11,40
Other Filamentous Phages of V. cholerae
Besides CTXφ, at least nine other filamentous phages or satellite filamentous phages that lack genes required for phage morphogenesis have been described. These include the RS1 and TLC satellite phages, and KSF-1, VGJ, VSK, VSKK, 493, fs1 and fs2 phages. Some of these phages are also involved in horizontal gene transfer (Table 1). Filamentous phages such as KSF-1φ and VGJφ can supply capsid, assembly and packaging proteins for RS1 and possible other satellite phage genomes and thus facilitate the horizontal transfer of the satellite phage genomes. With increasing numbers of different V. cholerae genomes being sequenced, the number of filamentous phage genome sequences and identification of novel prophages are likely to increase dramatically.
Table 1. Phages associated with horizontal gene transfer among V. cholerae.
| Designation | Genome size (bp) | Receptor | Description and function |
|---|---|---|---|
| CTXφ |
6,900 |
TCP |
Filamentous bacteriophage that encodes cholera toxin. CTX phage can integrate in the chromosome of V. cholerae forming stable lysogens. |
| RS1φ |
3,000 |
TCP/MSHA |
A satellite phage that uses CTXφ-encoded proteins to form RS1 phage particles. RS1 carries the gene for an anti-repressor protein RstC that influences CTXφ replication and transmission. |
| KSF-1φ |
7,107 |
MSHA |
A filamentous phage capable of packaging RS1 and possibly other heterologous DNA of V. cholerae. |
| VGJφ |
7,542 |
MSHA |
VGJφ is able to integrate its genome into the same chromosomal attB site as CTXφ, entering into a lysogenic state. |
| fs-2φ |
8,651 |
MSHA |
Filamentous phage fs-2φ forms turbid plaques on V. cholerae O139 and O1 El Tor biotype strains. A 715 nucleotide fragment located in the large intergenic region of fs-2 was highly homologous to a part of region RS2 of CTXφ. This phage acts as a helper to produce satellite phage TLCφ particles. |
| TLCφ |
5,364 |
MSHA |
The TLC satellite phage genome carries a sequence similar to the dif recombination sequence which functions in chromosome dimer resolution during cell division. Lysogeny by TLC phage generates a functional dif site in dif defective strains. The dif site also overlaps with the attB site, which is essential for stable integration of CTXφ genome. |
| CP-T1 | ~43,500 | O-antigen | A generalized transducing phage of V. cholerae, which can laterally transfer chromosomal segments among V. cholerae strains. |
RS1 satellite phage
The satellite filamentous phage RS1φ encodes four open reading frames (ORF) namely rstR, rstA, rstB and rstC (Fig. 1). The first three of these are homologous to the corresponding genes of the RS2 region of CTXφ, whereas the additional ORF rstC encodes an antirepressor protein RstC which causes the RstR repressor to aggregate and prevent binding to the PrstA promoter.29,30 The RstC protein promotes transcription of CTXφ genes required for phage production, supporting the transmission of both RS1φ and CTXφ.30 On the other hand, RS1 being a satellite filamentous phage lacks genes required for phage morphogenesis and depends on CTXφ for the capsid proteins, packaging and transmission.29 Upon infection of recipient cells the RS1φ genome, similar to CTXφ, integrates site-specifically into the V. cholerae chromosome.29 The interactions and cooperation of RS1 and CTX prophages thus appear to promote the propagation of both these phage genomes.
The TLC satellite phage
In toxigenic V. cholerae, the CTX prophage is associated with a tandemly duplicated DNA sequence originally referred to as toxin linked cryptic (TLC) which is located upstream of the CTX prophage (Fig. 1). In most strains, TLC-related sequences also exist extrachromosomally as plasmids (pTLC). Each copy of chromosomally integrated TLC element was found to comprise of five ORFs. For example, in strain AL 33457, the two copies of TLC comprised of ORFs, VC1466 to VC1470 and VC1472 to VC1476 respectively, with VC1471 located in between the two copies of the element.14 The largest ORF in TLC encodes a protein named Cri replicase which is similar to the replication initiation protein (pII) of E. coli F-specific filamentous phages. The TLC also encodes TlcR, a protein that displays sequence similarity to RstR, the repressor controlling lysogeny of the filamentous CTXφ. It has been recently confirmed that the TLC is the genome of a satellite filamentous phage which uses morphogenesis proteins of another filamentous phage known as fs2φ to form infectious TLCφ particles.14
Interactions of V. cholerae with TLCφ appears to be important in the evolutionary biology of V. cholerae. The TLC phage genome carries a sequence similar to the dif recombination sequence which functions in chromosome dimer resolution during cell division. Dif sequences are required for XerC/XerD-mediated resolution of chromosome dimers in E. coli and V. cholerae but are also exploited by various filamentous phages for integration of their genomes into the host chromosome.41-46 The dif site (Fig. 1) overlaps with the attB sequence which is utilized by CTXφ and RS1φ for their chromosomal integration though XerC/XerD-mediated recombination with the corresponding dif/attP site formed by annealing of ssDNA derived from the phage genomes.45,46
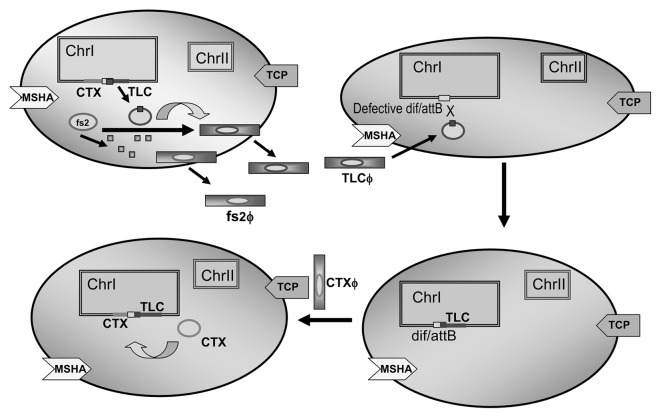
Bacterial cells defective in the dimer resolution often show a pathology of cell filamentation.41,42 It has been shown that TLCφ-related transducing phages can, upon chromosomal integration of their genome generate a functional dif sequence and correct aberrant filamentous morphology in cells of naturally occurring, TLC-negative strains that apparently exhibit defective dif/XerC/XerD-mediated chromosome dimer resolution. Lysogeny by such TLC phages also generates a chromosomal attB site which is essential for stable integration of CTXφ and conversion of V. cholerae to a toxigenic form (Fig. 2). Thus, TLCφ plays a critical role in improving not only the chromosome replication efficiency of V. cholerae but also its emergence as a human pathogen.
Figure 2. Role of TLCφ in CTXφ mediated toxigenic conversion of V. cholerae. Infectious TLCφ particles are produced from TLC satellite phage genome by using morphogenesis proteins of the helper phage fs2φ. The TLCφ infects recipient strains using MSHA pilus as the receptor. Upon chromosomal integration of the TLCφ genome, a functional dif/attB sequence is generated in TLC-negative strains that apparently exhibit defective dif/XerC/XerD-mediated chromosome dimer resolution. Lysogeny by such TLC phages also generates a chromosomal attB site which is essential for stable integration of CTXφ and conversion of V. cholerae to a toxigenic form. The CTXφ uses TCP as its receptor on the bacterial cell surface.
KSF-1φ
Besides CTXφ, the first filamentous phage found to act as a helper for the RS1 satellite phage was KSF-1φ, which supports the production of infectious RS1φ particles independently of CTXφ by providing the gene products required for capsid, assembly and packaging of RS1φ particles.47 KSF-1φ is a viable infectious phage that integrates into the host genome and contains an 18-bp att-like sequence. The phage particles have a length of 1.2 µm, and a width of 7 nm, and the particles contain a single-stranded DNA genome. The genome was found to comprise 7,107-bp with a GC content of 44% encoding 14 ORFs.48 The host cell receptor for KSF-1φ is the mannose sensitive hemagglutinin (MSHA) pilus. Since the receptor for KSF-1φ is the MSHA pilus, RS1φ particles produced by this phage can infect MSHA positive strains, and is independent of TCP for infecting recipient bacterial hosts. Thus, KSF-1φ is capable of facilitating the transfer of RS1 to strains that do not express toxin coregulated pilus. Given that RS1 can enhance coproduction of CTXφ particles, KSF-1φ-mediated dissemination of RS1 may indirectly promote the spread of toxin genes among V. cholerae strains.47
VGJφ
The filamentous phage VGJφ infects O1 and O139 strains, and integrate at the same chromosomal site as CTXφ.49 The genome of VGJφ is 7,542 bp with a GC content of 43% encoding 10 ORFs and is known to use the type IV pilus mannose-sensitive hemagglutinin (MSHA) as a receptor for infecting host cells.50 The genome sequence of VGJφ revealed the presence of two sites homologous to functional att sequences from other filamentous phages. These include V. cholerae phages CTXφ and VSK, and V. parahaemolyticus phage f237. VGJφ integrates through its own attP sequence into the same chromosomal attB site in V. cholerae where CTXφ integrates, and the integration is likely mediated by the XerCD recombinases of the host bacterium.49
The VGJφ also provides functions required for production and transmission of both CTXφ and RS1φ.50 In a novel type of specialized transduction, the site-specific co-integration of VGJφ and CTXφ (or RS1φ) genome results in single hybrid molecules that generate ssDNA hybrid genomes that are packaged into hybrid phages (VGJφ/CTXφ hybrid or VGJφ/RS1φ hybrid).50 Since the host cell receptor for VGJφ is the MSHA pilus, VGJφ is capable of transferring CTXφ to V. cholerae cells by a TCP-independent mechanism. Given that MSHA is present in a wide variety of V. cholerae strains and is presumed to express in the environment, diverse filamentous phages using this receptor are likely to contribute significantly to V. cholerae evolution.
Other MSHA dependent filamentous vibriophages
MSHA is used as the host cell receptor by at least five other filamentous phages, namely, VSK, 493, VSKK, fs1 and fs2.51-55 Phages fs1 and fs2 were initially isolated from certain V. cholerae O139 strains. These phages were later found to infect both V. cholerae O1 and O139 strains and the phage genome could also integrate into the host bacterial genome. Phage fs1 was also isolated from a number of clinical V. cholerae O1 isolates.56 This phage has a 6,340-bp ssDNA genome that encodes 15 ORFs, whereas the fs2φ genome size is 8,651 bp, containing nine ORFs and a large stem loop region.52,53 Almost all V. cholerae O139 and O1 El Tor biotype strains tested were sensitive to fs2φ, whereas most O1 classical biotype strains and non O1/nonO139 strains were found to be resistant.53 A 715 nucleotide fragment located in the large intergenic region of fs-2φ was highly homologous to a part of region RS2 of CTXφ. Recently, a phage similar to fs2φ has been shown to act as a helper for the TLC satellite phage by encoding functions leading to the morphogenesis of infectious TLCφ particles.14
The VSKφ was also isolated from a V. cholerae O139 strain. This phage has a genome of 6,880-bp with a GC content of 43%, and the phage genome integrates into the genome of the host bacterium.51 VSKφ genome has high sequence similarity to VGJφ (83%) and fs1φ (87%).51 Phage 493 was isolated from a 1994 isolate of V. cholerae O139, and was shown to inhibit pre-O139 El Tor biotype V. cholerae strains but not post-O139 El Tor strains in plaque assays.54 The emergence and decline of V. cholerae O139 in India and Bangladesh were suggested to have been influenced by the susceptibility and resistance of El Tor strains to 493φ.55 The 493φ has an approximate genome size of 9,300 bp with a 2,300-bp stem-loop structure, which may represent a transposable element.
In summary, most of the known filamentous phages of V. cholerae have genomes of a similar size (range 6 kb to 9.3 kb) and encode a similar number of ORFs (range 7 to 15). Distinct replication and morphogenesis modules are present in most of these phages. Most of the filamentous vibriophages studied integrate into the host bacterial genome, and probably use the XerD and XerC mechanism of integration since most of these phages contain att-like sites similar to that of CTXφ. The TCP pathogenicity island which has a significantly larger size (∼41 kb), and distinct from the known V. cholerae filamentous phage genomes was also reported to be the genome of a filamentous phage called VPIφ.57 However, a lack of reasonable similarity in sequence of the TCP island (alternatively known as VPI58 for Vibrio cholerae pathogenicity island) to the morphogenesis genes of canonical filamentous phages was one of the reasons for skepticism regarding the existence of the VPIφ. Subsequently it was shown that the initial study was incorrect and that VPI, is not itself a phage genome.59 However, the TCP island, which includes putative integrase and transposase genes and is flanked by att-like sequences, clearly resembles a horizontally acquired element.60
Possible role of transducing phages in horizontal gene transfer
Besides the TCP pathogenicity island, the V. cholerae genome is known to carry a number of other gene clusters with structural features, that are consistent with the ability of these genomic regions to move horizontally between strains.8 These islands include, VPI-2, encoding genes for neuraminidase (nanH) and amino sugar metabolism, and the Vibrio seventh pandemic islands VSP-1 and VSP-2.11,61 The VPI-2 is a 57 kb region that is required for growth of V. cholerae on sialic acid as a sole carbon source. In the mucus-rich environment of the gut, where sialic acid availability is extensive, the capacity to utilize sialic acid as a carbon and energy source by V. cholerae is believed to confer an advantage. V. cholerae non-O1/O139 pathogenic strains also contain VPI-2, which in addition to sialic acid catabolism genes also encodes a type 3 secretion system in these strains. Vibrio seventh pandemic islands VSP-1 and VSP-2 are unique to seventh pandemic El Tor and O139 isolates.61 VSP-1 spans a 16 kb region with a GC content of 40 %, in contrast to 47 % for the entire V. cholerae genome. The VSP-2 encompasses a 7.5 kb region with a GC content of 41%, and was found to show homology to a 43.4 kb genomic island in V. vulnificus.62 The precise functions of all genes in VSP-1 and VSP-2 have not been defined, but the presence of these islands in pandemic strains suggest their role in evolutionary fitness and spread of the seventh pandemic clone.
It has been suggested that the TCP island or VPI can excise from the chromosome and form circular DNA,63 and thus provides support to the possibility of horizontal transfer of this region. Similarly, VPI-2 and the two VSP islands were also shown to excise to form extrachromosomal products.64 Each of the pathogenicity islands VPI-2 and VSP-2 encodes a P4-like integrase adjacent to a tRNA locus, and the P4-like integrase was required for their excision.64,65 It seems possible that these pathogenicity islands are transmitted by phage transduction, and the excision from the chromosome is likely a first step in its horizontal transfer from cell to cell. For example, inter strain transfer of VPI mediated by a generalized transducing phage CP-T1 has been documented.66 Further studies directed toward identifying possible phages involved in the transfer of these pathogenicity islands will provide more insights into the contribution of phages in V. cholerae evolution.
Phage Predation and Selection of V. cholerae Strains
A distinctive epidemiological feature of cholera is its appearance with seasonal regularity in endemic areas, such as the Ganges Delta region of Bangladesh and India. These cholera epidemics are also known to be self-limiting, since the epidemics subside after reaching a peak, even without any active intervention. Among other factors, phages that can kill V. cholerae (cholera phages) have been demonstrated to play a significant role in modulating the course of epidemics.3,9,10 The dynamics of the interactions between V. cholerae and lytic phages was studied during cholera epidemics in Dhaka, Bangladesh between 2001 and 2004. These studies showed that the number of cholera patients increased whenever the number of lytic vibriophages in water decreased. Similarly, cholera epidemics tended to end concurrent with large increases in the concentration of these viruses in the water.
The incidence of phage excretion in stools of cholera patients during the entire course of an epidemic was also monitored. The buildup to the phage peak in the environment coincided with increasing excretion of the same phage in the stools of cholera patients.9 Further studies using animal models showed that the infectious dose (ID50) of V. cholerae cells in stools of cholera patients as well as that of laboratory-grown cells was higher in the presence of a phage, and ∼10-fold-more cells of V. cholerae would be required to cause a productive infection under the conditions in which cholera patients excrete V. cholerae together with phages in their stools.67 The increase of the required infectious dose due to the deleterious effect of co-ingesting phage with V. cholerae would therefore lead to a reduced number of patients. Thus phages antagonize the transmissibility of cholera during the late stage of a cholera epidemic,9,68 eventually leading to the collapse of the epidemic. Therefore, cholera phages in the environment, and their amplification in cholera patients remarkably influence the epidemiology of cholera, due mainly to phage mediated elimination of the causative epidemic strain. However, since the same V. cholerae strain often reemerges and causes a subsequent epidemic during the next cholera season, it is obvious that a proportion of the epidemic V. cholerae cells survive the phage attack. Therefore, factors or genetic changes which enhance the ability of the bacteria to resist phage predation may also be a strong driving force in the evolution of V. cholerae.
The resistance to phage may be manifested by the metabolic status of the V. cholerae cells, inadequate expression of phage receptors or other mutations that affect phage bacterial interactions. Under laboratory conditions, co-culture of a phage and V. cholerae or dilutions of phage-positive cholera stools has been shown to cause emergence of phage-resistant derivatives of the bacteria.67 Frequently these resistant derivatives were found to have lost their O1 antigen which often acts as the receptor for these phages. However, challenge studies in mice with a mixture of V. cholerae and phage suggested that the intestinal environment did not favor the emergence of phage-resistant derivatives that lost the O1 antigen.67 These results suggest that the outcome of phage bacterial interactions are more complex in nature and involves a multitude of factors including environmental factors, the lytic phages, and the role of the human host.69
Changing prevalence of vibriophages in the environment
The environmental surveillance system to monitor phages and V. cholerae in the aquatic environment in Bangladesh yielded various phage isolates and data on phage prevalence. Different phage strains3,7,15 (JSF1 through JSF16) which interact with V. cholerae have been identified and partially characterized (Table 2). A lytic vibriophage ICP1 was consistently prevalent in stools of cholera patients in Bangladesh.70,71 Genomic sequence analysis indicated that ICP1 is in fact identical to JSF4 and is closely related to JSF11 (W. Robins, S. Faruque and J. Mekalanos, unpublished results). Studies were also conducted to identify bacterial factors which mediate resistance to phage susceptibility, and thus provide a mechanism to survive predation by phages. As mentioned above, often phage resistant strains were found to have lost their O-antigen. Incorporation of mutations in the cyaA or crp genes encoding adenylate cyclase or cyclic AMP (cAMP) receptor protein (CRP) respectively, into V. cholerae O1 strains was also found to cause alterations in their phage susceptibility patterns, and the susceptibility correlated with the ability of the bacteria to adsorb these phages. These results suggested that cAMP-CRP-mediated downregulation of phage adsorption may contribute to a mechanism for the V. cholerae strains to survive predation by phages in the environment.15
Table 2. Lytic vibriophages isolated from surface water and cholera patients in Bangladesh3,7,15.
| Phage designation | Primary host strains | Alternative host strains | Plaque type | Isolation of lysogens |
|---|---|---|---|---|
| JSF-1 |
V. cholerae O1 |
Not found |
Clear |
- |
| JSF-2 |
V. cholerae O1 |
Not found |
Turbid |
+ |
| JSF-3 |
V. cholerae O139 |
Not found |
Clear |
+ |
| JSF-4 |
V. cholerae O1 |
Not found |
Clear |
+ |
| JSF-5 |
V. cholerae O1 |
Not found |
Clear |
- |
| JSF-6 |
V. cholerae O1 |
V. cholerae non-O1 non-O139 |
Clear |
- |
| JSF-7 |
V. cholerae O1 |
V. cholerae O141 strain V50; V. cholerae O139 strain AI1853 |
Clear on O1 strain; Clear/ turbid on non-O1 strains |
+ |
| JSF-8 |
V. cholerae O1 |
V. cholerae non-O1 non-O139 strains 3565, 3548; V. mimicus strains 957V1621, 778V1349, and 1016V1721 |
Clear on O1 strain; Clear/turbid on non-O1 strains |
+ |
| JSF-9 |
V. cholerae O1 |
V. cholerae O141 strain V50; non-O1 strains 79, 3565, 3548; V. mimicus strains 957V1621, 778V1349, and 1016V1721 |
Clear |
- |
| JSF10 |
V. cholerae O1 |
V. cholerae O139 strain Arg-3 V. cholerae O141 strains V46 and V47 |
Clear |
- |
| JSF-11 |
V. cholerae O1 |
Not found |
Clear |
- |
| JSF12 |
V. cholerae O1 |
V. cholerae non-O1 strains 79; V. mimicus strains 957V1621, 1016V1721 |
Clear |
- |
| JSF-13 |
V. cholerae O1 |
Not found |
Clear |
- |
| JSF-14 |
V. cholerae O1 |
Not found |
Clear |
- |
| JSF-15 |
V. cholerae O1 |
V. cholerae O141 strain V50; non-O1 strain 79, V. mimicus strains 957V1621, and 778V1349 |
Clear |
- |
| JSF-16 | V. cholerae O1 | V. cholerae O141 strain V50 | Clear/turbid | + |
Fluctuation in the prevalence of the different predatory phages have also been observed (unpublished data), with a temporal change in the most prevalent phage type (Table 3) which is a factor in the collapse of epidemics by phage predation.3 The significance of this change is not clear. Nevertheless, occasional change in the predominant phage appears to maintain susceptibility of the epidemic strain to the most prevalent phage responsible for ending epidemics in a self-limiting manner.
Table 3. Predominant phages isolated from surface water and cholera patients in Bangladesh during 2002–2011.
| Year | Abundant phages | Predominant phage |
|---|---|---|
| 2002–20043 |
JSF1 through JSF6 |
JSF4 |
| 2005 |
JSF1 through JSF9 |
JSF4 |
| 2006 |
JSF1, JSF4, JSF6 through JSF9 |
JSF4 |
| 2007 |
JSF1 through JSF11 |
JSF4/JSF11 |
| 2008 |
JSF1, JSF4 through JSF11 |
JSF11 |
| 2009 |
JSF1, JSF4 through JSF11 |
JSF11 |
| 2010 |
JSF1, JSF4 through JSF11 |
JSF11 |
| 2011 | JSF1, JSF4 through JSF16 | JSF11 |
Conclusion
Although there are multiple other factors which drive the evolution of V. cholerae, phages have been conclusively shown to contribute to horizontal gene transfer among V. cholerae strains, and in selection of particular clones. Complete genomic sequencing of a number of V. cholerae strains is beginning to reveal changes that the organism has undergone and that have been linked to its emergence as a pathogen. Some of these genetic changes define important virulence altering traits. Pathogenic strains typically carry genes for different virulence factors and the lipopolysaccharide O1 or O139 antigens that are typically absent in environmental strains.5,8 Thus, while there are over 200 serogroups of V. cholerae, only two serogroups (O1 and O139) are associated with epidemic and endemic disease. The O139 serogroup is a derivative of the highly successful CTX+ TCP+ O1 El Tor “7th pandemic clone,”31 and hence carry the CTX prophage and TCP pathogenicity island, but many O1 environmental strains do not. Both classical and El Tor biotypes of V. cholerae O1 carry CTX and TCP; so the explanation for the evolutionary success of the seventh pandemic clone over the preexisting sixth pandemic strain remains largely a mystery. Recent studies are beginning to suggest possible role of phages in this process as well.7
Similar to CTX prophage, the CTXφ receptor TCP is limited to subsets of strains within three species, V. cholerae, V. mimicus and V. fischeri, thus explaining the limited distribution of the CTX prophage. The bacterial cell surface receptor for VSK, VJG, KSF-1 and fs2 phages is MSHA, a type IV pilus that is widely present among Vibrio species, indicating the potential broad host range of these filamentous phages and their potential role in horizontal gene transfer across species boundaries. However, emergence of pathogenic strains of V. cholerae with endemic and pandemic potential has been exclusively linked to the acquisition of the TCP and CTX genes by only a limited spectrum of V. cholerae strains. This fact suggests that the precursors of these pandemic clones probably display traits that are lacking in typical environmentally adapted V. cholerae regardless of their serogroup. This is not to say that new pathogenic clones do not emerge from environmental V. cholerae. But the evolution of environmental strains to typical pathogenic strains would require more widespread gene transfer events than shown to occur through the known phages until now. While conjugative plasmids and a single generalized transducing phage for V. cholerae namely CPT-1 are known to exist,70 other studies are beginning to suggest more extensive role of possible additional vibriophages in the horizontal transfer of chromosomal segments as well as in selection of different V. cholerae clones. V. cholerae has been reported to become naturally competent to uptake exogenous DNA, while growing on a chitin substrate.71 More recently, it has been suggested that free DNA released in the aquatic environment by phage mediated lysis of V. cholerae can be harnessed and assimilated by surviving V. cholerae strains.72 Thus, phages contribute to bacterial evolution in numerous ways and the full potential of these mechanisms in the evolution of V. cholerae as a pathogen is yet to be appreciated.
Acknowledgments
Research in the laboratories of S.M.F. and J.J.M. were funded in part by National Institutes of Health grants 2RO1-GM068851, and AI070963-01A1 under different sub-agreements between the Harvard Medical School and the ICDDR,B. The ICDDR,B is supported by countries and agencies that share its concern for the health problems of developing countries.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/22351
References
- 1.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–4. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 3.Faruque SM, Naser IB, Islam MJ, Faruque ASG, Ghosh AN, Nair GB, et al. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci U S A. 2005;102:1702–7. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaper JB, Morris JG, Jr., Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–14. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddique AK, Nair GB, Alam M, Sack DA, Huq A, Nizam A, et al. El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol Infect. 2009;138:347–52. doi: 10.1017/S0950268809990550. [DOI] [PubMed] [Google Scholar]
- 7.Zahid MSH, Waise Z, Kamruzzaman M, Ghosh AN, Nair GB, Khairul Bashar SA, et al. An experimental study of phage mediated bactericidal selection & emergence of the El Tor Vibrio cholerae. Indian J Med Res. 2011;133:218–24. [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque SM, Mekalanos JJ. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 2003;11:505–10. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Faruque SM, Islam MJ, Ahmad QS, Faruque ASG, Sack DA, Nair GB, et al. Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage. Proc Natl Acad Sci U S A. 2005;102:6119–24. doi: 10.1073/pnas.0502069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen MA, Faruque SM, Mekalanos JJ, Levin BR. Modeling the role of bacteriophage in the control of cholera outbreaks. Proc Natl Acad Sci U S A. 2006;103:4652–7. doi: 10.1073/pnas.0600166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd F. Filamentous phages of Vibrio cholerae In: Faruque SM, Nair GB ed. Vibrio cholerae: Genomics and Molecular Biology. Horizon Scientific Press, Ltd. UK, 2008:pp 49-66. [Google Scholar]
- 12.Sulakvelidze A, Alavidze Z, Morris JG., Jr Bacteriophage therapy. Antimicrob Agents Chemother. 2001;45:649–59. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hankin EH. L’action bactericide des eaux de la Jumna et du Gange sur le vibrion du cholera. Ann Inst Pasteur (Paris) 1896;10:511. [Google Scholar]
- 14.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010;467:982–5. doi: 10.1038/nature09469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahid MSH, Waise TM, Kamruzzaman M, Ghosh AN, Nair GB, Mekalanos JJ, et al. The cyclic AMP (cAMP)-cAMP receptor protein signaling system mediates resistance of Vibrio cholerae O1 strains to multiple environmental bacteriophages. Appl Environ Microbiol. 2010;76:4233–40. doi: 10.1128/AEM.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldor MK, Rubin EJ, Pearson GD, Kimsey H, Mekalanos JJ. Regulation, replication, and integration functions of the Vibrio cholerae CTXphi are encoded by region RS2. Mol Microbiol. 1997;24:917–26. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 17.Russel M. Moving through the membrane with filamentous phages. Trends Microbiol. 1995;3:223–8. doi: 10.1016/S0966-842X(00)88929-5. [DOI] [PubMed] [Google Scholar]
- 18.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417:656–9. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 19.Heilpern AJ, Waldor MK. pIIICTX, a predicted CTXphi minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J Bacteriol. 2003;185:1037–44. doi: 10.1128/JB.185.3.1037-1044.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol Cell. 2005;19:559–66. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Davis BM, Lawson EH, Sandkvist M, Ali A, Sozhamannan S, Waldor MK. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science. 2000;288:333–5. doi: 10.1126/science.288.5464.333. [DOI] [PubMed] [Google Scholar]
- 22.Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol Lett. 2006;255:175–86. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 23.Boyd EF, Heilpern AJ, Waldor MK. Molecular analyses of a putative CTXphi precursor and evidence for independent acquisition of distinct CTX(φ)s by toxigenic Vibrio cholerae. J Bacteriol. 2000;182:5530–8. doi: 10.1128/JB.182.19.5530-5538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimsey HH, Nair GB, Ghosh A, Waldor MK. Diverse CTXphis and evolution of new pathogenic Vibrio cholerae. Lancet. 1998;352:457–8. doi: 10.1016/S0140-6736(05)79193-5. [DOI] [PubMed] [Google Scholar]
- 25.Davis BM, Kimsey HH, Chang W, Waldor MK. The Vibrio cholerae O139 Calcutta bacteriophage CTXphi is infectious and encodes a novel repressor. J Bacteriol. 1999;181:6779–87. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faruque SM, Chowdhury N, Kamruzzaman M, Ahmad QS, Faruque AS, Salam MA, et al. Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerg Infect Dis. 2003;9:1116–22. doi: 10.3201/eid0909.020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya T, Chatterjee S, Maiti D, Bhadra RK, Takeda Y, Nair GB, et al. Molecular analysis of the rstR and orfU genes of the CTX prophages integrated in the small chromosomes of environmental Vibrio cholerae non-O1, non-O139 strains. Environ Microbiol. 2006;8:526–634. doi: 10.1111/j.1462-2920.2005.00932.x. [DOI] [PubMed] [Google Scholar]
- 28.Mekalanos JJ. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–63. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 29.Faruque SM, Asadulghani, Kamruzzaman M, Nandi RK, Ghosh AN, Nair GB, et al. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXphi. Infect Immun. 2002;70:163–70. doi: 10.1128/IAI.70.1.163-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis BM, Kimsey HH, Kane AV, Waldor MK. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 2002;21:4240–9. doi: 10.1093/emboj/cdf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldor MK, Mekalanos JJ. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J Infect Dis. 1994;170:278–83. doi: 10.1093/infdis/170.2.278. [DOI] [PubMed] [Google Scholar]
- 32.Davis BM, Waldor MK. CTXphi contains a hybrid genome derived from tandemly integrated elements. Proc Natl Acad Sci U S A. 2000;97:8572–7. doi: 10.1073/pnas.140109997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis BM, Moyer KE, Boyd EF, Waldor MK. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J Bacteriol. 2000;182:6992–8. doi: 10.1128/JB.182.24.6992-6998.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhadra RK, Roychoudhury S, Banerjee RK, Kar S, Majumdar R, Sengupta S, et al. Cholera toxin (CTX) genetic element in Vibrio cholerae O139. Microbiology. 1995;141:1977–83. doi: 10.1099/13500872-141-8-1977. [DOI] [PubMed] [Google Scholar]
- 35.Khetawat G, Bhadra RK, Nandi S, Das J. Resurgent Vibrio cholerae O139: rearrangement of cholera toxin genetic elements and amplification of rrn operon. Infect Immun. 1999;67:148–54. doi: 10.1128/iai.67.1.148-154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faruque SM, Tam VC, Chowdhury N, Diraphat P, Dziejman M, Heidelberg JF, et al. Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci U S A. 2007;104:5151–6. doi: 10.1073/pnas.0700365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faruque SM, Asadulghani, Saha MN, Alim AR, Albert MJ, Islam KM, et al. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXPhi: molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66:5819–25. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faruque SM, Sack DA, Sack RB, Colwell RR, Takeda Y, Nair GB. Emergence and evolution of Vibrio cholerae O139. Proc Natl Acad Sci U S A. 2003;100:1304–9. doi: 10.1073/pnas.0337468100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Shea YA, Reen FJ, Quirke AM, Boyd EF. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J Clin Microbiol. 2004;42:4657–71. doi: 10.1128/JCM.42.10.4657-4671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyd EF, Moyer KE, Shi L, Waldor MK. Infectious CTXPhi and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun. 2000;68:1507–13. doi: 10.1128/IAI.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417:656–9. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 42.Blakely G, May G, McCulloch R, Arciszewska LK, Burke M, Lovett ST, et al. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–61. doi: 10.1016/0092-8674(93)80076-Q. [DOI] [PubMed] [Google Scholar]
- 43.Iida T, Makino K, Nasu H, Yokoyama K, Tagomori K, Hattori A, et al. Filamentous bacteriophages of vibrios are integrated into the dif-like site of the host chromosome. J Bacteriol. 2002;184:4933–5. doi: 10.1128/JB.184.17.4933-4935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLeod SM, Waldor MK. Characterization of XerC- and XerD-dependent CTX phage integration in Vibrio cholerae. Mol Microbiol. 2004;54:935–47. doi: 10.1111/j.1365-2958.2004.04309.x. [DOI] [PubMed] [Google Scholar]
- 45.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol Cell. 2005;19:559–66. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Das B, Bischerour J, Val ME, Barre FX. Molecular keys of the tropism of integration of the cholera toxin phage. Proc Natl Acad Sci U S A. 2010;107:4377–82. doi: 10.1073/pnas.0910212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faruque SM, Kamruzzaman M, Asadulghani, Sack DA, Mekalanos JJ, Nair GB. CTXφ-independent production of the RS1 satellite phage by Vibrio cholerae. Proc Natl Acad Sci USA. 2003;100:1280–5. doi: 10.1073/pnas.0237385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faruque SM, Bin Naser I, Fujihara K, Diraphat P, Chowdhury N, Kamruzzaman M, et al. Genomic sequence and receptor for the Vibrio cholerae phage KSF-1φ: evolutionary divergence among filamentous vibriophages mediating lateral gene transfer. J Bacteriol. 2005;187:4095–103. doi: 10.1128/JB.187.12.4095-4103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campos J, Martínez E, Suzarte E, Rodríguez BL, Marrero K, Silva Y, et al. VGJ phi, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTX phi. J Bacteriol. 2003;185:5685–96. doi: 10.1128/JB.185.19.5685-5696.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campos J, Martínez E, Marrero K, Silva Y, Rodríguez BL, Suzarte E, et al. Novel type of specialized transduction for CTX phi or its satellite phage RS1 mediated by filamentous phage VGJ phi in Vibrio cholerae. J Bacteriol. 2003;185:7231–40. doi: 10.1128/JB.185.24.7231-7240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kar S, Ghosh RK, Ghosh AN, Ghosh A. Integration of the DNA of a novel filamentous bacteriophage VSK from Vibrio cholerae 0139 into the host chromosomal DNA. FEMS Microbiol Lett. 1996;145:17–22. doi: 10.1111/j.1574-6968.1996.tb08550.x. [DOI] [PubMed] [Google Scholar]
- 52.Honma Y, Ikema M, Toma C, Ehara M, Iwanaga M. Molecular analysis of a filamentous phage (fsl) of Vibrio cholerae O139. Biochim Biophys Acta. 1997;1362:109–15. doi: 10.1016/S0925-4439(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 53.Ikema M, Honma Y. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology. 1998;144:1901–6. doi: 10.1099/00221287-144-7-1901. [DOI] [PubMed] [Google Scholar]
- 54.Jouravleva EA, McDonald GA, Garon CF, Boesman-Finkelstein M, Finkelstein RA. Characterization and possible functions of a new filamentous bacteriophage from Vibrio cholerae O139. Microbiology. 1998;144:315–24. doi: 10.1099/00221287-144-2-315. [DOI] [PubMed] [Google Scholar]
- 55.Jouravleva EA, McDonald GA, Marsh JW, Taylor RK, Boesman-Finkelstein M, Finkelstein RA. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect Immun. 1998;66:2535–9. doi: 10.1128/iai.66.6.2535-2539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehara M, Shimodori S, Kojima F, Ichinose Y, Hirayama T, Albert MJ, et al. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol Lett. 1997;154:293–301. doi: 10.1016/S0378-1097(97)00345-5. [DOI] [PubMed] [Google Scholar]
- 57.Karaolis DK, Somara S, Maneval DR, Jr., Johnson JA, Jr., Kaper JB. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–9. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 58.Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, Reeves PR. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci U S A. 1998;95:3134–9. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faruque SM, Zhu J, Asadulghani, Kamruzzaman M, Mekalanos JJ. Examination of diverse toxin-coregulated pilus-positive Vibrio cholerae strains fails to demonstrate evidence for Vibrio pathogenicity island phage. Infect Immun. 2003;71:2993–9. doi: 10.1128/IAI.71.6.2993-2999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovach ME, Shaffer MD, Peterson KM. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–74. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 61.Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A. 2002;99:1556–61. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Shea YA, Finnan S, Reen FJ, Morrissey JP, O’Gara F, Boyd EF. The Vibrio seventh pandemic island-II is a 26.9 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 43.4 kb genomic island in V. vulnificus. Microbiology. 2004;150:4053–63. doi: 10.1099/mic.0.27172-0. [DOI] [PubMed] [Google Scholar]
- 63.Rajanna C, Wang J, Zhang D, Xu Z, Ali A, Hou YM, et al. The vibrio pathogenicity island of epidemic Vibrio cholerae forms precise extrachromosomal circular excision products. J Bacteriol. 2003;185:6893–901. doi: 10.1128/JB.185.23.6893-6901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murphy RA, Boyd EF. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol. 2008;190:636–47. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Almagro-Moreno S, Napolitano MG, Boyd EF. Excision dynamics of Vibrio pathogenicity island-2 from Vibrio cholerae: role of a recombination directionality factor VefA. BMC Microbiol. 2010;10:306–15. doi: 10.1186/1471-2180-10-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Shea YA, Boyd EF. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol Lett. 2002;214:153–7. doi: 10.1111/j.1574-6968.2002.tb11339.x. [DOI] [PubMed] [Google Scholar]
- 67.Zahid MSH, Udden SM, Faruque ASG, Calderwood SB, Mekalanos JJ, Faruque SM. Effect of phage on the infectivity of Vibrio cholerae and emergence of genetic variants. Infect Immun. 2008;76:5266–73. doi: 10.1128/IAI.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nelson EJ, Chowdhury A, Flynn J, Schild S, Bourassa L, Shao Y, et al. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 2008;4:e1000187. doi: 10.1371/journal.ppat.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson EJ, Harris JB, Morris JG, Jr., Calderwood SB, Camilli A. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol. 2009;7:693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seed KD, Bodi KL, Kropinski AM, Ackermann HW, Calderwood SB, Qadri F, et al. Evidence of a dominant lineage of Vibrio cholerae-specific lytic bacteriophages shed by cholera patients over a 10-year period in Dhaka, Bangladesh. MBio. 2011;2:e0033r–10. doi: 10.1128/mBio.003344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seed KD, Faruque SM, Mekalanos JJ, Calderwood SB, Qadri F, Camilli A. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog. 2012;8:e1002917. doi: 10.1371/journal.ppat.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogg JE, Timme TL, Alemohammad MM. General transduction in Vibrio cholerae. Infect Immun. 1981;31:737–41. doi: 10.1128/iai.31.2.737-741.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–7. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 74.Udden SMN, Zahid MSH, Biswas K, Ahmad QS, Cravioto A, Nair GB, et al. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc Natl Acad Sci U S A. 2008;105:11951–6. doi: 10.1073/pnas.0805560105. [DOI] [PMC free article] [PubMed] [Google Scholar]