Abstract
When treated with the steroid hormone 20-hydroxyecdysone (20E), C7-10 cells from the mosquito, Aedes albopictus, arrest in the G1 phase of the cell cycle. To explore whether 20E-mediated cell cycle arrest proceeds through increased levels of cell cycle inhibitor (CKI) proteins, we cloned the Ae. albopictus homolog of dacapo, the single member of the Cip/Kip family of CKI proteins known from Drosophila melanogaster. The Ae. albopictus dacapo cDNA encoded a 261-amino acid homolog of the Aedes aegypti protein XP_001651102.1, which is encoded by an ~23 kb gene containing three exons. Like dacapo from D. melanogaster, the ~27 kDa protein from Aedes and Culex mosquitoes contained several S/TXXE/D motifs corresponding to potential protein kinase CK2 phosphorylation sites, and a binding site for proliferating cell nuclear antigen (PCNA). When extracts from cells treated with 20E were analyzed by western blotting, using a primary antibody to synthetic peptides from the mosquito dacapo protein, up-regulation of an ~27 kDa protein was observed within 24 h, and the abundance of the protein further increased by 48 h after hormone treatment. This is the first investigation of a cell cycle inhibitory protein in mosquitoes. The results reinforce growing evidence that 20E affects expression of proteins that regulate cell cycle progression.
Keywords: mosquito, insect cell line, cell cycle, dacapo, 20-hydroxyecdysone, G1 arrest
INTRODUCTION
Proliferation of 20-hydroxyecdysone (20E)-responsive insect cell lines is typically depressed in response to hormone treatment. In the Kc cell line from Drosophila melanogaster, and in some lepidopteran cells, 20E-treated cells arrest in the G2 phase of the cell cycle, after DNA synthesis has been completed (Besson et al., 1987; Auzoux-Bordenave et al., 2002; Siaussat et al., 2005, 2008). In contrast, 20E arrests proliferation of the C7-10 cell line from the mosquito, Aedes albopictus, in G1, and levels of cyclin A, which is required for progression into the S phase of the cycle, decrease in 20E-treated cells (Gerenday and Fallon, 2004). To better understand how 20E interacts with the cell cycle, we have begun to investigate proteins that participate in cell cycle control and are likely to be up-regulated in C7-10 cells after treatment with 20E.
In mammalian cells, G1 arrest is accompanied by accumulation of cyclin-dependent kinase inhibitor (CKI) proteins (Coats et al., 1996). CKIs that interact with cyclins D, E, A and B, and their associated cyclin dependent kinases (CDKs), belong to the Cip/Kip family, and include p21Cip/Waf1/Sdi1 (p21), p27Kip1 (p27) and p57Kip2 (p57), which were initially thought to be tumor suppressors (Besson et al., 2008). Cip/Kip proteins share a conserved N-terminal domain that interacts with cyclins and CDKs that participate in the G1/S transition (Liu et al., 2002). In mammalian cells, p27 modulates cell cycle arrest/reentry in a tissue specific manner, and changes in abundance of p27 have been noted in cells treated with estrogen and progesterone (Musgrove et al., 1998). As a basis for this study, we hypothesized that mosquito homologs of the mammalian Cip/Kip proteins become more abundant as C7-10 cells arrest in response to 20E.
The Drosophila melanogaster genome encodes a single Cip/Kip protein known as dacapo (CG1772; synonyms include dap, CDI4, p27Dap, and p21dacapo), which has a predicted mass of ~27kDa. Absence of dacapo results in an embryonic-lethal phenotype, and loss-of-function mutants have an extra division cycle in embryonic epidermal cells. Conversely, premature expression of dacapo causes precocious G1 arrest, supporting the suggestion that dacapo coordinates exit from the cell cycle with terminal differentiation (De Nooij et al., 1996; Lane et al., 1996). In Drosophila oocytes, dacapo maintains a state of arrest in meiotic prophase II, and oscillation of dacapo abundance relative to levels of cyclin E regulates endocycling of the nurse cells (Hong et al., 2007).
We took advantage of limited amino acid sequence homologies between D. melanogaster dacapo and a hypothetical Ae. aegypti protein, to obtain cDNA encoding the Ae. albopictus dacapo. We used the deduced amino acid sequence of Ae. albopictus dacapo to identify short conserved peptides for production of polyclonal antibody, showed that the antibody reacts with a ~27 kDa protein in extracts from Ae. albopictus cells, and demonstrated that the abundance of this protein increases in cells treated with 20E.
MATERIALS AND METHODS
Reagents
Deoxyribonucleotides and Taq DNA polymerase were from Promega (Madison, WI); Bio-Rad protein assay and Freeze “N” Squeeze DNA gel extraction spin columns were from Bio-Rad Laboratories (Hercules, CA), and Micro-Fast Track 2.0 kit, SuperScript® III First-Strand synthesis system, and pEXP5-NT/TOPO TA cloning vector and PCR primers were from Invitrogen Corporation (Carlsbad, CA). Primary antibody against Ae. albopictus dacapo was prepared by New England Peptide LLC (Gardner, MS) from rabbits immunized with a mixture of two synthetic peptides, Ac-VDKQESKKFIDRQLAC-amide and Ac-RITDFLKESKRLSPGS-amide. Western blots were probed with goat anti-rabbit IgG conjugated with horseradish peroxidase as the secondary antibody, and developed using Super Signal West Pico, from Thermo Scientific, Waltham, MA. 20E from Sigma-Aldrich (St. Louis, MO) was prepared in 10% ethanol as described previously (Gerenday and Fallon, 2004). Components for cell culture media were from Invitrogen, and fetal bovine serum from Atlanta Biologicals was heat-treated at 56°C.
Cells and Culture Conditions
Cells were maintained in Eagle’s minimal essential medium supplemented with vitamins, glutamine, nonessential amino acids, antibiotics, and 5% heat-inactivated fetal bovine serum as described by Shih et al. (1998). Medium with 5% serum is called E-5 medium. For expression experiments, medium was aspirated from monolayer cultures, and replaced with fresh medium, with or without 20E.
cDNA Synthesis
Total RNA was prepared from C7-10 cells using guanidine isothiocyanate (Davis et al., 1986), and mRNA was prepared from total RNA using the Micro-Fast Track 2.0 kit. cDNA was prepared using the Superscript III First-Strand Synthesis system following the manufacturer’s instructions. PCR products were recovered from agarose gels using Squeeze “N” freeze columns, and a portion of the flow through was sequenced at the University of Minnesota BioMedical Genomics center.
PCR Reactions
PCR reactions were carried out on a Techne TC-312 thermo cycler. Initial reactions with primer pairs F20/R23 and F23/Oligo(dT) included an initial denaturation at 94°C, followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 63°C for 30 sec, and elongation at 72°C for 2 min, with a final extension at 72°C for 10 min. Full length dacapo cDNA was obtained using Platinum Taq DNA Polymerase High Fidelity with an initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 65°C for 30 sec, and elongation at 68°C for 2 min, with a final extension at 68°C for 10 min. The product was recovered from an agarose gel and cloned into pEXP5-NT/TOPO TA vector.
Western Blots
For detection of dacapo by Western blotting, cells (2×106 in 12 ml) were plated in E-5 medium in 100 mm diameter tissue culture dishes. After 48 h, the medium was replaced with fresh E-5 with or without 20E at a final concentration of 2×10−6 M. Cells were collected 24 and 48 h later and lysed in RIPA buffer containing proteinase inhibitors as described previously (Gerenday and Fallon, 2004). Cell pellets were disrupted by sonication, and total protein was assayed using the Bio-Rad Protein Assay. Approximately 40 μg/lane was electrophoresed on 12% polyacrylamide minigels with 1.5 mm spacers.
Gels were blotted onto nitrocellulose membrane using a Mini-Genie Blotter (Idea Scientific, Minneapolis, MN) at 12 volts for 1 h at room temperature using Tris-glycine transfer buffer containing 3.03 g of Tris base, 14.4 g glycine, and 20% methanol. The membrane was incubated in blocking solution made by dissolving 5% dry milk in TBST (20 mM Tris-HCl, pH 7.5, 0.5M NaCl, 0.05% Tween-20) for 1 h at room temperature, after which primary antibody (1:2,500 dilution) was added directly to the blocking solution. Incubation was continued overnight at 10°C with gentle rotation. The membrane was briefly rinsed three times with TBST at room temperature, and washed in fresh TBST for three consecutive 5 min periods, followed by a fourth wash in TBST for 15 min. The membrane was incubated with secondary antibody in blocking solution for 1 h at room temperature, and the membrane was washed as described above. Blots were developed using Super Signal West Pico (Thermo Scientific) according to the manufacturer’s instructions. The image was visualized by exposure to Kodak Biomax Light Film for 3–5 min.
RESULTS
Ae. albopictus Dacapo cDNA
The D. melanogaster genome contains a single gene (CG1772) encoding a homolog of mammalian Cip/Kip CKI proteins. The CG1772 sequence spans 3.8 kb, and encodes a 2.4 kb mRNA, encoding a protein called dacapo. The dacapo gene has three exons, with a short 5′-intron separating exons 1 and 2, and an approximately 1.3 kb intron separating exons 2 and 3. Initial BLAST searches of the An. gambiae genome database with dacapo-based nucleotide or protein queries failed to reveal sufficient homology to support recovery of a mosquito homolog from C7-10 cells. After publication of the Ae. aegypti genome, a tblastn search recovered two separate peptides that allowed design of primers for PCR-based recovery of the Ae. albopictus homolog. These peptides (boxed in Fig. 1) lie within hypothetical Ae. aegypti protein XP_001651102. With introduction of gaps, XP_001651102 was 36% identical to dacapo from D. melanogaster, with 51% positive amino acids over 237 residues, and an E value of 7e–21. Residues 40 to 90 corresponded to a conserved CDI (CKI) domain in NCBI CDD pfam 02234.
Figure 1.

Alignment of dacapo from D. melanogaster and the hypothetical homolog XP_001651102 from Ae. aegypti. Boxes indicate conserved peptides used to design PCR primers based on the Ae. aegypti nucleotide sequence (see Fig. 2).
We used primers based on Ae. aegypti nucleotide sequences and oligo(dT) to obtain the dacapo cDNA from Ae. albopictus (Fig. 2). Initially, PCR products from primer pairs F20/R23 and F23/oligo(dT) were excised from agarose gels, sequenced, and compared with the genomic DNA sequence from Ae. aegypti. To synthesize the full length cDNA from Ae. albopictus, we used primer F20 with the specific reverse primer R20, based on nucleotide sequence downstream of the stop codon (Fig. 2).
Figure 2.
Alignment of dacapo cDNAs from Ae. Albopictus, Ae. aegypti, and Culex pipiens. Arrows indicate PCR primers used to obtain the full-length Ae. albopictus cDNA. The boxed TAG is the stop codon.
A search with the full-length Ae. albopictus cDNA against sequences in the insect nr database (taxid 50557) by discontinuous megablast (Altschul et al., 1997) gave matches to partial mRNA XM_001651052.1 from Ae. aegypti (E value 0.0), and XM_001847110.1 from Culex quinquefasciatus (9e–56), but failed to find matches with nucleotide sequence data from Anopheles mosquitoes. As expected, nucleotide identities were higher between the two Aedes mosquitoes, relative to Cx. quinquefasciatus, and most of the short insertions in the Culex sequence were multiples of three nucleotides. Two stretches of Culex sequence (nt 1–403 and nt 571–658) are shown in Figure 2. With TBLASTX we identified XM_001651052.1 from Ae. aegypti (2e–84), and XM_001847110.1 from Cx. quinquefasciatus (5e–48), as well as several matches with members of the Drosophilidae, and with the human body louse (4e–5).
Aedes albopictus Dacapo Protein
The conceptual Ae. albopictus dacapo protein contained 261 amino acids, with 76% identity, 84% similarity to its Ae. aegypti homolog; 55% identity, 72% similarity to the Cx. quinquefasciatus homolog; 37% identity, 53% similarity to D. melanogaster, and 36% identity, 52% similarity to Chymomyza costata protein sequences. Note that C. costata is in the family Drosophilidae, and that several dacapo homologs in various species of Drosophila are not included in the alignment.
Our search for Dipteran homologs of Ae. albopictus dacapo uncovered hypothetical protein AND-15659 from Anopheles darlingi. Although this protein has N-terminal sequence indicative of a member of the CDI superfamily, its limited similarity to dacapo proteins from either the Culicidae or the Drosophilidae (Fig. 3) suggest that it is not the Anopheles homolog of the Ae. albopictus dacapo described here. Likewise, generation of a tree from all dacapo sequences described for Insecta (taxid 50557) with the NCBI multiple alignment tool, COBALT, showed that the An. darlingi protein had less homology to the other mosquito proteins than the multiple dacapo proteins in various Drosophila species (Fig. 4). This inconsistency with the known phylogeny of the Diptera raises the possibility that mosquitoes express an additional CKI protein reminiscent of the multiple Cip/Kip proteins in mammals. Outside the diptera, putative dacapo homologs from the beetle, Tribolium castaneum (XP_976393.1) and from An. gambiae (XP_307826.3) were too short to align accurately.
Figure 3.
Alignment of dacapo protein sequences from Ae. albopictus, Ae. aegypti (XP_001651102.1), Culex quinquefasciatus (XP_001847162.1), D. melanogaster (gbAAB36557.1), Chymomyza costata (gbACT79565.1) and putative dacapo homologs from An. gambiae (XP_307826.3) and An. darlingi (AND_15659). The alignment was done with ClustalW (1.81) at the KEGG (Kyoto University Bioinformatics Center) website using the slow/ accurate method and the following parameters: Pairwise settings: Gap open penalty 35; gap extension penalty 0.3; Multiple alignment settings: gap open penalty 15; gap extension penalty 0.3 with weight transition and hydrophilic gaps off. Dots were substituted to represent identities to the Ae. albopictus sequence at top and dashes indicate gaps. Open boxes designate S/TXXE/D CD2 motifs. The shaded box indicates a PCNA binding domain.
Figure 4.
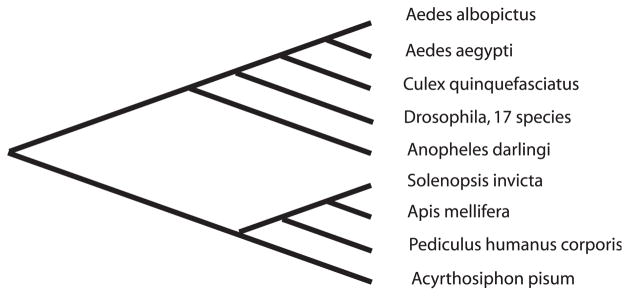
Relationships among decapo proteins from the Insecta (Taxid 50577). Proteins were obtained by BLASTP at the NCBI website and the tree was produced using COBALT (Papadopoulos and Agarwala, 2007). Accessions for the Diptera are given in the legend to Figure 3; the An. gambiae sequence was not retrieved in this search and 17 Drosophila sequences show as a single branch on the tree. Accessions were EFZ13437 for the fire ant Solenopsis invicta, XP_003251288 for the honeybee Apis mellifera, XP_002424234 for the body louse Pediculus humanus corporis, and XP_001947107 for the aphid Acyrthosiphon pisum.
In contrast to mammalian p27 proteins, which share a single strict consensus CD2 phosphorylation site (S/TXXE/D) at serine-83 (Tapia et al., 2004), Ae. albopictus dacapo contains four S/TXXE/D motifs that are potentially phosphorylated by protein kinase CK2. These lie at residues 10–13 and 116–119; two tandem motifs occur at residues 160–163; 164–168. With the exception of An. gambiae and An. darlingi, the tandem motifs are conserved among the Diptera, and in the Drosophilidae, a shared motif is displaced by only three amino acids from the 116–119 motif in Aedes and Culex mosquitoes (Fig. 3). In general, however, the positions of additional S/TXXE/D boxes in Drosophila and Chymomyza differ from those of the mosquito homologs. The abundance of potential CK2 phosphorylation sites in the Dipteran proteins, relative to mammalian p27, may reflect a multiplicity of functions in a single Cip/Kip protein, relative to three Cip/Kip proteins in mammals. For example, in Drosophila, dacapo is involved in embryonic functions that resemble those of mammalian p57.
Finally, dacapo proteins from both Culicidae and Drosophilidae contain a proliferating cell nuclear antigen (PCNA) binding motif toward the C-terminal end of the protein (Fig. 3; Warbrick et al., 1988). Among the mammalian CKIs, PCNA binding has been observed only with p21, but not with p27 and p57 (Parekh et al., 1997). The presence of this site in dacapo proteins supports the possibility that it has multiple functions, relative to mammalian p27.
Dacapo Gene Structure in Ae. aegypti
We used the Ae. albopictus cDNA sequence to deduce the exon–intron organization of the 23 kb gene encoding Ae. aegypti XP_001651102.1. Because annotation of the Ae. aegypti gene is available in the VB-2011-02 release of VectorBase, under Ae. aegypti transcript AAEL005580-RA, the details of our analysis will not be described here. Note, however, that the introns in the Ae. aegypti gene measure ~6 and 16 kb, and are substantially longer than the corresponding introns in the D. melanogaster gene. As would be expected based on the cDNA alignment (Fig. 2), the highest level of conservation occurs within the first exon and 5′-half of second exon, whereas the third exon shows more divergence. Ae. albopictus nt 1–152 also recovered a 72% identity in the Culex pipiens genome, corresponding to a hypothetical mRNA with an estimated size greater than 5 kb, encoding hypothetical protein XP_001847162, shown in Figure 3. Matches to other Dipteran sequences, including those of An. gambiae, were short, with only 3–8% coverage.
Dacapo Expression in 20E-Treated Cells
We used Western blots to examine whether treatment with 20E affected levels of Ae. albopictus dacapo protein. In preliminary experiments, we established that antibody to Ae. albopictus dacapo synthetic peptides detected a band at ~27 kDa, consistent with the calculated mass (27,061 kDa) for the translation product. In pilot studies (not shown), we verified that signal intensity increased with protein amount in the range of 40–70 μg of total protein per lane on mini-gels. With 40 μg samples, dacapo signal increased in normally growing cells as they approached confluency, and also increased when cells were incubated in nutrient-depleted medium. Likewise, dacapo signal decreased when nutrient-starved cells were refed. Thus, in the absence of 20E, dacapo signal intensity reflected growth conditions, increased with conditions associated with cell cycle inhibition (age of the culture), and decreased with cell cycle reentry (nutrient replenishment).
We evaluated decapo expression using cells grown in hormone-free medium for 48 h, then treated with 2×10−6M 20E. Hormone was added directly to the original culture medium (Fig. 5, lanes 2 and 6), and control cells were maintained without hormone (Fig. 5, lanes 1 and 5). To ensure that dacapo expression represented a bona fide response to 20E, rather than nutrient depletion, a second set of cells was refed with fresh medium containing 20E (Fig. 5, lanes 4 and 8) or lacking 20E (Fig. 5, lanes 3 and 7). Cells were harvested and assayed for dacapo expression by western blot after 24 (Fig. 5, lanes 1–4) or 48 h (Fig. 5, lanes 5–8). Dacapo levels were low but detectable in the cells without 20E, with a slight increase between 24 and 48 h. In the presence of 20E, dacapo levels showed a consistent increase, which was enhanced after 48, relative to 24 h of treatment. The amount of dacapo protein was similar regardless of medium replacement, indicating that the cells were growing exponentially, in the presence of adequate nutrients.
Figure 5.
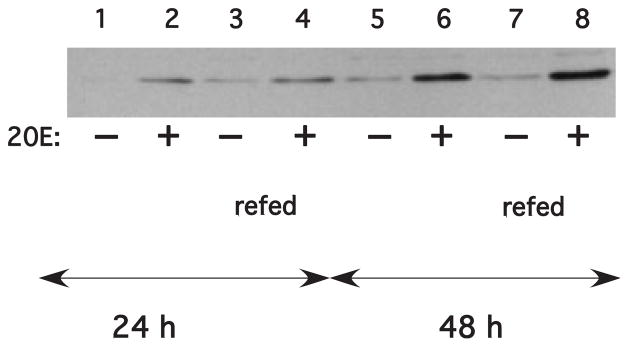
Effect of 20E on dacapo abundance. Cells were seeded as detailed in the Materials and Methods and incubated in the presence (+) or absence (−) of 2×10−6M 20E for 24 or 48 h. For cells in lanes 1, 2, 5, and 6 20E was added to the growth medium; in lanes 3, 4, 7, and 8 the medium was replaced with fresh E-5 medium containing 2×10−6 M 20E.
DISCUSSION
Ecdysone Responses in Mosquito Cells
Although their responses differ in detail, proliferation of 20E-responsive insect cell lines is invariably inhibited by hormone treatment. Consistent with its negative effect on the cell cycle, only a small number of proteins have been shown to be up-regulated after 20E treatment (for a review, see Echalier, 1997), and among different cell lines, identities of ecdysone-inducible proteins (EIPs) fail to converge on consensus regulatory molecules/pathways that provide a paradigm for understanding how the 20E response engages the cell cycle machinery.
Pathways that link steroid hormone receptor to cell cycle machinery are best understood in mammalian cells that respond to estrogen and progestin in the context of breast cancer, where detailed exploration of regulatory protein expression has potential therapeutic implications. Insofar as 20E-treated C7-10 mosquito cells complete the ongoing cycle and divide before arresting in G1 (Gerenday and Fallon, 2004; Fallon and Gerenday, 2010), their response to 20E resembles the biphasic effects of progesterone (Musgrove et al., 1991) more closely than the stimulatory effects of estrogen (for a review, see Butt et al., 2008). The diversity of phenotypes exhibited by steroid-responsive mammalian cell lines underscores the potential complexity of 20E-response pathways in insects and provides a context for understanding differences between the few insect cell lines in which these responses have been investigated.
Among 11 EIPs detected in our earlier analysis of Ae. albopictus cells (Lan et al., 1993), a ~26 kDa protein from total and cytoplasmic extracts migrated to a position consistent with the pI (9.8) calculated from the Ae. albopictus dacapo cDNA described in this study. Because we excluded the possibility that this protein was one of the small heat shock proteins that are induced in some 20E-responsive Drosophila cell lines (Ireland and Berger, 1982), it will be of interest to reexamine whether this protein is dacapo. Likewise, given that Ae. albopictus dacapo has a PCNA binding site, a ~29 kDa protein recovered by co-immunoprecipitation with PCNA (Ma et al., 2006) also merits re-investigation using antibodies to dacapo.
A second up-regulated EIP in Ae. albopictus cells is a 52 kDa protein homolog of D. melanogaster gene CG17337, which is conserved in An. gambiae, as well as in more distantly related organisms (Eccleston et al., 2002). Although little more has been learned of this protein’s function, its putative role in proteolysis reduces the likelihood that 52 kDa EIP is a second mosquito CKI or a core component of the cell cycle machinery.
Dacapo in Anopheles Mosquitoes
It remains unclear whether a homolog of Ae. albopictus dacapo has been annotated in Anopheles mosquitoes. Based on evolutionary considerations, we expected to uncover evidence for an Anopheles protein similar to dacapo from D. melanogaster, but with a sequence more closely related to the Aedes and Culex proteins, relative to the Drosophila homolog. The closest Anopheles gambiae candidate was a partial, 81 amino acid peptide (XP_307826.3) with good homology with the dacapo PCNA binding domain near the C-terminal end of the protein (Fig. 3). However, this protein fragment did not include upstream sequence corresponding to the better-conserved N-terminal domain of CKI proteins. The closest full-length match to dacapo from Aedes and Culex was hypothetical protein AND_15659 from An. darlingi (Fig. 3), a 366 amino acid protein with ~40% amino acid identity in a CDI domain spanning residues 40–90. Inspection of the alignment in Figure 3 indicates that this An. darlingi protein is ~100 residues longer than Ae. albopictus dacapo, and lacks the PCNA binding motif. This poor alignment between the An. darlingi protein and the other dacapo proteins (Figs. 3 and 4) raises the possibility that there may be multiple CKIs in mosquitoes.
A tblastn search with this An. darlingi query further uncovered a 1,410-nucleotide mRNA sequence in An. funestris (Afun003500). Using this mRNA sequence as query in a discontiguous megablast against Est_others, we pulled out short sequences with ~85% identity to An stephensi (Accession FL 484363) and An. gambiae (BM635733, corresponding to XM_310511.2), which encodes a 61 amino acid protein with two ~15 amino acid stretches that align (with gaps) to the Ae. albopictus dacapo upstream of the 81 amino acid An. gambiae sequence (XP_307826.3) containing the PCNA binding domain shown in Figure 3. Although the An. gambiae database assigns these short homologies to different proteins on chromosomes X (XM_310511.2) and 3L (XP_307826.3) respectively, these short matches support the likelihood that Anopheles genomes encode a dacapo homolog, perhaps in a region of genomic DNA for which annotation is incomplete. Finally, we note that the gene roughex, which encodes a putative cyclin A inhibitor protein with limited structural similarity to dacapo, is evolving rapidly in Drosophila (Avedisov et al., 2001), and no clear homologies to the roughex protein have yet been described in mosquitoes.
Dacapo and the Cell Cycle
Aside from efforts to elucidate the structure and evolution of dacapo genes in insects, the involvement of dacapo and other cell cycle regulatory proteins in insect growth and metamorphosis remain to be explored. Before embryogenesis, dacapo is involved in mitotic to endocycle transitions in ovarian follicle cells (Shcherbata et al., 2004) as well as in the oocyte and nurse cells (Hong et al., 2003, 2007). During Drosophila embryogenesis, regulation of dacapo involves a complex assembly of cis-acting factors, and its expression in the embryo is not necessarily coupled with cell cycle progression (Meyer et al., 2002). Because maternal effects may influence embryogenesis, it will be of interest to examine expression of cell cycle regulatory proteins in imaginal discs or larval epidermis, particularly during their response to 20E. Insofar as cell cycling is influenced by nutrient availability, we note that Terashima et al. (2005) describe a correlation between nutritional deprivation and 20E-mediated apoptosis in Drosophila ovaries, and Gu and Lin (2009) provide evidence for DNA synthesis preceding an increase in ecdysteroidogenesis in Bombyx mori prothoracic glands. As additional systems are developed, we anticipate that a wide range of cell cycle regulatory proteins will be found to participate in hormone-mediated events essential to insect growth and development.
Acknowledgments
This work was supported in part by grant AI 43791 from the National Institutes of Health, Bethesda, MD and by the University of Minnesota Agricultural Experiment Station, St. Paul, MN.
Grant sponsor: National Institutes of Health; Grant number: AI 43791; Grant sponsor: University of Minnesota Agricultural Experiment Station.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auzoux-Bordenave S, Hatt PJ, Porcheron P. Anti-proliferative effect of 20-hydroxyecdysone in a lepidopteran cell line. Insect Biochem Mol Biol. 2002;32:217–223. doi: 10.1016/s0965-1748(01)00096-0. [DOI] [PubMed] [Google Scholar]
- Avedisov SN, Rogozin IB, Koonin EV, Thomas BJ. Rapid evolution of a cyclin A inhibitor gene roughex in Drosophila. Mol Biol Evol. 2001;18:2110–2118. doi: 10.1093/oxfordjournals.molbev.a003752. [DOI] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Besson MT, Cordier G, Quennedey B, Quennedey A, Delachambre J. Variability of ecdysteroid-induced cell cycle alterations in Drosophila Kc cell lines. Cell Tissue Kinetics. 1987;20:413–425. doi: 10.1111/j.1365-2184.1987.tb01326.x. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Caldon CE, McNeil CM, Swarbrick A, Musgrove EA, Sutherland RL. Cell cycle machinery: links with genesis and treatment of breast cancer. In: Berstein LM, Santen RJ, editors. Innovative endocrinology of cancer. Boston: Landes Bioscience and Springer Science + Business Media; 2008. pp. 189–205. [DOI] [PubMed] [Google Scholar]
- Coats S, Flanagan WM, Nourse J, Roberts JM. Requirements of p27Kip1 for restriction point control of the fibroblast cell cycle. Science. 1996;272:877–880. doi: 10.1126/science.272.5263.877. [DOI] [PubMed] [Google Scholar]
- Davis LG, Dibner MD, Battey JF. Basic methods in molecular biology. New York: Elsevier; 1986. pp. 130–135. [Google Scholar]
- De Nooij JC, Letendre MA, Hariharan IK. A cyclin-dependent kinase inhibitor Dacapo is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- Eccleston ED, Gerenday A, Fallon AM. Leveraging genomic databases: from an Aedes albopictus mosquito cell line to the malaria vector Anopheles gambiae via the Drosophila genome project. Insect Mol Biol. 2002;11:187–195. doi: 10.1046/j.1365-2583.2002.00324.x. [DOI] [PubMed] [Google Scholar]
- Echalier G. Drosophila cells in culture. New York: Academic Press; 1997. Experimental models of gene regulation: 2 Cell responses to hormone; pp. 397–438. [Google Scholar]
- Fallon AM, Gerenday A. Ecdysone and the cell cycle: investigations in a mosquito cell line. J Insect Physiol. 2010;56:1396–1401. doi: 10.1016/j.jinsphys.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerenday A, Fallon AM. Ecdysone-induced accumulation of mosquito cells in the G1 phase of the cell cycle. J Insect Physiol. 2004;50:831–838. doi: 10.1016/j.jinsphys.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Gu S-H, Lin J-L. Recent studies on prothoracic gland cell growth and ecdysteroidogenesis in the silkworm Bombyx mori. In: Smagghe G, editor. Ecdysone: structures and functions. Dordrecht, Netherlands: Springer Science + Business Media BV; 2009. pp. 271–281. [Google Scholar]
- Hong A, Lee-Kong S, Iida T, Sugimura I, Lilly MA. The p27cip/kip ortholog dacapo maintains the Drosophila oocyte in prophase of meiosis I. Development. 2003;130:1235–1242. doi: 10.1242/dev.00352. [DOI] [PubMed] [Google Scholar]
- Hong A, Narbonne-Reveau K, Riesgo-Escovar J, Fu H, Aladjem MI, Lilly MA. The cyclin-dependent kinase inhibitor Dacapo promotes replication licensing during Drosophila endocycles. EMBO J. 2007;26:2071–2082. doi: 10.1038/sj.emboj.7601648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland RC, Berger EM. Synthesis of low molecular weight heat shock peptides stimulated by ecdysterone in a cultured Drosophila cell line. Proc Natl Acad Sci USA. 1982;79:855–859. doi: 10.1073/pnas.79.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Gerenday A, Fallon AM. Cultured Aedes albopictus mosquito cells synthesize hormone-inducible proteins. In Vitro Cell Dev Biol. 1993;29A:813–818. doi: 10.1007/BF02634349. [DOI] [PubMed] [Google Scholar]
- Liu T-H, Li L, Vaessin H. Transcription of the Drosophila CKI gene dacapo is regulated by a modular array of cis-regulatory sequences. Mech Dev. 2002;112:25–36. doi: 10.1016/s0925-4773(01)00626-8. [DOI] [PubMed] [Google Scholar]
- Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo a cyclin-dependent kinase inhibitor stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- Ma L, Gerenday A, Coley KM, Fallon AM. Co-immunoprecipitation of putative proteins that interact with mosquito proliferating cell nuclear antigen. Insect Mol Biol. 2006;15:197–205. doi: 10.1111/j.1365-2583.2006.00628.x. [DOI] [PubMed] [Google Scholar]
- Meyer CA, Kramer I, Dittrich R, Marzodko S, Emmerich J, Lehner CF. Drosophila p27Dacapo expression during embryogenesis is controlled by a complex regulatory region independent of cell cycle progression. Development. 2002;129:319–328. doi: 10.1242/dev.129.2.319. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Lee CS, Sutherland RL. Progestins both stimulate and inhibit breast cancer cell cycle progression while increasing expression of transforming growth factor α epidermal growth factor receptor c-fos and c-myc genes. Mol Cell Biol. 1991;11:5032–5043. doi: 10.1128/mcb.11.10.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA, Swarbrick A, Lee CSL, Cornish AL, Sutherland RL. Mechanisms of cyclin-dependent kinase inactivation by progestins. Mol Cell Biol. 1998;18:1812–1825. doi: 10.1128/mcb.18.4.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Parekh H, Pillarisetti K, Kunapuli S, Simpkins H. Isolation of a hamster cDNA homologous to the mouse and human cyclin kinase inhibitory protein p27Kip1. Somatic Cell Mol Genet. 1997;23:147–151. doi: 10.1007/BF02679973. [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Althauser C, Findley SD, Ruohola-Baker H. The mitotic-to-endocycle switch in Drosophila follicle cells in executed by Notch-dependent regulation of G1/S G2/M and M/G1 cell-cycle transitions. Development. 2004;131:3169–3181. doi: 10.1242/dev.01172. [DOI] [PubMed] [Google Scholar]
- Shih KM, Gerenday A, Fallon AM. Culture of mosquito cells in Eagle’s medium. In Vitro Cell Dev Biol—Anim. 1998;34:629–630. doi: 10.1007/s11626-996-0010-1. [DOI] [PubMed] [Google Scholar]
- Siaussat D, Bozzolan F, Queguiner I, Porcheron P, Debernard S. Cell cycle profiles of EcR USP HR3 and B cyclin mRNAs associated to 20E-induced G2 arrest of Plodia interpunctella imaginal wing cells. Insect Mol Biol. 2005;14:151–161. doi: 10.1111/j.1365-2583.2004.00540.x. [DOI] [PubMed] [Google Scholar]
- Siaussat D, Bozzolan F, Porcheron P, Debernard S. The 20-hydroxyecdysone-induced signaling pathway in G2/M arrest of Plodia interpunctella imaginal wing cells. Insect Biochem Mol Biol. 2008;38:529–539. doi: 10.1016/j.ibmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Terashima J, Takaki K, Sakurai S, Bownes M. Nutritional status affects 20-hydroxyecdysone concentration and progression of oogenesis in Drosophila melanogaster. J Endocrinol. 2005;187:69–79. doi: 10.1677/joe.1.06220. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Heatherington W, Lane DP, Glover DM. PCNA binding proteins in Drosophila melanogaster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res. 1988;26:3925–3932. doi: 10.1093/nar/26.17.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]




