Abstract
Background
Marek’s disease virus (MDV) is a highly cell-associated oncogenic α-herpesvirus that causes a disease characterised by T-cell lymphomas. The pathogenesis, or the nature of the interaction of the virus and the host, in the thymus are still unclear.
Results
In this study, we identified 119 differentially expressed proteins using two-dimensional electrophoresis and mass spectrometry from the thymuses of chickens infected with the RB1B strain of MDV. These differentially expressed proteins were found mainly at 21, 28 and 35 days post-infection. More than 20 of the differentially expressed proteins were directly associated with immunity, apoptosis, tumour development and viral infection and replication. Five of these proteins, ANXA1, MIF, NPM1, OP18 and VIM, were further confirmed using real-time PCR. The functional associations and roles in oncogenesis of these proteins are discussed.
Conclusions
This work provides a proteomic profiling of host responses to MDV in the thymus of chickens and further characterises proteins related to the mechanisms of MDV oncogenesis and pathogenesis.
Background
Marek’s disease virus (MDV) is a highly cell-associated oncogenic α-herpesvirus that leads to serious economic losses in the poultry industry [1,2]. Marek’s disease (MD) is a lymphoproliferative disease characterised by immunosuppression, neurological disorders, and neoplastic T-cell lymphomas in chickens. MD was the first tumour disease to be prevented by vaccination, and thus, provides an important animal model for the study of viral cancer development and immunity [3].
The primary target cells for MDV infection in the chicken in vivo are B cells, then T cells, and eventually the formation of a T-cell lymphoma occurs [4]. The mechanism of lymphoma formation is very complex and has not yet been clarified. In recent years, the dynamics of host-protein expression in chicken immune organs have been studied at different phases of MDV infection using two-dimensional polyacrylamide gel electrophoresis (2-DE) followed by the identification and characterisation of the proteins by mass spectrometry (MS) [5-7]. In the spleens of MDV-infected chickens, 61 protein spots representing 48 host proteins have been detected. These proteins are involved in a variety of cellular processes, including antigen processing and presentation, ubiquitin-proteasome protein degradation (UPP), formation of the cytoskeleton, cellular metabolism, signal transduction, and translation regulation [5]. In the bursa of Fabricius, 24 differentially expressed proteins associated mainly with tumour biology, protein folding, signal transduction, immunology, cell proliferation and apoptosis have been successfully identified, and the tumour-associated proteins were significantly increased at 14 and 21 days post-infection (dpi) [6]. Furthermore, 20 proteins have been found to be differentially expressed in the spleen when comparing MD-susceptible B19 and MD-resistant B21 chickens [7]. These studies have characterised the proteomic profiles of the host response to MDV in chickens and are the basis for illustrating the mechanism of MD lymphoma formation. In addition, Niikura et al. found that a lytic infection with MDV up-regulates the cell surface expression of MHC class II infected cells [8] but down-regulates the expression of MHC class II cells in the spleens of MDV-infected chickens [9].
The thymus is the specific organ in which the maturation and differentiation of avian T lymphocytes take place. T-cell immune suppression and lymphomas due to MDV infection and latent infections are predominantly related to activated CD4+ T lymphocytes. Morimura et al. reported that MDV can induce apoptosis and the down-regulation of CD8 molecules on peripheral CD4+ T cells and the thymus, which could contribute to immune suppression [10-12]. However, all the changes in the protein profiles that occur after MDV infection have not been reported. Such changes may indicate the nature of the interaction between MDV and host and provide some clues to the pathogenesis of MDV. In this study, 119 proteins that were differentially expressed in the thymus in response to MDV infection were identified by two-dimensional electrophoresis and mass spectrometry techniques at 4, 7, 14, 21, 28, 35 and 42 dpi. These proteins are associated with a wide range of biological processes, including metabolism, immunity, apoptosis, death, tumour development and virus infection and replication. These proteins provide some information for further interpretations of the pathogenesis and oncogenesis of MDV.
Results
Pathological changes associated with MDV infection at different stages
The thymus of each chicken displayed severe atrophy at 21, 28 and 35 dpi with RB1B, and the size of the thymus gradually returned to normal at 42 dpi (Figure 1A). Compared with the control group, the thymus and body weight parameters of the MDV-infected chickens displayed significant differences at 7, 21, 28 and 35 dpi (Figure 1B).
Figure 1.
(A) The macroscopic appearance of the thymus at different stages of infection and in the control group. (B) The differences in the thymus and body weight between the two groups at the different stages of infection.
Differential expression in the MDV-infected chicken thymuses at different time points
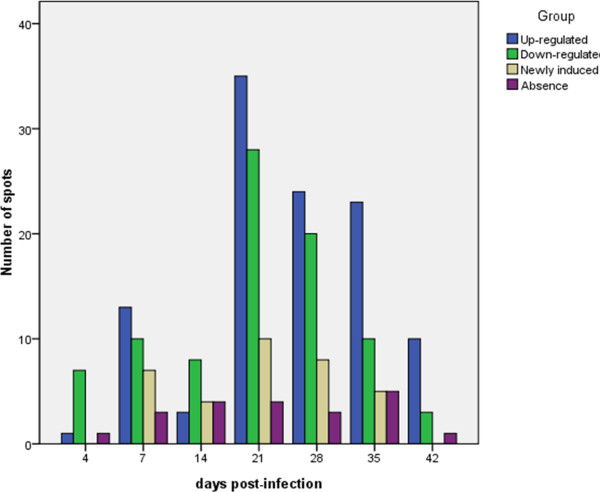
To obtain protein expression profiles at different time points in the thymus from the MDV-infected and uninfected control chickens, total protein was collected from the thymus specimens, separated by two-dimensional electrophoresis and analysed using PDQuest 8.0.1 software. More than 1000 protein spots could be detected in each gel (Figure 2). In total, 250 protein spots were detected as either quantitatively (p≤0.05 and fold change≥2) or qualitatively differentially expressed (35 that were novel and 20 that were no longer visible) in the thymus.These spots were predominantly identified at 21, 28 and 35 dpi, which was consistent with the pathological observations (Figure 3).
Figure 2.
The analysis of the chicken thymus proteins by two-dimensional gel electrophoresis. Above images represent the proteome of chicken thymus at 21 dpi. The differentially expressed proteins (p ≤ 0.05 and fold change ≥ 2) are marked on the gel. A. Uninfected control thymus. B. MDV-infected thymus.
Figure 3.
Comparison of the total numbers of protein spots that were significantly differentially expressed in the MDV-infected thymus at the different time points. A total of 250 protein spots were detected, among which 9, 33, 19, 77, 55, 43 and 14 were detected at 4, 7, 14, 21, 28, 35 and 42 dpi, respectively. Notably, some of the protein spots were detected at least once during the different sampling points, and thus, only 119 protein identities required MS analyses.
MS analysis and GO annotations
Some protein spots were detected more than once differentially expressed during experiment, resulting less number of identities than the total number of spots. In total, 119 differentially expressed proteins were successfully identified by MALDI-TOF/TOF (Table 1). These proteins are involved in a wide range of biological processes, including metabolism, immunity, apoptosis, death, tumour development, virus infection and replication. More than 20 of the differentially expressed proteins were directly associated with immunity, apoptosis, tumour development and viral infection and replication, including macrophage migration inhibitory factor (MIF), heat shock protein 90 alpha (Hsp90alpha), and annexin-A1 (Anx-A1).
Table 1.
Fundamental information of 119 differentially expressed protein spots identified by MS
|
SSP |
Protein Name |
Fold change in expression |
Accession number |
MW |
PI |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4dpi | 7dpi | 14dpi | 21dpi | 28dpi | 35dpi | 42dpi | |||||
|
Actin cytoskeleton and cellular structural proteins | |||||||||||
|
3908 |
collagen alpha-1(VI) chain precursor |
−1.05 |
1.57 |
1.44 |
4.53** |
3.47** |
1.14 |
−1.65 |
gi|49225581 |
107916.8 |
5.63 |
|
3910 |
collagen alpha-1(VI) chain precursor |
1.32 |
2.92** |
−1.66 |
3.77** |
3.72* |
−1.73 |
1.06 |
gi|49225581 |
107916.8 |
5.63 |
|
3914 |
collagen alpha-2(VI) chain precursor |
1.01 |
1.79 |
1.54 |
4.11** |
−1.2 |
1.35 |
−1.01 |
gi|45384382 |
109108.4 |
5.66 |
|
3915 |
collagen alpha-2(VI) chain precursor |
−1.33 |
2.24** |
1.4 |
3.58** |
1.71 |
1.89 |
1.09 |
gi|45384382 |
109108.4 |
5.66 |
|
508 |
keratin, type I cytoskeletal 15 |
−1.22 |
−1.17 |
1.06 |
2.67** |
1.28 |
−1.02 |
−1.03 |
gi|47604932 |
48060.8 |
5.11 |
|
6713 |
keratin, type II cytoskeletal 1 |
−1.03 |
−1.68 |
1.23 |
−2.25** |
−1.43 |
−1.2 |
−1.69 |
gi|119395750 |
65999 |
8.15 |
|
4502 |
keratin, type II cytoskeletal cochleal |
1.69 |
−1.15 |
−2.55** |
−1.2 |
1.64 |
1.42 |
−1.23 |
gi|45384378 |
53770.5 |
5.97 |
|
106 |
Chain B, Refined 1.8 Angstroms Resolution Crystal Structure Of Porcine Epsilon-Trypsin |
1.34 |
−1.31 |
−1.14 |
−2.62** |
−2.28** |
−2.63** |
−1.29 |
gi|999627 |
8813.5 |
6.67 |
|
9222 |
PREDICTED: LOW QUALITY PROTEIN: keratin, type II cytoskeletal 1-like |
AB |
IN |
AB |
AB |
AB |
AB |
AB |
gi|296211766 |
66379 |
8.16 |
|
6206 |
REDICTED: similar to Chain D, Crystal Structure Of Arp23 COMPLEX |
1.08 |
1.34 |
1.07 |
−1.04 |
−1.08 |
1.21 |
2.14** |
gi|118093746 |
34379.6 |
6.84 |
|
6211 |
PREDICTED: similar to Chain D, Crystal Structure Of Arp23 COMPLEX |
−2.11** |
3.72** |
1.09 |
4.76** |
2.22** |
4.28** |
1.73 |
gi|118093746 |
34379.6 |
6.84 |
|
314 |
beta-tropomyosin |
−1.19 |
1.04 |
1.37 |
1.82 |
1.66 |
2.86** |
1.43 |
gi|212809 |
32779.7 |
4.77 |
|
6601 |
fibrinogen beta chain precursor |
−1.37 |
1.28 |
−1.01 |
1.88 |
2.33** |
−1.2 |
−1.44 |
gi|267844833 |
54545.7 |
7.84 |
|
2606 |
plastin-2 |
−1.65 |
2.04** |
1.23 |
2.44** |
2.24* |
2.13** |
1.27 |
gi|56605886 |
69704.5 |
5.16 |
|
2206 |
beta-actin |
−1.1 |
1.87 |
1.08 |
1.06 |
1.53 |
2.08** |
2.2** |
gi|154818367 |
41812.8 |
5.32 |
|
3906 |
collagen alpha-1(VI) chain precursor |
−1.07 |
1.67 |
1.51 |
4.29** |
1.68 |
1.55 |
−1.61 |
gi|49225581 |
107916.8 |
5.63 |
|
Enzymes | |||||||||||
|
4406 |
adenylosuccinate synthetase isozyme 2 |
−1.29 |
1.1 |
−1.39 |
2.19* |
2.2* |
−1.46 |
−1.16 |
gi|71895783 |
49423.4 |
5.93 |
|
4410 |
argininosuccinate synthase |
1.48 |
1.56 |
1.49 |
IN |
3.99 |
2.64** |
−1.2 |
gi|61657937 |
46873.8 |
6.1 |
|
5402 |
argininosuccinate synthase |
−1.63 |
4.04** |
1.03 |
1.15 |
1 |
1 |
−1.45 |
gi|61657937 |
46873.8 |
6.1 |
|
5904 |
argininosuccinate synthase |
1.35 |
2.49** |
1.22 |
1.82 |
1.83 |
−1.23 |
−1.67 |
gi|61657937 |
46873.8 |
6.1 |
|
6705 |
ATP-dependent RNA helicase DDX3X |
−1.03 |
−1.33 |
1.15 |
−2.2** |
−2.27** |
1 |
−1.55 |
gi|71895253 |
72004.5 |
6.54 |
|
3414 |
B-creatine kinase |
−1.32 |
1.73 |
−1.3 |
2.09** |
1.76 |
1.34 |
−1.08 |
gi|211235 |
42240.4 |
5.78 |
|
7213 |
L-lactate dehydrogenase A chain |
−1.08 |
1.71 |
−1.03 |
2.36** |
2.63** |
2.43** |
−1.4 |
gi|45384208 |
36491.2 |
7.75 |
|
6606 |
phosphoglucomutase 2 |
AB |
REP |
REP |
−1.36 |
AB |
−1.4 |
1.29 |
gi|71897287 |
67900.1 |
6.56 |
|
9221 |
PREDICTED: similar to 2,4-dienoyl-CoA reductase |
AB |
IN |
AB |
AB |
AB |
AB |
AB |
gi|50731694 |
35723.5 |
9.46 |
|
3205 |
PREDICTED: similar to pyridoxal kinase |
AB |
1.56 |
1.45 |
2.74* |
2.02** |
2.45** |
−1.04 |
gi|118083963 |
43075.8 |
8.36 |
|
3207 |
RecName: Full=3-mercaptopyruvate sulfurtransferase; Short=MST |
AB |
AB |
IN |
AB |
AB |
AB |
AB |
gi|90110410 |
33002.5 |
6.11 |
|
6107 |
RecName: Full=Carbonic anhydrase 2; AltName: |
−1.13 |
−1.45 |
1.13 |
1.38 |
3.04 |
1.19 |
2.22* |
gi|115454 |
29388.4 |
6.56 |
|
Full=Carbonate dehydratase II; AltName: Full=Carboni anhydrase II; Short=CA-II | |||||||||||
|
2622 |
PREDICTED: similar to serine/threonine kinase isoform 1 |
AB |
1.17 |
1.44 |
REP |
−1.22 |
−1.43 |
−2.46** |
gi|118094908 |
57883.8 |
6.04 |
|
4117 |
PREDICTED: similar to Glyoxylase 1 |
REP |
AB |
AB |
AB |
AB |
AB |
AB |
gi|50740506 |
20540.2 |
6.1 |
|
6402 |
creatine kinase M-type |
AB |
1.16 |
1.32 |
2.64* |
2.68** |
2.47** |
−1.04 |
gi|45382875 |
43301 |
6.5 |
|
6408 |
creatine kinase M-type |
−1.39 |
2.07* |
1.75 |
6.7** |
1.14 |
2.27 |
−1.06 |
gi|45382875 |
43301 |
6.5 |
|
6414 |
enolase |
1.24 |
1.18 |
1.28 |
1.59 |
2.38** |
1.4 |
1.25 |
gi|116248308 |
41047.2 |
5.62 |
|
9101 |
glutathione S-transferase |
1.04 |
1.99 |
1.39 |
2.47** |
2.7** |
2.58** |
−1.13 |
gi|49169816 |
25282.4 |
8.86 |
|
4210 |
PREDICTED: gemin 4 |
−1.58 |
1.18 |
1.56 |
−1.24 |
1.07 |
−2.17** |
−1.37 |
gi|224076562 |
30154.9 |
4.96 |
|
1109 |
cathepsin B precursor |
1.02 |
−1.03 |
1.04 |
2.27** |
2.39** |
2.86** |
1.04 |
gi|46195455 |
37562.7 |
5.74 |
|
5607 |
peptidase D |
−1.39 |
1.49 |
IN |
1.73 |
IN |
2.08** |
−1.21 |
gi|169139269 |
55101.6 |
6.07 |
|
9111 |
peptidyl-prolyl cis-trans isomerase B precursor |
1.15 |
1.55 |
1.12 |
2** |
1.78 |
1.88 |
−1.17 |
gi|45382027 |
22398.8 |
9.4 |
|
6111 |
PREDICTED: similar to Glutathione S-transferase theta 1 isoform 3 |
1.55 |
−1.1 |
−1.59 |
−2.38** |
−1.56 |
−3.36** |
1.13 |
gi|118098704 |
27843.7 |
6.25 |
|
Immunity, apoptosis, tumor development and viral infection and replication | |||||||||||
|
6507 |
septin-6 |
−1.15 |
−1.27 |
−6.03** |
−1.82 |
−1.35 |
−1.39 |
1.2 |
gi|71895629 |
48656.9 |
6.67 |
|
6521 |
septin-6 |
1.1 |
1.25 |
2.42** |
−1.3 |
−1.05 |
−1.15 |
−1.55 |
gi|71895629 |
43021.1 |
6.63 |
|
7310 |
septin-9 |
−1.08 |
−1.38 |
−1.08 |
−2.87** |
−2.09* |
−2.33* |
−1.25 |
gi|71897123 |
65482.8 |
8.42 |
|
3203 |
PREDICTED: similar to PTB-associated splicing factor |
AB |
AB |
−2.1** |
AB |
−2.75** |
−1.09 |
2.06** |
gi|118101662 |
69173.9 |
9.43 |
|
7116 |
PREDICTED: heterogeneous nuclear ribonucleoprotein A3 |
−1.97 |
2.16** |
−1.67 |
11.96** |
2.33** |
2.4** |
1.4 |
gi|118093536 |
40603.2 |
9.24 |
|
8310 |
PREDICTED: heterogeneous nuclear ribonucleoprotein A3 |
AB |
AB |
AB |
−6.86** |
AB |
AB |
AB |
gi|118093536 |
40603.2 |
9.24 |
|
1310 |
PREDICTED: heterogeneous nuclearribonucleoproteins C1/C2-like |
−1.22 |
−1.19 |
−1.04 |
−3.14** |
−2.43* |
−1.36 |
1.29 |
gi|296224780 |
23551.2 |
9.87 |
|
8304 |
heterogeneous nuclear ribonucleoprotein A/B |
1.02 |
1.29 |
−1.92 |
−2.39** |
−2.58** |
−2.11** |
−1.18 |
gi|45384514 |
31841.8 |
8.62 |
|
2613 |
heterogeneous nuclear ribonucleoprotein H |
−1.14 |
−1.78 |
1.87 |
−2.13** |
−3.36 |
−2.48 |
1.05 |
gi|45383173 |
56530.6 |
5.38 |
|
2610 |
heterogeneous nuclear ribonucleoprotein H |
1.22 |
−2.25* |
−1.31 |
AB |
−2.11** |
−1.82 |
−2.91 |
gi|45383173 |
56530.6 |
5.38 |
|
3101 |
heterogeneous nuclear ribonucleoproteins A2/B1 |
1.69 |
1.46 |
−1.39 |
−1.6 |
−2.00** |
1.79 |
1.07 |
gi|71896753 |
36962.4 |
8.67 |
|
8211 |
heterogeneous nuclear ribonucleoproteins A2/B1 |
−1.1 |
−1.11 |
1.06 |
−2** |
−2** |
−1.68 |
−1.12 |
gi|71896753 |
36962.4 |
8.67 |
|
8213 |
heterogeneous nuclear ribonucleoproteins A2/B1 |
1.04 |
−1.07 |
−1.46 |
−2.53** |
−1.41 |
−2.01** |
−1.1 |
gi|71896753 |
36962.4 |
8.67 |
|
9205 |
heterogeneous nuclear ribonucleoproteins A2/B1 |
−1.08 |
1.07 |
−1.41 |
−2.47** |
−2.19** |
−1.57 |
−1.12 |
gi|71896753 |
36962.4 |
8.67 |
|
7006 |
macrophage migration inhibitory factor |
1.35 |
−1.03 |
−2.04** |
−1.64 |
−1.77 |
−1.84 |
1.74 |
gi|212258 |
12704.2 |
6.88 |
|
2905 |
heat shock protein HSP 90-alpha |
−1.37 |
−1.55 |
−1.23 |
−3.73** |
−1.58 |
−1.63 |
−1.67 |
gi|157954047 |
84006.5 |
5.01 |
|
7215 |
annexin A1 |
AB |
IN |
1 |
IN |
IN |
IN |
AB |
gi|46195459 |
38475.9 |
7.05 |
|
7703 |
far upstream element-binding protein 1 |
1.03 |
−2.56 |
1.34 |
−2.53** |
−2.38* |
−1.85 |
−1.48 |
gi|83320094 |
67155.3 |
7.18 |
|
3014 |
stathmin |
1.19 |
1 |
−1.16 |
−4.27** |
−7.72** |
−7.18** |
1.05 |
gi|50053682 |
17071.9 |
6.18 |
|
4003 |
stathmin |
1.1 |
−2.1** |
−1.1 |
−4.91** |
−4.49** |
−5.08** |
−1.1 |
gi|50053682 |
17071.9 |
6.18 |
|
405 |
vimentin |
−1.5 |
3.27* |
1.56 |
3.31** |
1.36 |
1.06 |
1.04 |
gi|114326309 |
53109.6 |
5.09 |
|
1605 |
vimentin |
1.62 |
1.76 |
1.77 |
2.9** |
−1.35 |
−1.08 |
−1.01 |
gi|114326309 |
53109.6 |
5.09 |
|
409 |
vimentin |
1.23 |
1.64 |
1.42 |
2.26** |
1.47 |
1.31 |
1 |
gi|114326309 |
53109.6 |
5.09 |
|
205 |
nucleolar protein B23/No38 |
1.28 |
1.76 |
−1.51 |
1.54 |
1.58 |
1.65 |
6.95** |
gi|212456 |
10716.2 |
4.38 |
|
6003 |
beta-galactoside-binding lectin |
1.07 |
2.22** |
1.56 |
2.7** |
2.66** |
2.4** |
−1.45 |
gi|45382785 |
15053.5 |
6.58 |
|
3113 |
PREDICTED: similar to natural killer cell enhancing factor isoform 4 |
1.63 |
1.12 |
1.63* |
1.58 |
2.5** |
2.28** |
−1.27 |
gi|50751518 |
22300.5 |
8.24 |
|
5304 |
unnamed protein product |
1.23 |
1.88 |
1.45 |
−1.49 |
−1.26 |
1.28 |
2.11** |
gi|74142813 |
50433.1 |
6.17 |
|
1104 |
PREDICTED: similar to interferon, gamma-inducible protein 30 |
1.14 |
−1.05 |
−1.04 |
−2.27** |
−1.29 |
−1.12 |
1.19 |
gi|50761132 |
14269.9 |
5.73 |
|
401 |
mCG49244 |
1.23 |
−1.56 |
1.27 |
−2.25* |
−2.04** |
−1.63 |
−1.73 |
gi|148694498 |
21747.1 |
5.71 |
|
114 |
SET |
1.62 |
5.16** |
1.18 |
−1.01 |
−1.07 |
1.01 |
1.51 |
gi|3953617 |
24348 |
4.97 |
|
Mitosis, replication and translation | |||||||||||
|
1402 |
put. beta-actin (aa 27–375) |
1.01 |
−1.27 |
−2.74** |
−13.22** |
−14.21** |
−1.1 |
−1.07 |
gi|49868 |
39160.6 |
5.78 |
|
2303 |
suppressor of G2 allele of SKP1 homolog |
AB |
−2.33* |
−1.1 |
REP |
REP |
REP |
REP |
gi|71895155 |
20497.5 |
6.35 |
|
6307 |
mitotic checkpoint protein BUB3 |
AB |
−3.53** |
1.61 |
REP |
REP |
REP |
−2.97** |
gi|57529813 |
37256.3 |
6.5 |
|
3501 |
PREDICTED: similar to eukaryotic translation initiation factor 4H isoform 1 |
1.26 |
−2.31* |
−1.33 |
−1.99 |
−1.45 |
−1.84 |
1.02 |
gi|126314438 |
27406.4 |
6.78 |
|
8202 |
PREDICTED: similar to eukaryotic translation initiation factor 4H isoform 1 |
1.17 |
−3.81** |
1.13 |
−2.86** |
−1.72 |
−1.38 |
−2.65** |
gi|126314438 |
27406.4 |
6.78 |
|
2013 |
PREDICTED: similar to histone H2B |
3.97* |
REP |
REP |
REP |
REP |
REP |
8.73** |
gi|149617840 |
29704.7 |
9.68 |
|
7117 |
single-strand binding protein |
AB |
AB |
AB |
IN |
IN |
IN |
AB |
gi|42523087 |
15766.8 |
6.13 |
|
5603 |
pre-mRNA-processing factor 19 |
1.61 |
−1.34 |
−2.62** |
−1.23 |
−1.09 |
−1.32 |
−1.16 |
gi|86129600 |
55100.2 |
6.19 |
|
Signal transduction | |||||||||||
|
3616 |
Amb7 |
1.22 |
1.9 |
1.94 |
4.4** |
1.87 |
IN |
AB |
gi|117168610 |
225516.8 |
5.61 |
|
5518 |
coronin-1C |
AB |
AB |
AB |
IN |
AB |
AB |
AB |
gi|86129440 |
53174.1 |
6.22 |
|
6102 |
coronin-1C |
−1.1 |
1.32 |
−1.33 |
2.41** |
3.78* |
3.37** |
3.76** |
gi|86129440 |
53174.1 |
6.22 |
|
3015 |
fatty acid-binding protein, adipocyte |
−2.05** |
2.38* |
2.34** |
4.91** |
4.73** |
2.39** |
1.11 |
gi|45383556 |
14884.6 |
6.34 |
|
3012 |
fatty acid-binding protein, heart |
−1.23 |
−1.49 |
AB |
9.97** |
1.5 |
1.62 |
1.77 |
gi|71894843 |
14806.6 |
5.92 |
|
3709 |
Lipocalin precursor |
1.75 |
−1.47 |
1.09 |
4.11** |
6.62** |
3.26* |
1.47 |
gi|225716896 |
20257.1 |
7.6 |
|
6213 |
PREDICTED: similar to GADS protein |
−1.46 |
−1.24 |
1.56 |
−2.49** |
1.7 |
−1.08 |
1.19 |
gi|118082689 |
33178.5 |
6.23 |
|
5205 |
PREDICTED: similar to GADS protein |
−1.11 |
−2.14** |
−1.15 |
−3.84** |
1.04 |
−1.16 |
−1.45 |
gi|118082689 |
33178.5 |
6.23 |
|
6310 |
PREDICTED: similar to nucleic acid binding protein isoform 2 |
1.42 |
−1.6 |
−1.03 |
−1.41 |
−2.07** |
−1.59 |
−1.63 |
gi|126345445 |
35784.3 |
7 |
|
6011 |
PREDICTED: similar to retinoid binding protein 7 |
−4.75** |
IN |
IN |
IN |
IN |
4.34** |
AB |
gi|118101075 |
14701.2 |
5.31 |
|
1008 |
thioredoxin |
−1.26 |
−1.11 |
−1.02 |
2.46* |
1.28 |
1.7 |
1.07 |
gi|45382053 |
11692.8 |
5.1 |
|
Transport | |||||||||||
|
6517 |
ferritin heavy chain |
−1.14 |
2.81** |
−1.44 |
2.13** |
4.11** |
2.25** |
1.02 |
gi|45384172 |
21078.3 |
5.78 |
|
7203 |
ferritin heavy chain |
−4.57** |
1.42 |
−1.18 |
3.89** |
2.12** |
2.49** |
1.03 |
gi|45384172 |
21078.3 |
5.78 |
|
8018 |
hemoglobin alpha-A chain |
−3.13** |
1.13 |
1.41 |
1.37 |
1.2 |
1.4 |
1.26 |
gi|4894665 |
10914.7 |
7.19 |
|
8003 |
hemoglobin alpha-A chain |
−2.54** |
−1.15 |
−1.12 |
−1.03 |
1.28 |
−1.14 |
1.06 |
gi|4894665 |
10914.7 |
7.19 |
|
6009 |
hemoglobin subunit alpha-D |
−1.08 |
−1.07 |
3.59** |
1.2 |
1.02 |
1.04 |
1.03 |
gi|52138645 |
15685 |
7.01 |
|
6804 |
ovotransferrin BB type |
1.03 |
1.32 |
−1.27 |
2.71* |
1.82 |
1.78 |
1.03 |
gi|71274075 |
77781.4 |
6.85 |
|
6810 |
ovotransferrin BB type |
AB |
1.95 |
REP |
20.2* |
IN |
IN |
AB |
gi|71274075 |
77781.4 |
6.85 |
|
6801 |
ovotransferrin CC type |
1.56 |
1.27 |
1.19 |
2.52** |
2.89* |
2.48* |
−1.17 |
gi|71274077 |
77750.4 |
7.08 |
|
7201 |
mediator of cell motility 1 |
1.37 |
−1.06 |
1.02 |
−2.93** |
−1.09 |
1.54 |
−1.24 |
gi|300676818 |
31517.5 |
6.49 |
|
Others proteins | |||||||||||
|
6104 |
603815993F1 CSEQCHN52 Gallus gallus cDNA clone ChEST809j23 5′, mRNA sequence |
1.14 |
−1.17 |
−1.57 |
−2.53** |
−2.12** |
−1.94 |
1.19 |
gi|25541234 |
23338.9 |
6.03 |
|
304 |
alpha-tropomyosin (partial) |
−1.32 |
1.18 |
1.12 |
2.37** |
2.38** |
2.46** |
1.71 |
gi|212815 |
30045.3 |
4.64 |
|
5815 |
conalbumin |
−1.17 |
1.11 |
−1.05 |
5.97* |
3.44** |
1.9 |
−1.12 |
gi|295721 |
79551.1 |
6.85 |
|
4012 |
cytoChrome c subfamily, putative |
AB |
IN |
AB |
IN |
IN |
26.4 |
AB |
gi|124005227 |
24128.2 |
9.21 |
|
307 |
putative nucleophosmin 1 variant 1 |
1.2 |
−2.66* |
1.23 |
−2.09** |
−3.13** |
−1.19 |
1.06 |
gi|45383996 |
32612 |
4.66 |
|
4412 |
putative protein product of Nbla10058 |
AB |
IN |
AB |
AB |
AB |
AB |
AB |
gi|76879893 |
48587.2 |
5.85 |
|
204 |
PREDICTED: similar to Acidic leucine-rich nuclear |
1.3 |
−1.71 |
−1.6 |
−2.22* |
−3.2** |
−2.45** |
−1.14 |
gi|118096008 |
32075.9 |
3.98 |
|
4420 |
PREDICTED: similar to cyclophilin |
AB |
AB |
AB |
IN |
IN |
AB |
AB |
gi|118089782 |
39402.9 |
5.61 |
|
6304 |
PREDICTED: similar to KIAA0089 |
1.01 |
−1.19 |
−1.45 |
−1.43 |
−2.6** |
−1.8 |
1.2 |
gi|50732786 |
38564.1 |
6.55 |
|
4116 |
PREDICTED: similar to LOC129607 protein |
−1.25 |
IN |
AB |
IN |
IN |
IN |
AB |
gi|118088982 |
31865.5 |
8.45 |
|
4114 |
PREDICTED: similar to MGC84496 protein |
−1.43 |
−1.32 |
−1.37 |
1.17 |
1.64 |
1.98 |
2.77** |
gi|118101652 |
22693.7 |
6.07 |
|
8002 |
protein S100-A10 |
−1.27 |
1.37 |
1.11 |
3.15* |
1.27 |
1.3 |
1 |
gi|45382861 |
11282.5 |
6.82 |
|
1006 |
transthyretin precursor |
−1.12 |
−1.01 |
1.35 |
−2.64** |
−3.31** |
−1.57 |
−1.09 |
gi|45384444 |
16299.1 |
5.11 |
|
Hypothetical proteins | |||||||||||
|
3507 |
PREDICTED: hypothetical protein |
1.52 |
−1.52 |
−1.24 |
−3.08** |
−2.04* |
−1.49 |
−1.31 |
gi|50762370 |
47124.5 |
5.64 |
|
2203 |
PREDICTED: hypothetical protein |
−1.37 |
−1.72 |
−1.72 |
−6.61** |
−2.27** |
−3.83** |
−1.02 |
gi|118087111 |
30122.5 |
5.51 |
|
2209 |
PREDICTED: hypothetical protein |
−1.12 |
−1.22 |
−2.19** |
−1.3 |
−1.27 |
1.14 |
1.43 |
gi|118092623 |
32616.1 |
5.53 |
|
3110 |
PREDICTED: hypothetical protein |
AB |
−2.73** |
IN |
AB |
−3.04** |
−2* |
−1.22 |
gi|118084734 |
18881.6 |
6.08 |
|
5108 |
PREDICTED: hypothetical protein |
−5.55** |
REP |
REP |
−1.2 |
IN |
REP |
1.75 |
gi|118084734 |
18881.6 |
6.08 |
|
7214 |
PREDICTED: hypothetical protein |
−1.48 |
1.03 |
−2.13** |
−1.46 |
1.88 |
1.06 |
−1.04 |
gi|50762370 |
47124.5 |
5.64 |
|
2307 |
hypothetical protein |
−1.24 |
1.82 |
1.47 |
1.39 |
1.55 |
2.28* |
2.19** |
gi|53126859 |
42107.9 |
5.31 |
|
4013 |
hypothetical protein Bsuib36_09931 |
AB |
AB |
AB |
IN |
AB |
AB |
AB |
gi|254704229 |
122393.8 |
8.07 |
| 3210 | hypothetical protein PANDA_003704 | AB | AB | AB | IN | AB | AB | AB | gi|281343027 | 31307.8 | 5.44 |
Fold change = infected/control. Positive indicates up regulation, negative indicates down-regulation.
IN = induced, detected only in infected group, REP = repressed, detected only in the control group, AB = absent in both groups, dpi = days post-infection.
Asterisks (*) indicates statistically significant difference. (*) indicates p<0.05 and (**) indicates p<0.01.
Based on the GO annotations, more than 50% of the associated biological processes were metabolic processes (GO:0008152, 23.4%), regulations of biological processes (GO:0050789, 17.3%) and responses to stimuli (GO:0050896, 12.7%). In addition, the majority of the associations, with respect to molecular function, were with GO terms such as protein binding (GO:0005515, 27.2%), nucleic acid binding (GO:0003676, 13.6%) and hydrolase activity (GO:0016787, 12.3%). Meanwhile, 7.0% of the proteins were associated with functions such as signal transduction, antioxidation, transcription regulation and receptor binding.
Analysis of the protein functional interaction networks
The STRING Database was used to establish possible interactions among the detected proteins. In this analysis, 16 tumour proteins were identified by interaction analysis, and a network map with the tumour protein p53 (TP53), myc proto-oncogene protein (MYC) and hypoxia-inducible factor 1-alpha (HIF1A) at its core was obtained (Figure 4).
Figure 4.
Based on the STRING database, the network of protein–protein interactions in the MDV-infected thymuses. Sixteen differentially expressed proteins formed a network map with the tumour protein p53 (TP53), myc proto-oncogene protein (MYC) and hypoxia-inducible factor 1-alpha (HIF1A) at its core.
Validation of the mRNA expression levels using real-time PCR
To validate the results of the proteomic analysis, the expression levels of ANXA1, MIF, NPM1, OP18 and VIM in the chicken thymus were determined using real-time PCR. The mRNA expression levels of these proteins were down-regulated at 4 dpi and then remained down-regulated for MIF and OP18 and up-regulated for ANXA1 and VIM (Figure 5). These results are consistent with the results of our proteomic analysis (see Table 1 and Figure 5).
Figure 5.
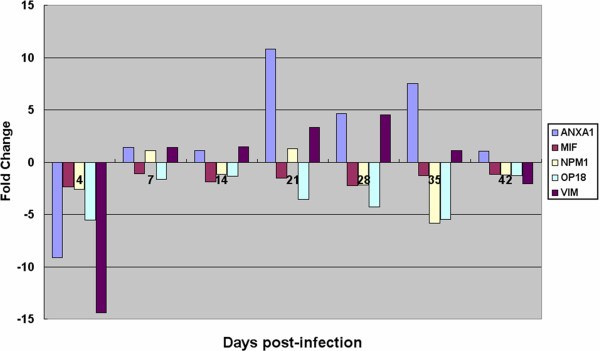
mRNA changes for the five proteins that were significantly altered in response to the MDV infection at the different stages. The y-axis is the fold change, with the positive values representing up-regulation and the negative values representing down-regulation. The abscissa axes represent different stages.
Viral infection levels in the thymuses of the MDV-infected chickens
The MDV infection levels in the thymus at seven points were detected using real-time PCR and are shown in Figure 6. The expression level of gB was transiently increased at 4 and 7 dpi and decreased at 14 dpi, the latent infection phase. The gB expression level was increased again at 21 dpi, suggesting the occurrence of MDV reactivation and transformation, and decreased at 28 dpi. These changes were consistent with the numbers of differentially expressed proteins. After 28 dpi, the mRNA level of gB in the MDV-infected chickens displayed a rapid decrease, and little mRNA was detected at 42 dpi. These changes coincided with the Meq expression levels.
Figure 6.

mRNA levels of the MDV transcripts in the chicken thymuses infected with RB1B. The expression levels of Meq and gB mRNA were detected using real-time PCR and calculated using the 2 –ΔΔCT method.
Discussion
The thymus is the specific organ in which the maturation and differentiation of avian T-lymphocytes take place. T lymphocytes, or T cells, are of a key importance to the immune system and are at the core of adaptive immunity. Chickens infected with MDV display thymic atrophy (Figure 1A) and eventually the formation of T-cell lymphoma. However, little is known about the underlying mechanisms of this phenomenon. In the present study, 119 proteins differentially expressed in thymus specimens from chickens infected with MDV were identified by two-dimensional electrophoresis and mass spectrometry at seven time points. The chicken thymuses displayed severe atrophy at 21, 28 and 35 days after infection with RB1B, and their size gradually returned to normal at 42 dpi. The change in thymus size during the course of the MDV infection may have influenced the expression of cellular structural proteins, enzymes and cytoskeleton proteins. However, we should also note that the 2-DE and real-time PCR analyses were conducted using the same amounts of protein and mRNA, which might exclude the effect of thymus size to some extent. In addition, the change in the cellular composition of the thymus could be affected the total proteome. The thymic atrophy in the chickens infected with MDV has a significant influence on host immune suppression. Permanent immunosuppression tends to correlate with the eventual development of tumours, which enhances our understanding of the mechanisms of T-lymphoma formation.
To better understand host responses to MDV infection, the expression of viral genes (gB and Meq) was detected using real-time PCR (Figure 6). We found a good correlation between viral gene product and host differentially expressed proteins. The expression levels of gB and Meq were transiently increased at 4 and 7 dpi and decreased at 14 dpi during the latent infection period. The gB and Meq expression levels were again increased at 21 dpi, suggesting the occurrence of MDV reactivation and transformation (Figure 6). At 21 dpi, the over-expressed viral gene products led to serious changes in protein expression levels and thymic atrophy (Figures 1, 3 and 6). These changes could be due to the disease pathology induced by MDV infection. Consistent with the MDV pathogenesis, early cytolytic infection occurred at 3 to 7 dpi and then entered latency, followed by the proliferative/transformation phase, leading to lymphoma formation at approximately 21 to 28 dpi. After 28 dpi, the mRNA levels of gB and Meq in the MDV-infected chickens displayed a rapid decrease, and little mRNA was detected at 42 dpi. This change may be due to the cellular response of lesion regression. It has been reported that lymphomas can occur at any time from approximately 3~4 weeks, and lesion regression can occur after lymphomas [13].
More interestingly, more than 20 differentially expressed proteins were directly associated with immunity, apoptosis, tumour development and viral infection and replication (Table 1). Notably, at least nine proteins were identified for the first time in this study: macrophage migration inhibitory factor, heat shock protein 90-alpha, annexin A1, far upstream element-binding protein 1, septin-6, septin-9, beta-galactoside-binding lectin, mCG49244 and an unnamed protein product. Among these proteins, some formed a network map encompassing TP53, MYC and HIF1A at its core and are directly associated with immunity, apoptosis, tumour development and viral infection and replication (Figure 4). Within this map, we found that MIF, HSP90AA1, NPM1, STMN1(OP18) and VIM can interact with the tumour suppressor protein TP53. It has been reported that the Meq oncoprotein directly interacts with p53 and inhibits p53-mediated transcriptional activity and apoptosis [14]. This scenario could provide an interesting link between MEQ and these proteins and enhance our understanding of MDV pathogenesis. The mRNA levels of these proteins were confirmed by real-time PCR (Figure 5), and some of these proteins are further discussed in this study.
Roles of the differentially expressed proteins in MDV infection
After MDV infection, the expression of the MIF protein displayed a slight increase at 4 dpi, followed by a down-regulation and subsequent increase at 42 dpi. The mRNA expression level of MIF was down-regulated at all stages, which may have resulted for two reasons. First, different mechanisms control the transcription and translation of the MIF gene in chicken thymus, and the mRNA abundance is not always consistent with the protein level. Second, it is possible that the transcription level of the MIF gene is easily influenced by MDV during the early stages of the infection. However, MIF is a pro-inflammatory factor, and its translational level is enhanced by the host’s regulatory mechanisms. Notably, the differences in the expression levels of the MIF mRNA and protein could be involved in the MDV infection. It is believed that macrophages transfer the virus to B cells, which are the primary target cells that are infected between 3 and 6 days post-infection [4]. A heavy infiltration of lymphocytes and macrophages occurs around blood vessels at 8–10 days post-MDV infection [15], and a study has demonstrated that some new MDV strains can replicate in macrophages, which leads to increased macrophage death [16]. MIF is involved in virus infection, and its target cells are mainly macrophages. It has been reported that human cytomegalovirus (HCMV), a herpesvirus, paralyses macrophage motility through the down-regulation of chemokine receptors, reorganisation of the cytoskeleton, and release of MIF [17]. MIF also promotes HIV-1 replication through the activation of HIV-1 long terminal repeats (LTR) [18]. Increased MIF at 4 dpi may promote MDV replication. The down-regulation of MIF indicates a strong macrophage migration activity, and macrophages that carry MDV could spread the MDV infection to other cells. Another protein, Anx-A1, was induced at 7, 21, 28 and 35 dpi and may be associated with MDV infection. Research studies suggest that the annexin family members are involved in the viral replication cycle, which may integrate the regulation of virus infection by forming networks [19]. Nucleolar protein B23 (NPM1), an acidic nucleolar protein, was up-regulated in the thymus of chickens infected with MDV and may indicate the nuclear importation of MDV. It has been demonstrated that NPM1 stimulates the nuclear importation of the HIV-1 Rev protein [20], and NPM1 is also an important factor for the nucleolar localisation of the HIV protein Tat [21]. The reduction in the Hsp90alpha level during early MDV infection might be related to a latent infection. Hsp90 plays an important role in the replication and infectivity of some herpesviruses, such as herpes simplex virus type 1 [22], Epstein–Barr virus (EBV) [23] and Kaposi’s sarcoma-associated herpesvirus (KSHV) [24], and is required for the folding, stability and intracellular transport of multiple viral proteins and for the activity of viral polymerases. Taken together, our observations suggest that changes in the levels of these proteins influence MDV replication and infection. However, the specific mechanisms are unknown and will require further study.
The chicken immune response to MDV infection
Suppression of the immune response by MDV infection is a critical feature of the disease. MIF down-regulation may contribute to the inhibition of the immune response. MIF, a classic pro-inflammatory cytokine and a pivotal regulator of innate immunity, promotes innate and adaptive immune responses through the activation of macrophages and T cells [25,26]. Moreover, it directly inhibits the immunosuppressive actions of glucocorticoids [27] through the suppression of mitogen-activated protein kinase (MAPK) phosphatase-1 (MKP-1) [28-30]. MKP-1, which is induced by glucocorticoids, inactivates the proinflammatory ERK1/2, JNK, and p38 pathways. Anx-A1, a pivotal regulator of the innate and adaptive immune systems, also promotes immunosuppression [31]. This protein was induced at 7, 21, 28 and 35 dpi and has a powerful suppressive effect on the innate immune system. Recent investigations on the role of this protein in the adaptive immune response have revealed a previously unknown ‘dark side’ to this protein, that is, it is a positive modulator of T cell activation [32]. High levels of Anx-A1 influence the differentiation of T cells in vivo, and hence, may contribute to the development of T-cell-driven autoimmune diseases.
While heat shock proteins are molecular chaperones, they have also been implicated in the stimulation of the innate and adaptive immune systems [33,34]. Recent biochemical evidence highlights the role of Hsp90alpha in the endogenous processing of MHC class I antigens, and the absence of Hsp90alpha results in the decreased surface expression of MHC I [35]. Levy et al. [36] reported that RB1B was able to markedly decrease MHC class I expression. In addition, those authors found that MDV pUL49.5 directly down-regulates MHC class I expression [37], and beta2 microglobulin was also decreased in the bursa of Fabricius of chickens infected with RB1B [6]. A subsequent gene expression study indicated a down-regulation of MHC class II expression in the spleens of MDV-infected chickens [9]. The specific relationship between the down-regulation of HSP90A and MHC class I expression during the course of MDV infection is not yet clear. However, these findings indicate that a potential mechanism of immune evasion mediated by MHC expression on cell surfaces might be employed by MDV. In addition, the immune suppression that occurs after MDV infection is not only related to changes in immune-related proteins but may also be associated with immune evasion.
Apoptosis and thymic atrophy after MDV infection
The thymic atrophy of chickens infected with MDV is the most significant manifestation of immune suppression and might be related to apoptosis. Morimura et al. reported that MDV can induce apoptosis and the down-regulation of CD8 molecules on peripheral CD4+ T cells and in the thymus, which could contribute to immune suppression [10-12]. In this study, although we did not detect cell apoptosis by flow cytometry, changes in some proteins indicated apoptosis in the thymuses of the chickens. According to the network of protein–protein interactions in the thymuses infected with MDV, five differentially expressed proteins can interact with P53 (Figure 6). The decreased levels of stathmin/oncoprotein18 (Op18) and MIF may increase the level of p53 and promote p53-mediated apoptosis before T-cell lymphoma formation. It has been demonstrated that MIF suppresses the expression of p53 and its activity [26,38]. MIF increases resistance to apoptosis by activating the nuclear factor-kappa B (NF-κB) system and repressing the function of p53 [39]. Changes in Op18 may be related to the increase in P53 because p53 is associated with the negative regulation of stathmin expression [40-42]. In the thymus, this protein was mainly down-regulated and was significantly differentially expressed at 21, 28 and 35 dpi. Lu et al. [6] also reported that Op18 was significantly reduced at 4, 7 and 21 dpi in the bursa of Fabricius of chickens infected with RB1B. In addition, ANX-A1 is relevant to the regulation of cell growth and apoptosis [43,44], and ANXA1 overexpression has been shown to promote apoptosis [45,46]. It has also been reported that ANXA1 expression in leukaemic cells mediates the engulfment of apoptotic cells by macrophages [47]. In short, MDV infection results in immune suppression and the induction of apoptosis, which eventually leads to thymic atrophy.
T-cell lymphoma formation during MDV infection
The Meq oncoprotein of the Marek’s disease virus is the major oncogene involved in the induction of tumours and inhibits p53 transcriptional and apoptotic activities by interacting with p53 [14]. Permanent immunosuppression tends to correlate with the eventual development of tumours. The role of host proteins associated with tumour growth and metastasis in the formation of T-cell lymphoma cannot be ignored. MIF, a negative regulator of the important tumour suppressor p53, is involved in tumour occurrence and evolution [48-50]. A significant amount of evidence indicates that MIF influences several important biological mechanisms and processes by which tumours thrive and spread. One of the most important of these mechanisms is the modulation of hypoxic adaptation within the tumour microenvironment through the direct promotion of the hypoxia-induced stabilisation of HIF-1α [51]. We speculate that the increased level of MIF protein observed at 42 dpi may indicate hypoxic adaptation within the tumour microenvironment. Anx-A1 is directly related to tumour development [52]. This protein was induced at 7, 21, 28 and 35 dpi and could be a key host factor that enhances the formation of T-cell lymphomas. Strong evidence for this process is provided by the finding that the increased expression of Anx-A1 promotes tumour growth, invasion and metastasis in gastric carcinoma [53], melanoma [54], breast cancer [55] and colorectal cancer [56]. Using AnxA1- knock out (KO) mice, it has been determined that tumour growth and metastasis are significantly decreased, whereas rodent survival and tumour necrosis are significantly increased when tumours grow in AnxA1-KO mice [57]. In addition, the up-regulation of several cytoskeletal network proteins, e.g., vimentin, beta-actin and keratin (type I cytoskeletal 15), also promotes tumour growth and metastasis. Research suggests that vimentin, a major intermediate filament (IF) protein of mesenchymal cells, is very important for tumour growth and metastasis [58,59]. Beta-actin specifically controls cell growth and migration [60], and an increase in beta-actin levels correlates with a higher level of invasiveness for a select hepatoma in Morris 5123 cells [61]. Changes in these proteins indicate that MDV infection and T-cell lymphoma formation involve the host cytoskeleton. Nucleolar protein B23 (NPM1) plays multiple roles in cell growth and proliferation [62]. Thanthrige-Don et al. also reported an up-regulation of beta-actin and NPM1 in the spleens of MDV-infected chickens [5]. Interestingly, a report has shown that the interaction between Meq and Hsp70 is significant during MDV oncogenesis [63]. However, the biological consequences of the Meq–Hsp70 interaction are not clear. Finally, HSP90A was reduced at 21 dpi; however, whether HSP90 can interact with Meq and affect Meq carcinogenicity remains to be studied.
In addition, septin 9 and septin 6 levels were significantly decreased in the thymuses of chickens infected with MDV. Septins are a highly conserved family of GTP-binding cytoskeletal proteins implicated in oncogenesis [64,65]. Septin 9 (SEPT9), a DNA methylation-based biomarker, has been functionally linked with oncogenesis through its activation of the hypoxia-inducible factor-1 (HIF-1) pathway, which promotes tumour progression, and the c-Jun-N-terminal kinase (JNK) pathway, which plays an important role in cell proliferation, cell transformation, and tumour progression [66,67]. The differential expression of heterogeneous nuclear ribonucleoproteins (hnRNPs), including HnRNP A / B, HnRNP A2/B1, HnRNP H, and HnRNP C1/C2, was also detected in this study. These proteins play key roles in tumour development and progression [68]. Studies have shown that hnRNP A2 / B1 and hnRNP A1 can combine their telomere DNA sequences and that their encoded protein isoforms can interact with the telomerase. Tumour cells fail to enter senescence due to their telomere lengths, which are maintained by telomere-bound proteins that recruit the enzyme telomerase [68,69]. Currently, the roles of septins and hnRNPs in MD tumour progression are not clear and require further study.
Conclusions
This paper provides a proteomic profiling of host responses to MDV in the thymus of chickens. Changes in protein levels partially elicited the mechanisms involved in MDV oncogenesis and pathogenesis. The functions of these proteins will be verified in our future works.
Methods
Experimental animals and virus
All chickens used in this study were 1-day-old specific-pathogen-free white Leghorns obtained from Merial Vital (Laboratory Animal Technology Co., Ltd., Beijing, China). The chickens were housed in an isolation facility at the College of Veterinary Medicine, Yangzhou University. The RB1B strain of very virulent MDV was maintained in the laboratory.
Experimental design
The experimental work was performed as reported previously [6]. Briefly, a total of 48, 1-day-old birds were randomly divided into infected and uninfected control groups and kept in separate units under similar environmental conditions. The infected group (n = 24) was given 800 plaque-forming units of the RB1B virus intraperitoneally at 1 day old. At 4, 7, 14, 21, 28, 35 and 42 dpi, six chickens (three infected and three uninfected control birds) were sacrificed, and the whole thymus was excised rapidly, rinsed with ice cold phosphate-buffered saline (PBS, pH 7.4) to remove blood contaminants and immediately stored in liquid nitrogen until the proteomic and real-time PCR analyses. The animal experiments were conducted in accordance with the guidelines provided by the Chinese Council on Animal Care. All experiments complied with institutional animal care guidelines and were approved by the University of Yangzhou Animal Care Committee (protocol number 06R015).
Sample preparation
The samples of thymus protein were prepared as previously described [6]. Frozen thymus tissue was ground into a fine powder in liquid nitrogen with a pre-chilled mortar and pestle. This ground tissue was used for both the protein and real-time PCR analyses. Five milligrams of ground tissue was dissolved directly in 1.0 ml of extraction buffer (8 M urea, 2 M thiourea, 2% CHAPS, 60 mM DTT (dithiothreitol), 0.2% Bio-Lyte 3∕10 ampholyte, 0.1% Bio-Lyte 5∕8 ampholyte and 0.001% bromophenol blue) and shaken on ice for 2 h. A cocktail of protease inhibitors (Sigma) was added every two hours during sample preparation to protect the proteins from degradation. The homogenate was sonicated, gently shaken on ice for 4 h and subsequently centrifuged at 16,000 g (5415 R Eppendorf) for 70 min (16°C). The supernatant was collected and stored at −20°C. Protein concentrations were determined using the Bradford method [70].
2-DE
Protein sample (300 μg) was incubated for 30 min at 20°C in 400 μl rehydration buffer (8 M urea, 2 M thiourea, 2% CHAPS, 60 mM DTT, 0.2% Bio-Lyte 5∕8 ampholyte, 0.2% Bio-Lyte 3∕10 ampholyte and 0.001% bromophenol blue) and centrifuged at 16,000 g (5804 R Eppendorf) for 10 min (16°C). The supernatant was collected, and ReadyStrip IPG strips (17 cm, pH 3–10 nonlinear gradient, Bio-Rad) were rehydrated overnight at 20°C in a PROTEAN IEF focusing tray (Bio-Rad, USA). After rehydration, isoelectric focusing (IEF) was performed at 20°C using a Protean IEF Cell (Bio-Rad, USA) and the following conditions: 250 V, slow, 0.5 h; 500 V, linear, 0.5 h; 1000 V, rapid, 1 h; 8000 V, linear, 4 h; 8,000 V, rapid, 55,000 V-hr; 500 V, rapid, any time. After IEF, the IPG strips were incubated in SDS-PAGE equilibration buffer I (6 M urea, 0.375 M Tris–HCl, pH 8.8, 2% (w/v) SDS, 20% (v/v) glycerol and 2% (w/v) DTT) and buffer II (the DTT was replaced with 2.5% (w/v) iodoacetamide), with a 10-min incubation for each buffer. The strips were then loaded onto 11% homogeneous acrylamide gels and sealed with 0.5% (w/v) agarose in SDS running buffer (25 mM Tris base, 192 mM glycine, 0.1% (w/v) SDS). The second dimension, the SDS-PAGE, was run at 20°C using a PROTEAN II Multi-Cell (Bio-Rad, USA). The run was first conducted at 16 mA/gel for 30 min followed by 24 mA/gel for 5 h. The gels were stained using the colloidal Coomassie stain method [71] and Brilliant Blue G-250.
Image analysis
The stained gels were scanned at a 600 dpi (dots per inch) resolution using a PowerLook 2100XL scanner (Umax, USA). Automated detection and matching, quantification and annotation of spots were performed using the PDQuest software package (version 8.0.1, Bio-Rad). The 2-DE was performed with three infected and three uninfected control bird samples, and each sample was repeated 2~3 times. Finally, we selected three representative gels for the control group and three for the infected group, the reproducibility within each group was approximately 85% or better. The gel images were then normalised according to the total quantity in the analysis set. Protein spots (FC≥2) were compared using Student’s t-test and the Statistical Package for the Social Sciences (version 16.0). Spots that had both P≤0.05 and ≥2-fold difference in mean normalised volumes were considered significantly different.
Protein identification
Differentially expressed protein spots from fresh CCB-stained gels were excised and plated in 96-well microtitre plates. The excised spots were first destained twice with 60 μl of 50 mM NH4HCO3 and 50% acetonitrile and subsequently dried twice with 60 μl of acetonitrile. The dried pieces of gel were then incubated in ice-cold digestion solution (12.5 ng/μl trypsin and 20 mM NH4HCO3) for 20 min and transferred into a 37°C incubator for an overnight digestion. Finally, the peptides in the supernatant were collected after being extracted twice with 60 μl of the extraction solution (5% formic acid in 50% acetonitrile).
The peptide solution described above was dried under the protection of N2. A 0.8 μl aliquot of matrix solution (5 mg/ml α-cyano-4-hydroxy-cinnamic acid diluted in 0.1% TFA, 50% ACN) was pipetted to dissolve it and subsequently spotted onto a matrix-assisted laser desorption ionisation (MALDI) target plate (Applied Biosystems). The MS analysis of the peptides was performed using an ABI 4700 TOF-TOF Proteomics apparatus (Applied Biosystems). The ultraviolet (UV) laser was operated at a 200 Hz repetition rate with a wavelength of 355 nm. The accelerated voltage was operated at 20 kV, and the mass resolution was maximised at 1500 Da. Myoglobin digested with trypsin was used to calibrate the mass instrument using the internal calibration mode. All the spectra acquired from the samples were processed using 4700 ExploreTM Software (Applied Biosystems) in the default mode. The data were searched using GPS Explorer (v3.6) with the search engine MASCOT(2.1). The search parameters were as follows: the database, NCBInr; taxonomy, Viridiplantae (green plants); the protein molecular mass, from 700 to 4000 Da; trypsin digestion with one missing cleavage; MS tolerance, 100 ppm; and MS/MS tolerance, 0.6 Da. Proteins with scores greater than 71 or a best ion score (MS/MS) of more than 30 were considered to be significant (P<0.05). This work was performed in collaboration with the Proteome Research Center of Fudan University. Typically, ten identities from one spot were provided, and the proteins of Gallus gallus (species) and the highest scores were selected according to the criteria for the determination of proteins, as provided by the Proteome Research Center of Fudan University.
Gene Ontology (GO) annotation using the Agbase database
The proteins were analysed using the AgBase database ( http://www.agbase.msstate.edu). The list of accession numbers was entered into GORetriever to return all existing GO annotations available for that dataset. GORetriever also provides a list of proteins without GO annotation and enters this second list into GOanna to retrieve GO annotations assigned on the basis of sequence similarities. The resulting annotations were summarised on the basis of the GOA whole proteome GOSlim set using GOSlimViewer.
Protein functional interaction network analysis
The functional interaction networks of the proteins were analysed using STRING ( http://string.embl.de), which is a database of known and predicted protein interactions and includes direct (physical) and indirect (functional) associations. The list of protein names was entered into a table to provide the network of protein–protein interactions. The Occurrence and Coexpression programs provided the functional associations of the proteins. Functionally associated proteins often have similar phylogenetic profiles and/or display the phenomenon of co-expression.
Real-time PCR
The expression levels of ANXA1, MIF, OP18, VIM and NOM1 in the thymus of the chickens were determined using real-time PCR (7500 Real-Time PCR System, ABI). The sequences of the primers are provided in Table 2, and the primer sequences for MIF, GAPDH and Meq have been previously reported [72,73]. The primers were synthesised by Shenergy Biocolor Bioscience & Technology Company (Shanghai, China). Total RNA was prepared from ground thymus tissue using the AxyPrep™ Multisource Total RNA Miniprep kit (AXYGEN, USA). Total RNA (1 μg) was reverse transcribed into first-strand cDNA using the PrimeScript RT Master Mix (TaKaRa, USA) following the manufacturer’s instructions, and the synthesised cDNA was diluted 1:10 with nuclease-free water. Diluted cDNA (1 μl), 400 nM primers and 10 μl of SYBR Green Master Mix were used for the real-time PCR in a final volume of 20 μl. The amplification conditions were: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 34 s. Dissociation curves were generated to analyse the individual PCR products after 40 cycles. The expression levels of five proteins were normalised against the expression of chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The analyses of the data for relative gene expression were performed using the 2 –△△CT method [74].
Table 2.
Primers used for real-time PCR
| Gene Symbol | Nucleotide sequence | Product size (bp) | Accession number |
|---|---|---|---|
|
MIF |
F 5′-GCCCGCGCAGTACATAGC-3′ |
57 |
XM42_5824 |
|
R 5′-CCCCCGAAGGACATCATCT-3′ | |||
|
OP18 |
F 5′-TTGAGCTGATCCTTGGTCCC-3′ |
128 |
NM_001001858.1 |
|
R 5′-CTTGCGTCTCTCTTCTGC-3′ | |||
|
VIM |
F 5′-CAACACGGAGTTCAAGGCGA-3′ |
79 |
NM_001048076.1 |
|
R 5′-GATGTAGTTGGCGAAGCGGT-3′ | |||
|
NPM1 |
F 5′-GGTTACATTAGGGGCTGG-3′ |
73 |
NM_205267.1 |
|
R 5′-GTTGCCTTCGTAGTCCAGTG-3′ | |||
|
ANXA1 |
F 5′-AAAACTGCCTGACTGCCCTT-3′ |
90 |
NM_206906.1 |
|
R 5′-TTCCACTCCCCTTCATTGCC-3′ | |||
|
Meq |
F 5′-GTCCCCCCTCGATCTTTCTC −3′ |
184 |
NC_002229.3 |
|
R 5′-CGTCTGCTTCCTGCGTCTTC-3′ | |||
|
gB |
F 5′-ACCCCATTCGGTGGCTTTTC-3′ |
122 |
NC_002229.3 |
|
R 5′-GCGTCCAGTTGTCTGAGG-3′ | |||
|
GAPDH |
F 5′-AGGGTGGTGCTAAGCGTGTTA-3′ |
78 |
NM_204305 |
| R 5′-TCTCATGGTTGACACCCATCA-3′ |
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AQ supervised all the experiments and revised the manuscript. XH performed the experiments and prepared the manuscript. HX, WX and JM assistant research. KQ. provided a critical review of the manuscript. CY helped to conduct the real-time PCR for MIF. All the authors read and approved the final manuscript.
Contributor Information
Xuming Hu, Email: huxuming999@126.com.
Aijian Qin, Email: aijian@yzu.edu.cn.
Kun Qian, Email: qiankun@yzu.edu.cn.
Hongxia Shao, Email: hxshao@yzu.edu.cn.
Chuan Yu, Email: yu_chuan1985@163.com.
Wencai Xu, Email: 123334897@qq.com.
Ji Miao, Email: miaoji.chn@gmail.com.
Acknowledgements
The research was supported by the Major Basic Research of Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant No. 12KJA23001),the National Natural Science Foundation of China (31272560,31072135) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT0978).
References
- Witter RL, Schat KA. Marek’s disease. Ames, Iowa: Iowa state University Press; 2003. [Google Scholar]
- Davison F, Nair V. Use of Marek’s disease vaccines: could they be driving the virus to increasing virulence? Expert Rev Vaccines. 2005;4:77–88. doi: 10.1586/14760584.4.1.77. [DOI] [PubMed] [Google Scholar]
- Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. Marek’s disease virus: from miasma to model. Nat Rev Microbiol. 2006;4:283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- Jarosinski KW, Tischer BK, Trapp S, Osterrieder N. Marek’s disease virus: lytic replication, oncogenesis and control. Expert Rev Vaccines. 2006;5:761–772. doi: 10.1586/14760584.5.6.761. [DOI] [PubMed] [Google Scholar]
- Thanthrige-Don N, Abdul-Careem MF, Shack LA, Burgess SC, Sharif S. Analyses of the spleen proteome of chickens infected with Marek’s disease virus. Virology. 2009;390:356–367. doi: 10.1016/j.virol.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Qin A, Qian K, Chen X, Jin W, Zhu Y, Eltahir YM. Proteomic analysis of the host response in the bursa of Fabricius of chickens infected with Marek’s disease virus. Virus Res. 2010;153:250–257. doi: 10.1016/j.virusres.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Thanthrige-Don N, Parvizi P, Sarson AJ, Shack LA, Burgess SC, Sharif S. Proteomic analysis of host responses to Marek’s disease virus infection in spleens of genetically resistant and susceptible chickens. Dev Comp Immunol. 2010;34:699–704. doi: 10.1016/j.dci.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Niikura M, Kim T, Hunt HD, Burnside J, Morgan RW, Dodgson JB, Cheng HH. Marek’s disease virus up-regulates major histocompatibility complex class II cell surface expression in infected cells. Virology. 2007;359:212–219. doi: 10.1016/j.virol.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Thanthrige-Don N, Read LR, Abdul-Careem MF, Mohammadi H, Mallick AI, Sharif S. Marek’s disease virus influences the expression of genes associated with IFN-gamma-inducible MHC class II expression. Viral Immunol. 2010;23:227–232. doi: 10.1089/vim.2009.0092. [DOI] [PubMed] [Google Scholar]
- Morimura T, Hattori M, Ohashi K, Sugimoto C, Onuma M. Immunomodulation of peripheral T cells in chickens infected with Marek’s disease virus: involvement in immunosuppression. J Gen Virol. 1995;76(Pt 12):2979–2985. doi: 10.1099/0022-1317-76-12-2979. [DOI] [PubMed] [Google Scholar]
- Morimura T, Ohashi K, Kon Y, Hattori M, Sugimoto C, Onuma M. Apoptosis and CD8-down-regulation in the thymus of chickens infected with Marek’s disease virus. Arch Virol. 1996;141:2243–2249. doi: 10.1007/BF01718230. [DOI] [PubMed] [Google Scholar]
- Morimura T, Ohashi K, Kon Y, Hattori M, Sugimoto C, Onuma M. Apoptosis in peripheral CD4+T cells and thymocytes by Marek’s disease virus-infection. Leukemia. 1997;11(Suppl 3):206–208. [PubMed] [Google Scholar]
- Sharma JM, Witter RL, Burmester BR. Pathogenesis of Marek’s disease in old chickens: lesion regression as the basis for age-related resistance. Infect Immun. 1973;8:715–724. doi: 10.1128/iai.8.5.715-724.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Li X, Shen Y, Qiu Y, Shi Z, Shao D, Jin Y, Chen H, Ding C, Li L. et al. The Meq oncoprotein of Marek’s disease virus interacts with p53 and inhibits its transcriptional and apoptotic activities. Virol J. 2010;7:348. doi: 10.1186/1743-422X-7-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno IM, Witter RL, Hunt HD, Lee LF, Reddy SM, Neumann U. Marek’s disease virus infection in the brain: virus replication, cellular infiltration, and major histocompatibility complex antigen expression. Vet Pathol. 2001;38:491–503. doi: 10.1354/vp.38-5-491. [DOI] [PubMed] [Google Scholar]
- Barrow AD, Burgess SC, Baigent SJ, Howes K, Nair VK. Infection of macrophages by a lymphotropic herpesvirus: a new tropism for Marek’s disease virus. J Gen Virol. 2003;84:2635–2645. doi: 10.1099/vir.0.19206-0. [DOI] [PubMed] [Google Scholar]
- Frascaroli G, Varani S, Blankenhorn N, Pretsch R, Bacher M, Leng L, Bucala R, Landini MP, Mertens T. Human cytomegalovirus paralyzes macrophage motility through down-regulation of chemokine receptors, reorganization of the cytoskeleton, and release of macrophage migration inhibitory factor. J Immunol. 2009;182:477–488. doi: 10.4049/jimmunol.182.1.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regis EG, Barreto-de-Souza V, Morgado MG, Bozza MT, Leng L, Bucala R, Bou-Habib DC. Elevated levels of macrophage migration inhibitory factor (MIF) in the plasma of HIV-1-infected patients and in HIV-1-infected cell cultures: a relevant role on viral replication. Virology. 2010;399:31–38. doi: 10.1016/j.virol.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. Role of annexin A2 in the production of infectious hepatitis C virus particles. J Virol. 2010;84:5775–5789. doi: 10.1128/JVI.02343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni A, Mehrotra B, Baumann A, Adam SA, Wingfield PT, Olson MO. Nucleolar protein B23 stimulates nuclear import of the HIV-1 Rev protein and NLS-conjugated albumin. Biochemistry. 1997;36:3941–3949. doi: 10.1021/bi9627931. [DOI] [PubMed] [Google Scholar]
- Li YP. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J Virol. 1997;71:4098–4102. doi: 10.1128/jvi.71.5.4098-4102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch AD, Weller SK. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J Virol. 2005;79:10740–10749. doi: 10.1128/JVI.79.16.10740-10749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Barlow EA, Ma S, Hagemeier SR, Duellman SJ, Burgess RR, Tellam J, Khanna R, Kenney SC. Hsp90 inhibitors block outgrowth of EBV-infected malignant cells in vitro and in vivo through an EBNA1-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:3146–3151. doi: 10.1073/pnas.0910717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen KW, Damania B. Hsp90 and Hsp40/Erdj3 are required for the expression and anti-apoptotic function of KSHV K1. Oncogene. 2010;29:3532–3544. doi: 10.1038/onc.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher M, Metz CN, Calandra T, Mayer K, Chesney J, Lohoff M, Gemsa D, Donnelly T, Bucala R. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc Natl Acad Sci U S A. 1996;93:7849–7854. doi: 10.1073/pnas.93.15.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaster H, Bernhagen J, Calandra T, Bucala R. The macrophage migration inhibitory factor-glucocorticoid dyad: regulation of inflammation and immunity. Mol Endocrinol. 2007;21:1267–1280. doi: 10.1210/me.2007-0065. [DOI] [PubMed] [Google Scholar]
- Roger T, Chanson AL, Knaup-Reymond M, Calandra T. Macrophage migration inhibitory factor promotes innate immune responses by suppressing glucocorticoid-induced expression of mitogen-activated protein kinase phosphatase-1. Eur J Immunol. 2005;35:3405–3413. doi: 10.1002/eji.200535413. [DOI] [PubMed] [Google Scholar]
- Aeberli D, Yang Y, Mansell A, Santos L, Leech M, Morand EF. Endogenous macrophage migration inhibitory factor modulates glucocorticoid sensitivity in macrophages via effects on MAP kinase phosphatase-1 and p38 MAP kinase. FEBS Lett. 2006;580:974–981. doi: 10.1016/j.febslet.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Toh ML, Aeberli D, Lacey D, Yang Y, Santos LL, Clarkson M, Sharma L, Clyne C, Morand EF. Regulation of IL-1 and TNF receptor expression and function by endogenous macrophage migration inhibitory factor. J Immunol. 2006;177:4818–4825. doi: 10.4049/jimmunol.177.7.4818. [DOI] [PubMed] [Google Scholar]
- D’Acquisto F, Perretti M, Flower RJ. Annexin-A1: a pivotal regulator of the innate and adaptive immune systems. Br J Pharmacol. 2008;155:152–169. doi: 10.1038/bjp.2008.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Acquisto F. On the adaptive nature of annexin-A1. Curr Opin Pharmacol. 2009;9:521–528. doi: 10.1016/j.coph.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu Rev Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- Javid B, MacAry PA, Lehner PJ. Structure and function: heat shock proteins and adaptive immunity. J Immunol. 2007;179:2035–2040. doi: 10.4049/jimmunol.179.4.2035. [DOI] [PubMed] [Google Scholar]
- Kunisawa J, Shastri N. Hsp90alpha chaperones large C-terminally extended proteolytic intermediates in the MHC class I antigen processing pathway. Immunity. 2006;24:523–534. doi: 10.1016/j.immuni.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Levy AM, Davidson I, Burgess SC, Dan Heller E. Major histocompatibility complex class I is downregulated in Marek’s disease virus infected chicken embryo fibroblasts and corrected by chicken interferon. Comp Immunol Microbiol Infect Dis. 2003;26:189–198. doi: 10.1016/S0147-9571(02)00055-3. [DOI] [PubMed] [Google Scholar]
- Jarosinski KW, Hunt HD, Osterrieder N. Down-regulation of MHC class I by the Marek’s disease virus (MDV) UL49.5 gene product mildly affects virulence in a haplotype-specific fashion. Virology. 2010;405:457–463. doi: 10.1016/j.virol.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Mitchell RA, Liao H, Chesney J, Fingerle-Rowson G, Baugh J, David J, Bucala R. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci U S A. 2002;99:345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. Control of p53 and NF-kappaB signaling by WIP1 and MIF: role in cellular senescence and organismal aging. Cell Signal. 2011;23:747–752. doi: 10.1016/j.cellsig.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Venugopal K, Payne LN. Molecular pathogenesis of Marek’s disease-recent developments. Avian Pathol. 1995;24:597–609. doi: 10.1080/03079459508419100. [DOI] [PubMed] [Google Scholar]
- Johnsen JI, Aurelio ON, Kwaja Z, Jorgensen GE, Pellegata NS, Plattner R, Stanbridge EJ, Cajot JF. p53-mediated negative regulation of stathmin/Op18 expression is associated with G(2)/M cell-cycle arrest. Int J Cancer. 2000;88:685–691. doi: 10.1002/1097-0215(20001201)88:5<685::AID-IJC1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Fang L, Min L, Lin Y, Ping G, Rui W, Ying Z, Xi W, Ting H, Li L, Ke D. et al. Downregulation of stathmin expression is mediated directly by Egr1 and associated with p53 activity in lung cancer cell line A549. Cell Signal. 2010;22:166–173. doi: 10.1016/j.cellsig.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Arur S, Uche UE, Rezaul K, Fong M, Scranton V, Cowan AE, Mohler W, Han DK. Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev Cell. 2003;4:587–598. doi: 10.1016/S1534-5807(03)00090-X. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Murata H, Sonegawa H, Sakaguchi Y, Futami J, Kitazoe M, Yamada H, Huh NH. Truncation of annexin A1 is a regulatory lever for linking epidermal growth factor signaling with cytosolic phospholipase A2 in normal and malignant squamous epithelial cells. J Biol Chem. 2007;282:35679–35686. doi: 10.1074/jbc.M707538200. [DOI] [PubMed] [Google Scholar]
- Solito E, de Coupade C, Canaider S, Goulding NJ, Perretti M. Transfection of annexin 1 in monocytic cells produces a high degree of spontaneous and stimulated apoptosis associated with caspase-3 activation. Br J Pharmacol. 2001;133:217–228. doi: 10.1038/sj.bjp.0704054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debret R, El Btaouri H, Duca L, Rahman I, Radke S, Haye B, Sallenave JM, Antonicelli F. Annexin A1 processing is associated with caspase-dependent apoptosis in BZR cells. FEBS Lett. 2003;546:195–202. doi: 10.1016/S0014-5793(03)00570-2. [DOI] [PubMed] [Google Scholar]
- Tabe Y, Jin L, Contractor R, Gold D, Ruvolo P, Radke S, Xu Y, Tsutusmi-Ishii Y, Miyake K, Miyake N. et al. Novel role of HDAC inhibitors in AML1/ETO AML cells: activation of apoptosis and phagocytosis through induction of annexin A1. Cell Death Differ. 2007;14:1443–1456. doi: 10.1038/sj.cdd.4402139. [DOI] [PubMed] [Google Scholar]
- Mitchell RA, Bucala R. Tumor growth-promoting properties of macrophage migration inhibitory factor (MIF) Semin Cancer Biol. 2000;10:359–366. doi: 10.1006/scbi.2000.0328. [DOI] [PubMed] [Google Scholar]
- Bifulco C, McDaniel K, Leng L, Bucala R. Tumor growth-promoting properties of macrophage migration inhibitory factor. Curr Pharm Des. 2008;14:3790–3801. doi: 10.2174/138161208786898608. [DOI] [PubMed] [Google Scholar]
- Conroy H, Mawhinney L, Donnelly SC. Inflammation and cancer: macrophage migration inhibitory factor (MIF)–the potential missing link. QJM. 2010;103:831–836. doi: 10.1093/qjmed/hcq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendon BE, Willer SS, Zundel W, Mitchell RA. Mechanisms of macrophage migration inhibitory factor (MIF)-dependent tumor microenvironmental adaptation. Exp Mol Pathol. 2009;86:180–185. doi: 10.1016/j.yexmp.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussunoor S, Murray GI. The role of annexins in tumour development and progression. J Pathol. 2008;216:131–140. doi: 10.1002/path.2400. [DOI] [PubMed] [Google Scholar]
- Zhu F, Xu C, Jiang Z, Jin M, Wang L, Zeng S, Teng L, Cao J. Nuclear localization of annexin A1 correlates with advanced disease and peritoneal dissemination in patients with gastric carcinoma. Anat Rec (Hoboken) 2010;293:1310–1314. doi: 10.1002/ar.21176. [DOI] [PubMed] [Google Scholar]
- Rondepierre F, Bouchon B, Papon J, Bonnet-Duquennoy M, Kintossou R, Moins N, Maublant J, Madelmont JC, D’Incan M, Degoul F. Proteomic studies of B16 lines: involvement of annexin A1 in melanoma dissemination. Biochim Biophys Acta. 2009;1794:61–69. doi: 10.1016/j.bbapap.2008.09.014. [DOI] [PubMed] [Google Scholar]
- de Graauw M, van Miltenburg MH, Schmidt MK, Pont C, Lalai R, Kartopawiro J, Pardali E, Le Devedec SE, Smit VT, van der Wal A. et al. Annexin A1 regulates TGF-beta signaling and promotes metastasis formation of basal-like breast cancer cells. Proc Natl Acad Sci U S A. 2010;107:6340–6345. doi: 10.1073/pnas.0913360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Xu XY, Chen H, Gao WC, Ruan CP, Wang Q, Sun YP. Increased expression of annexin A1 is correlated with K-ras mutation in colorectal cancer. Tohoku J Exp Med. 2010;222:243–250. doi: 10.1620/tjem.222.243. [DOI] [PubMed] [Google Scholar]
- Yi M, Schnitzer JE. Impaired tumor growth, metastasis, angiogenesis and wound healing in annexin A1-null mice. Proc Natl Acad Sci U S A. 2009;106:17886–17891. doi: 10.1073/pnas.0901324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Dutsch-Wicherek M. RCAS1, MT, and vimentin as potential markers of tumor microenvironment remodeling. Am J Reprod Immunol. 2010;63:181–188. doi: 10.1111/j.1600-0897.2009.00803.x. [DOI] [PubMed] [Google Scholar]
- Bunnell TM, Burbach BJ, Shimizu Y, Ervasti JM. beta-Actin specifically controls cell growth, migration and the G-actin pool. Mol Biol Cell. 2011;22:4047–4058. doi: 10.1091/mbc.E11-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow A, Nowak D, Malicka-Blaszkiewicz M. Actin cytoskeleton and beta-actin expression in correlation with higher invasiveness of selected hepatoma Morris 5123 cells. J Physiol Pharmacol. 2006;57(Suppl 7):111–123. [PubMed] [Google Scholar]
- Okuwaki M. The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein. J Biochem. 2008;143:441–448. doi: 10.1093/jb/mvm222. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kurian D, Xu H, Petherbridge L, Smith LP, Hunt L, Nair V. Interaction of Marek’s disease virus oncoprotein Meq with heat-shock protein 70 in lymphoid tumour cells. J Gen Virol. 2009;90:2201–2208. doi: 10.1099/vir.0.012062-0. [DOI] [PubMed] [Google Scholar]
- Russell SE, Hall PA. Do septins have a role in cancer? Br J Cancer. 2005;93:499–503. doi: 10.1038/sj.bjc.6602753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeseler S, Sandrock K, Bartsch I, Zieger B. Septins, a novel group of GTP-binding proteins: relevance in hemostasis, neuropathology and oncogenesis. Klin Padiatr. 2009;221:150–155. doi: 10.1055/s-0029-1220706. [DOI] [PubMed] [Google Scholar]
- Amir S, Wang R, Simons JW, Mabjeesh NJ. SEPT9_v1 up-regulates hypoxia-inducible factor 1 by preventing its RACK1-mediated degradation. J Biol Chem. 2009;284:11142–11151. doi: 10.1074/jbc.M808348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Makarova O, Peterson EA, Privette LM, Petty EM. Up-regulation of SEPT9_v1 stabilizes c-Jun-N-terminal kinase and contributes to its pro-proliferative activity in mammary epithelial cells. Cell Signal. 2009;21:477–487. doi: 10.1016/j.cellsig.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B, MacKay C, Alnabulsi A, MacKay M, Telfer C, Melvin WT, Murray GI. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim Biophys Acta. 2006;1765:85–100. doi: 10.1016/j.bbcan.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Ford LP, Wright WE, Shay JW. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21:580–583. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Kim S, Miska KB, Jenkins MC, Fetterer RH, Cox CM, Stuard LH, Dalloul RA. Molecular cloning and functional characterization of the avian macrophage migration inhibitory factor (MIF) Dev Comp Immunol. 2010;34:1021–1032. doi: 10.1016/j.dci.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Abdul-Careem MF, Hunter BD, Nagy E, Read LR, Sanei B, Spencer JL, Sharif S. Development of a real-time PCR assay using SYBR Green chemistry for monitoring Marek’s disease virus genome load in feather tips. J Virol Methods. 2006;133:34–40. doi: 10.1016/j.jviromet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]