Applications of micro total analysis systems (μTAS) span basic-science research, clinical medicine, and field work. Assay devices designed for these applications offer improvements to existing methods or provide fundamentally new strategies. Both mature methods and novel techniques have benefited from the increased throughput, integration and miniaturization afforded by μTAS. Traditional assays such Western blots and binding assays are recapitulated in a μTAS format but with reduced reagent usage, decreased performance times and added capabilities. An increasingly vibrant area is the performance of drug screening and toxicology assays on-chip, enabling the efficient screening of very large numbers of molecules. Similarly, recent μTAS reactors demonstrate greater chemical synthetic yields and novel product synthesis compared to macro-systems, often as a result of accurate control over reaction conditions including precision reagent dispensing. These exciting systems are now enabling on-site production of short-lived radioactive compounds for medical applications. The greatest impact of μTAS may very well be the ability to perform massively parallel laboratory experiments, for example, the use millions of reaction vessels or the analysis of hundreds of thousands of single cells. Another strength of μTAS lies in the creation of multicellular communities, for example, the combination of many cell types into an interacting system to explore intercellular communication. Devices with multiple layers of co-cultured tissues benefit from precise placement of molecules, such as extracellular matrices or growth factors, in both space and time. Similarly, the complexity and variety of organ-on-chip and organism-on-chip technologies continues to escalate rapidly. Impressively, the types of organisms cultured on-chip now range from the simplest bacteria to complex animals such as fish.
Automation, reliability, and integration must all increase as a device moves from the specialist environment of a lab to usage by non-expert personnel in the outside world, for example, at a clinical point-of-care or in environmental monitoring. Key innovations in recent months result in devices that operate with minimal external equipment, error-free operation, and unambiguous readouts, all critical for operation by untrained personnel. Lightweight, portable devices are increasingly used to identify chemical and biological toxins in water, air and soil with applications in public health, defense, and homeland security. Perhaps most exciting is the development of μTAS with sufficient robustness for operation in challenging environments, such as the ocean and outer space. A central component of these systems is the ability to withstand the unexpected. These systems push the boundaries of current integration principles and spur rapid growth of new design philosophies.
This review focuses on advances in the area of μTAS or “lab-on-a-chip” systems over the time span of May 2011 through September 2012 with a focus on applications in basic research, clinical medicine and field usage. A range of journals with 2011 impact factors from 2.0 to 36.3 were screened to cover publications with highly specialized content as well as those directed at multidisciplinary audiences. These publications included discipline-specific journals such as Analytical Chemistry and Lab on a Chip as well as general scientific publications, e.g. Science and Nature. To identify material beyond the individually examined journals, extensive key word searches in databases such as PubMed, SciFinder, and Web of Science were performed. Recent reviews in the area of μTAS were also examined for appropriate references. Care was taken to identify impactful and exciting work from across the globe. Well over a thousand papers in the three target areas were identified and discussed. Due to space limitations, we were unable to include all papers but instead incorporated those most fitting into the review scheme and those reporting innovations in basic microdevice technology as well as in applications to biological, physical and engineering sciences. We apologize in advance for omitted papers and welcome feedback regarding any oversights on our part.
FUNDAMENTALS
This initial section focuses on the fundamental innovations in μTAS that underlie the development of new devices and assays for the research setting, clinical lab and field environment. Common goals for these innovations included improving device reliability and repeatability as well as increasing device functionality by incorporating technologies not commonly paired with μTAS. Many of these fundamental advances have led to miniaturized replicas of macroscopic, industrial phenomena such as oil recovery, uranium fate, and carbon dioxide solubility. Other applications included unique assays for the biological sciences. These studies in the physical and biological sciences were often impossible to perform with prior-art technologies and thus even the simplest applications revealed novel phenomena and insights.
Microtechnology Advances
A broad range of improvements and innovations in microtechnology have been reported in the areas of electronics, machinery, and fluidics. Here we focus on three aspects that are closely related to μTAS-based applications: design advances, device fabrication, and surface modification. Many recent advances have tackled universal challenges present in virtually all μTAS, e.g. gaseous obstructions or surface fouling. Other developments embraced the “green” movement to produce degradable systems to replace more commonly used long-lived plastics/polymers.
Design
Using existing materials and established fabrication methods, novel microfluidic designs can improve the ease-of-use of μTAS. For example, “reagent integrators,” filled with dried reagents, were merged into a larger pair of sample-containing channels for controlled reconstitution of the pre-deposited reagents.1 Other designs addressed unwanted air bubbles that negatively affected device operation and experimental outcomes. Phaseguides, which gradually advanced the liquid–air interface using meniscus pinning, effectively eliminated trapped air bubbles in complex microfluidic geometries such as corners and dead angles.2 To remove bubbles from a flow stream, a membrane-based debubbler was incorporated into microfluidic devices.3 Air bubbles were forced to discharge through the porous membrane to the ambient environment while liquid flow continued through the debubbler. These developments represent significant advances in eliminating a phenomenon that has long plagued microchannels and compartments, i.e. undesired gas accumulation due to failure to fully wet a device, insufficient degassing of fluids, or electrolytic gas generation.
Fabrication
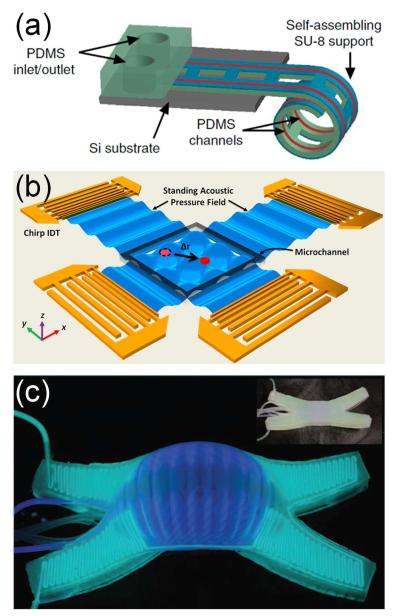
μTAS fabrication has matured, affording microstructure generation from a wide range of materials (e.g. glass, silicon, elastomers, plastics, thermosets, paper) and using an equally broad set of microfabrication methods (e.g. photolithography, soft lithography, injection molding, hot embossing, laser micromachining). Recent innovations in fabrication explored unconventional materials and fabrication strategies. Corn protein (zein) was processed by soft lithography and bonded to both a glass slide and another zein film by ethanol vapor deposition to form green microfluidic devices.4 An origami (paper folding) method was developed to fabricate three-dimensional (3D) paper devices from single sheets of flat paper in a single photolithographic step.5 Curved microfluidic networks were built from the self-assembly of differentially photo-crosslinked SU-8 films, which spontaneously and reversibly curled on film de-solvation and re-solvation (Figure 1a).6 Development of these unconventional materials and fabrication methods has expanded the portfolio for μTAS fabrication by offering the development of simple, low-cost, or biodegradable devices.
Figure 1.
Recent fundamental advances included (a) new microfabrication methods, (b) integration with acoustics, and (c) integration with biomimetic mechanical systems. (a) An illustration of a self-assembling microfluidic device with PDMS channels integrated with a differentially crosslinked SU-8 film attached to a silicon substrate. From ref 6. Reprinted with permission from Nature Publishing Group. (b) Acoustic tweezers with orthogonal pairs of chirped interdigital transducers for generating a standing surface acoustic wave field. Reprinted with permission from ref 17. Copyright 2012 National Academy of Sciences. (c) A soft robot glowing in the dark using chemiluminescence. (Inset) The same robot photographed in the light. From ref 29. Reprinted with permission from AAAS.
Surface Modification
Recent innovations explored new surface modification methods for specific μTAS applications. Perhaps the most common desired outcome for surface alteration is a reduction in biomolecule adsorption, a challenging goal yet to be fully addressed. To eliminate uncontrolled nonspecific bioadhesion on polydimethylsiloxane (PDMS) surfaces, biomimetic glycocalyx-like nanofilms were synthesized using the hydrosilylation click reaction and a methylated polysaccharide derivative (methylcellulose).7 This long-lasting, anti-adhesive coating may facilitate the use of PDMS in implanted biomedical devices. Fabrication of superhydrophobic PDMS microchannels from a PDMS-polytetrafluoroethylene (PTFE) composite was followed by isotropic etching of PDMS to excavate PTFE particles; these surfaces reduced drag and viscous forces in microfluidic applications.8
Integration with Other Technologies
μTAS have evolved from their primitive backbone platforms into highly advanced systems by incorporating technologies from other physical science fields, including optics, acoustics, electricity and magnetism, and mechanics. This integration benefits key functions in μTAS and can lead to interesting biomimetic devices, such as camouflaged robots. Microfluidics also enables new applications in other research areas by providing unique capabilities. The interface between microfluidics and other fields establishes new research areas, such as microfluidic electronics, which applies microfabrication strategies to produce flexible electronic devices.
Optics
Optical readout (including colorimetry, absorbance, scattering, fluorescence and luminescence) is the predominant detection method in μTAS, driving efforts to merge microfluidics with photonic elements. A multiple internal reflection photonic lab-on-a-chip for cell analysis integrated biconvex microlenses, self-alignment microchannels and air mirrors.9 Another compact device monolithically integrated a gallium nitride blue light emitting diode (LED) on a silicon substrate which served as a light source for the fluorescence analysis system.10 The integration of these micro-optical elements on-chip can reduce cost and enhance portability.
Integration of microfluidics and optical components has also yielded unique control of particles, fluids, and light. For example, optical forces can be used to precisely manipulate particles and cells. Recently, a chip composed of silicon microring resonators integrated with waveguides trapped particles and transferred them between rings.11 Optics can also dynamically control and manipulate fluid flow. Reconfigurable flow pathways and morphable channel structures (valves and traps) were created in seconds by illuminating an optical pattern over a photothermal absorbing substrate and thermorheological solution.12 Optofluidic designs use fluidics to control light paths for displays, tunable apertures and lenses, attenuators, switches, and lasers. Multilayer soft lithography produced optofluidic microlenses with tunable focal length and zooming power.13 This system has potential applications in portable microscopic imaging, bio-sensing, and laser configuration. A novel nanoliter-sized microfluidic laser combined microdroplets and a capillary-based optofluidic ring resonator.14 Tuning or switching of the lasing wavelength was achieved by merging two different dye droplets in the microfluidic channel. These advances in optofluidics have increased the flexibility of light-based sample manipulation and detection in chip-based platforms.
Acoustics
Acoustic waves in μTAS have been applied to cell and particle manipulation, fluid mixing and pumping. A pair of opposing surface acoustic waves induced rotation in a fluid droplet at rates up to 2250 rpm.15 This microfluidic “motor” provided a unique, miniaturized method for driving fast rotary motion that could easily be incorporated into microfluidics for a truly portable lab-on-a-chip device. Similarly, rotation of a 10-mm disc was driven by surface acoustic waves in a miniaturized centrifugal microfluidic platform with no mechanically moving parts.16 For particle manipulation, interdigital transducers controlled a standing surface acoustic wave field in real-time, producing acoustic tweezers that manipulated single microparticles, cells, and organisms (Figure 1b).17 Use of acoustic phenomena to control fluidic operations has tremendous potential since sounds waves readily traverse devices without requiring specialized entry points or mechanical components with complex moving parts.
Electricity and Magnetism
Electrical or magnetic control and detection have long been paired with microdevices due to the ease of integrating electrodes or external magnets. In particular, electrokinetic processes contribute diverse functionalities to μTAS such as fluid transport, sample handling, and separation. Electric readout (e.g. impedance, conductivity, and capacitance) is also a sensitive and versatile detection method in μTAS. For example, a droplet sensor recorded blood pressure by measuring the surface capacitance at the electrode–electrolyte interface.18 Other advances in electrowetting, such as unique tools to manipulate tiny volumes of liquids in digital microfluidics, are covered in the Droplet Manipulation section. Magnetism has also found increasing applications in fluid pumping,19 trapping, cell sorting and detection,20 and on-chip NMR.21
Electronics are widely used in off-chip instruments and have recently been combined with microfluidics in unique ways. For instance, on-chip integration of organic electronics, e.g. ion bipolar membrane diodes, has been demonstrated for controlling delivery of hydroxide ions into a receiving reservoir for pH control.22 Additionally, the combination of microfluidics with electronics opens a new, very early stage research area called microfluidic electronics. Microfluidic electronics aims to make or integrate electronic devices using microfluidic technologies. In one device, room temperature metal alloys were processed in PDMS microfluidic platforms to build integrated electronic devices, such as large-area strain sensors.23 These microfluidics-based, elastic electronics have appealing applications, for example, in clothing-based medical sensing systems.
Mechanics
Mechanical units are integrated in μTAS in the forms of valves, actuators and pumps to manipulate fluid flow or to enable precise detection of mechanical stimuli, such as pressure. A monolithic PDMS chip with integrated pneumatic lens arrays generated a 2D pressure map by detecting pressure changes from 2-15 psi based on changes in focal length.24 Microthrusters generated continuously variable thrust force up to 1 mN for a miniaturized propulsion system with potential applications in next-generation micro-/nanosatellites.25 Microfluidic technologies also contributed to novel mechanical devices. Sophisticated motions (undulation and crawling) of soft robots built exclusively from elastomer were controlled by pneumatic actuation of a series of pneu-net channels.26 The result was a unique class of locomotive robot without conventional mechanical joints, bearings or a hard skeleton, in which simple types of actuation produced complex motion.
Biomimetics
In a later section (Research Laboratory), we discuss cell-based organ mimics; however, microfluidic technology is also generating abiotic systems with functionality inspired by biology. One recent device mimicked fluid transport in plants, replicating the osmotic pumping effects used in plant vasculature.27 Magnetic artificial cilia were fabricated from photoreactive copolymer precursors and magnetic nanoparticles by a new photolithographic process. The cilia were integrated into a microchannel and operated using a rotating permanent magnet.19 While these biomimetics contributed new components to μTAS, other devices were built entirely by biomimetic design, for example, a 3D gas exchange unit based on vascularized lung-tissue.28 The soft robots/machines discussed above were inspired by invertebrate animals (e.g., squid, starfish, worms),26 and their further development mimicked the color-changing abilities of animals such as cephalopods, which can change their color, contrast, pattern, apparent shape, luminescence, and surface temperature for camouflage and display (Figure 1c).29 Finally, two recent microfluidic devices were used as tissue surrogates (phantoms) for calibration and validation of imaging methods, one standing in for human tissue in diffusion-tensor magnetic resonance imaging (MRI) measurements30 and the other for superficial vascular networks in biophotonic techniques such as laser speckle imaging or Doppler optical coherence tomography.31
Novel Physical Science
Fundamental advances in microfluidic technology enable new experimental studies, which may be inadequately executed by macroscopic instruments. In the physical sciences, μTAS have contributed to studies of porous media, emulsions and gas dissolution. In a reservoir engineering application, a traditional water-flooding experiment based on rock samples was replaced by a microfluidic device for studies of oil recovery.32 The device included a realistic pore network representative of reservoir rocks and allowed direct visualization of complex fluid flows and displacement mechanisms at the pore-scale. To study bacterial motility or growth in simulated subsurface environments, one device permitted microscopic examination of mixing in porous media mediated by bacterial motion;33 another device investigated the spatial controls exerted by biomass and iron phases on uranium fate and transport for biogeochemical cycling.34 Droplet microfluidics contributed to studies of emulsions and gas dissolution. The kinetic parameters governing demulsification were determined from the observation of thousands of individual coalescence events on an integrated microfluidic device.35 These results inform predictions of emulsion stability in industrial applications. Similarly, measurements of gas solubility, dissolution rates, and rheology have a broad range of industrial applications. Microfluidics provided fast gas-liquid reactions so that solubility data for carbon dioxide could be acquired systematically, rapidly and without the need for manual intervention.36 Another high-throughput microfluidic device was developed to generate a series of microrheology samples as droplets in an immiscible spacer fluid.37 The composition of the sample droplets was continuously varied over a wide range, maximizing the number of rheological measurements while simultaneously minimizing sample preparation time and amount of material. This method should therefore be particularly suited to the characterization of scarce or expensive materials. We expect many other macroscopic experiments to be miniaturized, but researchers should carefully consider possible limitations or inaccuracies caused by miniaturization of macroscopic phenomena.
Novel Biological Science
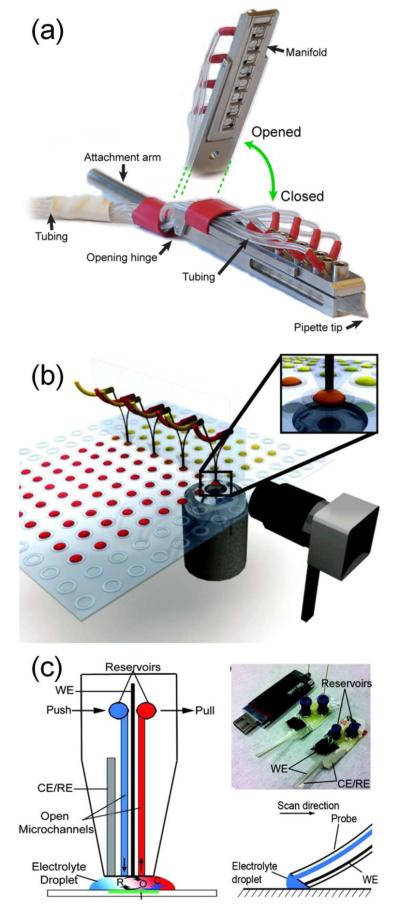
μTAS also enable collection of new biological data, either through novel devices or groundbreaking applications of commercial devices. Many applications take advantage of the ability of microfluidics to offer precise spatial and temporal control of chemical environments. For example, conventional methods to ascertain the IC50 of matrix metalloproteinases risk experimental error due to relatively small sample sizes, exacerbated by serial pipetting steps. Two recent devices addressed this limitation by using precise microfluidic methods to concentrate or dilute samples.38,39 Other examples of enhanced fluid handling in biological assays included high-resolution temporal characterization of cell lysis,40 sub-second hydrogen–deuterium exchange proteomics experiments,41 and single-molecule investigations of ATP hydrolysis42,43 and nitrite reductase kinetics.44 Some new devices used microfluidics to dispense ultra-low volumes off-chip, allowing precise application of reagents in cell culture environments (Figure 2a,b),45,46 for scanning electrochemical microscopy experiments (Figure 2c),47 and even to a living organism in flight.48 Novel biological materials and results were also obtained through microfluidic manipulation of lipid bilayers to produce, fuse, and otherwise assay giant vesicles.49,50 In addition to these novel device designs, other recent papers described novel biological results obtained from previously published43,44,51 or commercially available52-57 μTAS. While these studies did not describe fundamental advances in microtechnology, they demonstrated the critical contributions μTAS can make to diverse fields of biological inquiry, including enzymology,43 whole-genome sequencing,51 cancer biology,52,57 ion channel gating,53 immunology,54 signal transduction55 and virology.56
Figure 2.
Recent devices used microfluidics to dispense liquids accurately off-chip with applications in (a, b) cell-based assays and (c) scanning electrochemical microscopy. (a) This valve-less, single channel pipette sequentially dispensed capsaicin and calcium for signaling studies in single cells. Reproduced from ref 45 with permission of The Royal Society of Chemistry. (b) A multichannel dispenser applied reagents to low-volume cell cultures. Reprinted from ref 46. Copyright 2012 American Chemical Society. (c) A push-pull probe provided a continuously renewed droplet of redox mediators for scanning electrochemical microscopy. Reprinted from ref 47. Copyright 2011 American Chemical Society.
RESEARCH LABORATORY
Biochemical assays
Although some μTAS enable entirely new assays not performable by traditional methods, other devices improve on more established assays. The following sections provide examples of recent advances in the areas of drug screening and immunosensing.
Drug Screening
Microfluidic devices have enhanced drug screening by permitting detailed yet high-throughput studies of drug dissolution, membrane permeability, cellular toxicology, and therapeutic effectiveness. A recent device using post structures captured microparticles for dissolution studies in high flow rates that mimic in vivo conditions.58 A key advantage for μTAS in pharmaceutical applications is the potential to screen large numbers of drug compounds and formulations in small volumes, varying concentrations, and at low cost. For example, one system improved throughput by using droplets to screen large drug libraries against an important clinical target, protein tyrosine phosphatase 1B.59 μTAS technology has also been incorporated into drug toxicity studies using cells, tissues, organs, and organisms (with the latter two formats described in subsequent sections). An integrated microfluidic array plate performed gravity-driven cell capture, cell culture, and drug perturbation studies using real-time optical analysis,60 whereas another all-in-one device incorporated a colorimetric assay to evaluate cadmium toxicity in liver cells.61 Another device for studying liver cytotoxicity coupled microfluidics with ESI-Q-TOF mass spectrometry to simulate in vivo drug metabolism, performing rapid on-chip testing and sample pre-treatment before off-chip mass spectrometric analysis.62 Additionally, a high-throughput device utilized gradient generators to study pair-wise combinations of drugs on prostate cancer cells, providing information about sequential or simultaneous chemotherapies on a single population of cells.63
Similar multiplexed microfluidic devices have been applied to drug screening against infectious organisms, an important application as bacteria become increasingly resistant to a wide range of antibiotics. A pair of integrated droplet-based microfluidic devices permitted rapid drug screening by encapsulating bacteria and antibiotics into droplets followed by subsequent spectrophotometric analysis of cell growth within each droplet.64 An alternative device used droplets to evaluate the effect of indole signaling on the development of antibiotic resistance in bacterial colonies.65 Multiplexed microfluidic devices have also been used to rapidly screen the effects of antibiotics on heterogeneous bacterial colonies to identify resistant colonies using as few as 100 bacteria66 and to demonstrate how asymmetry in cell division can lead to differential antibiotic sensitivity.67 Other studies examined the effects of antibiotic concentrations and combinations; for example, microsegmented flows created a three-dimensional concentration space to study drug combination/dose response effects on E. coli.68
Immunosensing and Binding Assays
Immunosensing protocols often suffer from high reagent costs and lengthy incubation times, which limit their utility in clinical settings. Novel μTAS address these issues by decreasing reagent use and analysis time. For example, experimental conditions for immunofluorescence assays of lysosomal storage disorders were optimized using pneumatic valves for parallel staining.69 A Western Blot-on-a-chip device incorporated both gel electrophoresis (SDS-PAGE) and blotting steps on a single device, providing faster sample analysis, lower reagents costs, and a ‘renaturation’ step to remove SDS-PAGE reagents that could negatively impact affinity agent binding.70 Additionally, an enzyme-linked immunosorbent assay (ELISA)-on-a-chip was developed with comparable limits of detection (10 pg/mL) to traditional ELISA but greatly reduced antibody consumption and assay time.71 For immunoassays in tissues, a vertical microfluidic probe enabled staining of individual cores of tissue microarrays, diminishing antibody cross-reactivity and increasing the range of staining conditions applied to a single tissue section.72
Another major thrust of immunosensing in μTAS is the development of devices capable of rapid protein sensing with low detection limits. A recent device incorporated pH specific membranes to pre-concentrate, separate, and quantify glycoproteins directly from biological samples.73 Microfluidic devices employing magnetically-labeled cells also performed high-throughput analyses with low limits of detection. Recent examples included a giant magnetoresistive biosensor to evaluate protein-antibody binding74 and a micro-Hall detector to measure immunomagnetically tagged cells in whole blood.20 Antibody-functionalized microspheres provided another method to concentrate samples and improve detection limits. Microfluidic immunoassays based on polymer and magnetic microparticles were used to detect cytokines,75 cyanotoxins,76 and breast cancer biomarkers.77 In addition to low detection limits, many devices also featured high levels of automation,78 rapid analysis times (10-30 min),76,78 and adjustable dynamic ranges.77 Finally, the immunosensing capabilities of μTAS have also contributed to studies of key molecules in cell signaling pathways by probing for cell surface markers79 and molecular distributions of diffusible molecules,80 or by studying binding kinetics using surface plasmon resonance (SPR).81
High Throughput
μTAS are often highly automated, and as a result, increase assay speed. Additionally, micron-scale components expand the number of possible components on a single chip, facilitating multiplexed analysis. High-throughput μTAS have particularly benefited biological and biochemical assays. Automated proteomics assays in gel-based microfluidic devices had a 5- to 15-fold improvement in speed for prostate specific antigen detection compared to conventional Western blotting and a two-fold improvement over traditional capillary immunoblotting.82 Other systems provided rapid screening of millions of analytes, such as genes for engineered proteins based on directed evolution83 and affinity reagents for membrane-bound receptors on adherent cells.84 Microfluidic devices also facilitated rapid inspection of particles and cells. For example, a label-free nanoparticle analyzer with resistance-based detection determined the size and concentration of ~500,000 particles per second in blood-sample analysis,85 and a droplet-based system screened 300,000 hybridoma clones in less than a day.86 The small footprint of device components also facilitated high-density multiplexing on individual devices, for instance, a 16-channel device for chromatin immunoprecipitation.87 Advanced multiplexing has encompassed much larger numbers of components. A microfluidic flow cytometer used 384 parallel flow channels to investigate nuclear translocation events in models of disease states caused by protein misfolding.88 Another device included >900 micromechanical valves, creating a versatile device that could be controlled by software programming for a variety of functions.89
Synthesis
Precise control over small volumes and mixing has driven the use of microsystems as microreactors. Many of these systems have been used for the production of pharmaceuticals and imaging agents, and some even incorporated analyses of synthesized products on the same chip.
Microreactors
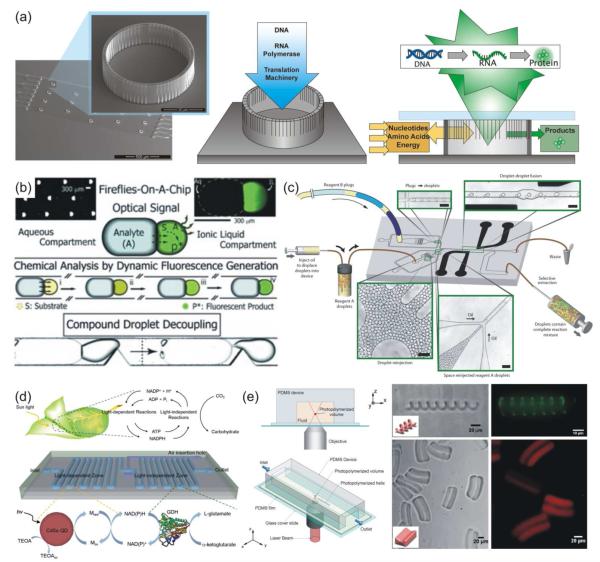
Chip-based systems have assembled a number of biomolecules, including proteins and DNA. Cell-free protein production was achieved using an array of cellular-scale porous containers housing transcription/translation reagents and a DNA template. Amino acids and other necessary reagents flowed through the system, and synthesized proteins were readily released from the porous vessels (Figure 3a),90 yielding twice the protein per volume compared to conventional batch production. Synthesis followed by subsequent analysis on a single device has also been demonstrated; for example, measurement of pH, temperature, and attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectra for reaction characterization.91 In many cases, microfluidic reactors also simplified chemical reaction steps. For example, the optical transparency and gas permeability of a PDMS device allowed photosensitized oxygenation reactions to be completed without gas presaturation, reducing reaction times from hours to minutes.92 A simple flow-based device coupled aqueous and ionic liquid droplets, allowing a soluble analyte to diffuse between droplets and catalyze a fluorescent reaction (Figure 3b),93 while a more complex device utilized electrocoalescence to perform synthesis by droplet fusion (Figure 3c).94 Another device even mimicked natural plant photosynthesis, performing photoenzymatic synthesis using quantum dots and redox enzymes on-chip (Figure 3d).95
Figure 3.
On-chip synthesis. Microfluidic systems allowed for (a) continuous protein production, (b) bicompartmental synthesis, (c) combinatorial synthesis, (d) mimicking of plant photosynthesis and (e) synthesis of 3D solids. (a) Flow-based continuous cell-free protein synthesis was performed in 40-μm diameter, 15-μm tall reaction vessels that contained DNA templates and translation components. Reactants were delivered via flow, and synthesized proteins were released through the container pores. From ref 90. Reproduced with permission of The Royal Society of Chemistry. (b) Bicompartmental droplets allowed analyte transport into an ionic liquid droplet to produce a fluorescent product that could be decoupled from the combined droplets. From ref 93. Copyright 2012 John Wiley & Sons, Inc. (c) A microfluidic device for combinatorial synthesis by droplet fusion utilized multiple droplet injection regions and electrocoalescence-based droplet fusion. From ref 94. Reproduced with permission of The Royal Society of Chemistry. (d) A microfluidic mimic of natural photosynthesis incorporated CdSe quantum dots for light-dependent reaction regions separate from light-independent reaction regions. From ref 95. Reproduced with permission of The Royal Society of Chemistry. (e) A microfluidic device utilized two-photon continuous flow lithography to produce 3D structures. From ref 104. Copyright 2012 John Wiley & Sons, Inc.
Numerous on-chip techniques have been devised to synthesize solids, including microspheres, nanoparticles and fibers. These devices yielded stable, uniformly sized particles, as in a recent synthesis of supramolecular microcapsules.96 Other on-chip syntheses formed products not possible in bulk reactions, such as the self-assembly of block copolymer micelles into kinetic cylinders, y-junctions, bilayers, and networks, while only spheres can be produced in bulk reactions.97 Improvements in microfluidic devices, such as the ability to withstand high pressures and temperatures, have permitted the use of supercritical microfluidics, allowing the synthesis of materials with advanced optical properties, such as ultraviolet (UV)-emitting zinc oxide (ZnO) nanocrystals.98 Some devices used microfluidic handling to prepare nanoparticle-containing microparticles, for instance, alginate microparticle barcodes doped with varying ratios of quantum dots.99 Not only can these solids be synthesized on-chip, but some can also be arranged, combined, and immobilized. A polymethylmethacrylate (PMMA) device fabricated chitosan microparticles, and then linked these particles together, controlling microparticle arrangement and chain length.100 Other devices produced and deposited ZnO nanowires101 or metal-organic frameworks102 on-chip. Novel devices have also formed continuous solids, such as fibers and complex, elongated structures. A two-layer device designed to mimic a silkworm gland controlled fiber properties of synthetic silk for characterization of sequence-structure relationships,103 and two-photon continuous flow lithography was used to produce extended 3D structures such as helices in a PDMS microdevice (Figure 3e).104
Pharmaceuticals and Imaging Agents
Microreactor platforms have frequently been applied to biomedical challenges, such as drug synthesis, packaging, and formulation. A microfluidic spray dryer produced amorphous drug nanoparticles of narrow size distribution from a drug-loaded spray,105 while a T-junction device controlled antisolvent precipitation for size-tunable formation of drug nanoparticles.106 The synthesis of therapeutic delivery devices is another active area of research. Recent microfluidic devices have packaged therapeutic nucleic acid materials, plasmid DNA cores107 and siRNA108 in lipid bilayer shells. Another system covalently linked drug molecules to delivery agents in an efficient synthesis of heparin-folic acid-retinoic acid bioconjugates.109 Additionally, advances in the synthesis of imaging agents were possible due to the use of droplet-based technology or on-site synthesis of short half-life reagents. Flow focusing chips produced phase-change perfluorocarbon droplets that act as acoustically activated contrast agents for MRI.110 Multiple devices also aided in the synthesis and labeling of fluorine-18 radiotracers for positron emission topography (PET) scanning, including an electrowetting on-demand device to synthesize fluorodeoxyglucose[18F]111 and a droplet generation device to optimize labeling of anti-prostate stem cell antigen diabodies with an 18F-containing small molecule.112 Microdevice-based synthesis of 18F-labeled molecules will enable a much broader application of this radioisotope since the molecules are produced on-demand and used prior to significant decay.
Digitization
As noted in the synthesis section, many μTAS take advantage of the precise volume control found in microfluidic systems. The ability to manipulate fluids as micro- or picoliter scale droplets, known as digitization, has opened the field to new and exciting applications.
Droplet Generation
Common droplet generation techniques use tapered microchannels to produce droplets from a continuous flow stream; however, several recent devices have used light sensitive reagents for droplet generation. UV light was used to solidify a hydrogel encapsulating an active compound-containing oil core.113 The encapsulated molecules were then released by simple hydration without the temperature stimulus commonly needed for hydrogel release. Another droplet generation system used a photo-sensitive surfactant to produce droplets. UV light released droplets from a continuous stream as a result of a change in surface energy.114 Other devices improved control of droplet properties. For example, a PMMA device utilized coaxial microfluidic channels to form gas-in-water-in-oil or gas-in-oil-in-water double emulsions with a high degree of control over encapsulated bubble size, number of encapsulated bubbles, and droplet size (Figure 4a).115
Figure 4.

Digitization advances included new means of droplet (a) generation and (b, c, d) manipulation as well as (e) high-density digital PCR. (a) A dual-coaxial microfluidic device produced gas/liquid/liquid double emulsions with a high degree of emulsion/droplet control. From ref 115. Reproduced with permission of The Royal Society of Chemistry. (b) A grid of electrodes allowed reagent actuation to the site of a dried blood spot for the quantification of amino acids in blood via in-line mass spectrometry. From ref 116. Reproduced with permission of The Royal Society of Chemistry. (c) Utilizing a silane-patterned open-surface microfluidics device, droplets containing an insoluble surfactant (green) were self-propelled along a sub-phase liquid (blue). From ref 122. Reproduced with permission of The Royal Society of Chemistry. (d) Electrowetting forces laterally spread droplets. Upon removal of electrowetting forces, the droplets converted stored energy to kinetic energy, causing them to “jump” off the surface. Reprinted with permission from Lee, S., Lee, S. & Kang, K. Droplet jumping by electrowetting and its application to the three-dimensional digital microfluidics. Appl. Phys. Lett.100, 081604. Copyright 2012, American Institute of Physics (ref 123). (e) High-density digitization of PCR samples into microscale compartments. This “megapixel” digital PCR device had reaction vessel densities exceeding 440,000 cm−2 and a dynamic range of 107. Reproduced from ref 127 with permission of the Nature Publishing Group.
Droplet Manipulation
Just as droplet generation techniques have progressed, new methods and applications for droplet manipulation have also been developed. An electrode grid moved individual droplets across an array to reagents stored either directly on the electrodes116 or on hydrogel disks.117 These devices were applied to dried blood spot analysis (Figure 4b),116 cell culture and evaluation,118 and proteomics studies.117 Other electrowetting systems enabled handling and crystallization of proteins for in situ matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) analysis.119 A recent improvement to electrode-based manipulation systems utilized a thin film transistor array to replace conventional patterned electrodes, allowing the fabrication of larger arrays (64 × 64) by eliminating individual electrode connections.120 Surface acoustic waves have been utilized in combination with a series of Fabry-Perot resonators for interaction-free droplet manipulation.121 In open devices, surfactant-containing droplets were self-propelled along patterned thin films of sodium dodecyl sulfate in glycerol without the need for external actuation (Figure 4c).122 Electrowetting techniques have even performed three-dimensional droplet manipulation. Energy was stored by stretching droplets using electrowetting forces, then released upon the removal of the forces, causing the droplets to ‘jump’ off a surface (Figure 4d).123
Droplet Analysis
Precise manipulation of large numbers of droplets facilitates screening over extended time periods. By storing a well-defined array of protein-containing aqueous droplets, time-lapse analysis of nucleation processes, such as the formation of amyloid bodies, was possible.124 The ability to perform this analysis will contribute to studies of Alzheimer’s, Parkinson’s, and diabetes mellitus type 2, which all involve pathological deposition of protein fibrils. In another system, oil-in-water droplets encapsulating cell lysates with antibody-functionalized beads were stored and analyzed on-chip for intracellular protein determinations.125 This system yielded faster results than conventional immunoblotting while requiring fewer cells.
PCR
μTAS platforms offer a variety of unique advantages for nucleic acid assays based on polymerase chain reaction (PCR). Massively parallel fabrication permits digital PCR in large numbers of reaction vessels while integration of sample processing and miniaturization benefit total assay performance by reducing sample and reagent consumption. Care must be taken, however, in the design of μTAS platforms because many common materials used in microfabrication, such as PMMA and SU-8 photoresist, have been shown to inhibit PCR reactions.126
Digital PCR
Traditional quantitative PCR (qPCR) assays suffer from limited precision and thus face challenges in quantifying absolute gene copy numbers. Digital PCR, which amplifies individual molecules in discrete reaction volumes, eliminates this shortcoming. While previous implementations of digital PCR in microfluidic reaction vessels achieved densities as high as 6,000 cm−2, valve-less implementations achieved up to 440,000 vessels per cm2 by filling the reagent loading channels with oil (Figure 4e).127 A robust implementation of digital PCR in a SlipChip format yielded quantitative results even with isothermal amplification at various temperatures.128 Another SlipChip multiplexed reverse-transcriptase PCR (RT-PCR) reactions in a multi-volume format for high dynamic range (from 1 to 1.2×105 copies) in a relatively small total number (880) of reaction vessels.129 The high dynamic range inherent in digital PCR has enabled the study of rare genomic events. This feature was exploited in the quantitation of rare KRAS mutants against a large background (1 in 200,000) of wild-type genes.130 With at least five commercial implementations of microfluidic digital PCR available on the market, this technique has become a μTAS success story.
Single-Cell PCR
Systems designed for digital PCR are optimized to isolate approximately 1 gene copy per vessel from diluted reaction mixtures. The technologies developed for digital PCR therefore translate readily to similar analyses on single cells. PCR on single bacteria isolated from the termite hindgut identified viral marker genes to explore phage-host interactions.131 A valve-based microfluidic platform was used to trap, lyse and perform RT-PCR on single mammalian cells for the quantitation of RNA expression in single cells.132 In addition to valve-based systems, a droplet-based microchip performed RT-PCR on canine kidney cells, creating the droplets, thermocycling and performing fluorescence readout by microscopy on-chip.133 These devices offered high sensitivity, wide dynamic ranges, and highly parallel designs, which allowed experiments to be conducted on hundreds to thousands of individual cells at once, a feature critical to the broader relevance of such devices.
Genotyping
From forensics to infectious diseases, rapid and accurate characterization of genetic samples has a variety of valuable applications. The high surface area-to-volume ratio of μTAS is particularly relevant in capture-based detection modalities, including giant magnetoresistive materials, which provide direct electrical readout of binding of biomolecules to tethered probes. Giant magnetoresistive sensors were recently used to genotype the four variants of hepatitis B virus, data that may be of clinical relevance in diagnosis and therapy.134 The ability of μTAS platforms to couple diverse functionalities has been well-demonstrated in genotyping applications. An excellent example incorporated continuous-flow PCR amplification, bioaffinity-based sample purification and concentration, and detection by capillary electrophoresis for gender and ethnicity determination of genetic samples.135 The combination of readily interpreted readout and functional automation have also made μTAS devices an appealing platform for developing world applications. A ligase detection reaction and conventional PCR amplification were integrated into a single-chip solution for the genotyping of drug-resistant M. tuberculosis.136The multiplexing advantages of the μTAS format allowed the integration of three controls and six target genes for detection into a single readout. For the characterization of complex and poorly understood systems, genome-wide screening is necessary in order to enable rapid genotyping, particularly in clinical samples. Gene expression profiles of non-small cell lung cancers were obtained using Fluidigm’s BioMark™ microfluidic platform and were used in combination with DNA methylation data to correctly differentiate between epithelial- and mesynchemal-derived cancers.57
Cell-Based Assays
Microfluidic devices have enabled many studies of biological processes including cell migration, cell proliferation, cell-to-cell signaling, and single-cell analysis. Low sample volumes, rapid incubation times, and more quantitative outputs have driven the development of new and improved devices. Many exciting developments in cell-based technology are still confined to the research laboratory but have the potential to enhance our understanding of essential biological pathways.
Microfluidic Cell Culture
Microfluidic devices provide several advantages over macroscale cell culture, such as lower reagent volumes, automation and controllable cell-cell interactions. Recent advances improved cell culture efficiencies by automating control over carbon dioxide and oxygen levels,137,138 temperature, and cell density over prolonged periods in both closed channels139 and digital systems.118,140 Multichamber cell culture devices recently demonstrated cell culture under low shear conditions141 or continuous perfusion.142 A gravity-driven device, using the principle of equivalent electric circuits, alternated the hydraulic resistance of channels to culture uniform 3D cell spheroids in < 24 h.143 Another device accurately evaluated cell layer integrity using transendothelial electrical resistance to measure impedance, providing an automated method to measure cellular confluency independent of visual inspection.144 Additionally, microfluidic devices have been developed to quantify proteins secreted from cells cultured on-chip. A novel quantitation method used mass spectrometric imaging to determine the distance secreted proteins travelled down a channel as a replacement for traditional, less quantitative MS peak intensities.145
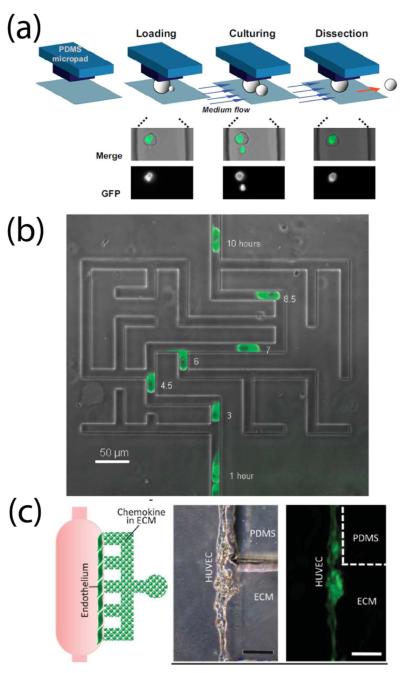
The ability to perform long-term cell culture under well-defined conditions while tracking individual cells has enabled important studies of cell aging and microbial communities. For example, a microfluidic device trapped 60 individual yeast cells while continually removing daughter cells over a three-day period,146 while another employed fluid flow to selectively release and track newly-budded daughter cells through several generations (Figure 5a).147 Cell aging and evolution of bacterial cells were also investigated by monitoring the formation of daughter E. coli cells as they developed within a confined microchannel.148 Microscale microbial communities have also yielded important information about their complex growth dynamics. Recent examples included a synthetic oscillator consisting of a dynamic switch upstream of a cell trapping region that controlled activation signals for a bacterial biological clock,149 algal cultures in discrete droplets that revealed heterogeneous cell populations,150 and a simple microfluidic device that gave insight into the trade-off between local competition and dispersal in a growing biofilm of Vibrio cholera.151
Figure 5.
Control of the cellular microenvironment. μTAS technology enabled new studies in a variety of biological systems. (a) Selective release and tracking of newly-budded yeast daughter cells across multiple generations controlled by microfluidic flow. Reprinted with permission from ref 147. Copyright 2012 National Academy of Sciences. (b) A microfluidic maze established tunable EGF gradients on the cellular level to study epithelial cell migration. From ref 153. Reproduced with permission of the Royal Society of Chemistry. (c) Chemokine-induced adenoid cystic carcinoma intravasation through a mock endothelial cell monolayer in microfluidic-based device. Scale bar, 200 μm. From ref 159. Adapted by permission from the Royal Society of Chemistry.
Microenvironment Control
A major advantage of on-chip cell-culture is the ability to control the cellular environment on the microscale. Consequently, μTAS have provided researchers with tools to investigate key biological processes, including cell migration and chemotaxis, adhesion, and cell-to-cell crosstalk. One device took advantage of two alternating inlet channels to elicit competing gradients, effectively trapping the slime mold Dictyostelium discoideum due to constant readjustment of cell polarity.152 In another device, epithelial cells “sensed” their way through a microfluidic maze following an epidermal growth factor (EGF) gradient (Figure 5b).153 A recent device utilized electrochemical impedance spectroscopy coupled with a traditional Boyden chamber to more quantitatively evaluate hepatocellular carcinoma migration in response to different extracellular matrix (ECM) proteins.154 Additionally, microfluidic control of the cellular microenvironment has elucidated the roles of essential proteins in cell behaviors, such as fibroblast chemotaxis155 and electrotaxis.156 Microfluidics also enabled studies of chemotropism, directed growth in response to a chemical gradient, through precise spatial and temporal control of pheromones to induce the mating response, or formation of “shmoos,” of Saccaromyces cerevisiae.157 Changes in interstitial flow through the extracellular matrix, common during inflammation, can also affect the morphology and migration of cells; however, the ability to directly visualize this response has been limited. A microfluidic system addressed this shortcoming and demonstrated that interstitial flow strength caused a directional bias of melanoma cell migration and affected the percentage of cells that became migratory.158
The cellular microenvironment often includes other cell types in addition to soluble factors and ECM. Several devices integrated soluble gradients and endothelial monolayers for transendothelial invasion studies. For example, adenoid cystic carcinoma cells migrated through an endothelial cell monolayer to a chemokine-loaded ECM (Figure 5c),159 and tumor intravasation was studied as a function of an external EGF gradient and endothelial paracrine signaling.160 Novel devices also elucidated the role of proteins such as selectins on neutrophil adhesion, a necessary step in inflammation.161 Cell migration is often the result of cellular crosstalk. Recent devices were used to evaluate kidney cell migration toward fibroblasts across a “wound” generated without mechanical tension on adjacent cells.162 Other μTAS have adjusted the microenvironment of the cell to study paracrine and autocrine signaling. Two recent devices used microfluidic flow to reduce the concentrations of secreted factors from mouse embryonic stem cells, providing valuable insight into signaling dynamics and differentiation.163,164
Organ-on-a-Chip
As μTAS cell culture continues to evolve, the designed devices more effectively recapitulate in vivo microenvironments. Recent microfluidic technology has generated on-chip mimics of angiogenesis, the cardiovascular system, and mechanical stimuli of organ systems. A ground-breaking device induced capillary formation by culturing human umbilical vein endothelial cells (HUVECs) on either side of a fibrin gel.165 Over time, HUVECs migrated into the gel and ultimately formed leak-free, perfusable vessels (Figure 6a). Another microfluidic vascular network demonstrated angiogenic remodeling, perivascular cell crosstalk, and the interaction between blood components and the endothelium.166 Other devices for angiogenesis studies examined leukocyte recruitment to new capillaries,167 the effects of matrix metalloproteinases on capillary diameter and elongation rates,168 and the effects of interstitial flow, a vascular endothelial growth factor gradient, and shear forces on endothelial sprouting in a collagen matrix.169 Fabrication of degradable glass filaments within endothelial cell cultures provided a better mimic of the vasculature network (Figure 6b).170 Other cardiovascular organ-on-a-chip systems focused on the electrical and mechanical stimulation of heart cells. A cardiomyocyte culture on an elastomeric membrane provided a system for monitoring applied stresses during cell contraction.171 A novel in vitro platform modeled cardiac muscle on a multielectrode array-based biochip and measured the electrical conductivity of laser-patterned stem cells bridging cardiomyocytes, an important factor in successful stem cell grafting to treat ischemia or infarction.172 Mechanical and shear stresses were also applied to systems modeling the colon173 and lung airways.174 While these systems have certainly advanced our understanding of cell interactions and fates, researchers should apply the “organ-on-a-chip” label selectively. True organ-on-chip systems must mimic organ microarchitecture (e.g. biophysical) and microenvironment (e.g. biochemical, mechanical) and generate organ-level physiological responses under external stimuli (e.g. toxins, bacteria, drugs etc.). Tumor cell lines should be avoided since their aberrant biology poorly reflects normal tissue physiology. Instead, primary cell lines and realistic co-cultures should be employed wherever possible.
Figure 6.
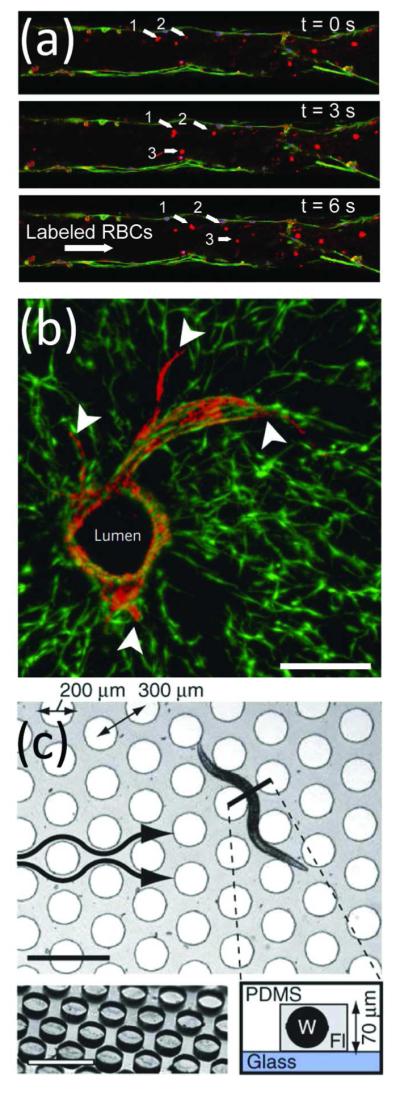
Organs and organisms-on-chip. Microfluidics allowed controlled studies of (a,b) cell-cell interactions and (c) whole organisms. (a) Time-lapse images of a red blood cells flowing through a capillary network developed from endothelial cells cultured in a microfluidic channel. From ref 165. Reproduced with permission of The Royal Society of Chemistry. (b) Sprouting of mCherry-expressing endothelial cells (arrowheads) from a central lumen was demonstrated within a co-culture of 10T1/2 cells expressing enhanced green fluorescent protein. Scale bar, 200 μm. Adapted by permission from Macmillan Publishers Ltd: Nature Materials, ref 170, copyright 2012. (c) A microfluidic device for examining behavioral responses of C. elegans to chemical changes. Scale bars, 500 μm. Adapted by permission from Macmillan Publishers Ltd: Nature Methods, ref 180, copyright 2012.
Organism-on-a-Chip
While whole organism studies might seem like an odd application for microscale technologies, μTAS devices have provided high-throughput and sensitive analysis of small organisms, such as plants, protozoa, zebrafish and worms. For example, directional pollen tube growth towards an unfertilized ovum in microfluidic channels provided controllable analysis of the mechanisms of plant reproduction.175 In addition to templating growth, microfluidics also aided in continuous monitoring of individual organisms over prolonged time periods. For example, microfluidic flow trapped individual Tetrahymena thermophila, a highly motile protozoan ciliate, for up to 40 hours.176 μTAS have also reduced analysis time by trapping organisms in parallel microcompartments, as exemplified by the development of 8-well and 48-well chambers for the study of zebrafish177 and Caenorhabditis elegans,178 respectively. Additionally, microfluidic devices have been used to examine the chemosensory crawling behavior of C. elegans, a difficult task for traditional technologies. One apparatus elucidated the role of interneurons on head movement in response to calcium stimuli,179 while another platform evaluated migratory turning dynamics in response to liquid-borne stimulation of the olfactory system (Figure 6c).180 Drug screening on intact model organisms has also been incorporated into microfluidic devices. Microfluidic platforms evaluated the effects of anthelmintic drugs on two nematode species, C. elegans and parasitic Oesophagotomum dentatum. These devices provided a variety of readouts, including real-time observation of pre-, concurrent, and post-exposure locomotion,181 muscular force measurements based on cantilever deflection,182 and non-invasive quantification of electrophysiological activity.183 The capabilities of μTAS systems to enable new biological assays and reproducibly screen high numbers of organisms will likely result in increased employment of these devices as they mature.
Single Cells
Recent technologies designed for analyzing individual cells have revealed inherent cellular heterogeneity. The biological importance of this heterogeneity has resulted in a plethora of strategies for manipulating, sorting and analyzing individual cells from mixed populations.
Single-Cell Analysis
Because population-based studies do not yield an accurate picture of cellular heterogeneity, microfluidic devices have been developed to isolate single cells and analyze them in a high-throughput fashion. Recent techniques applied cell trapping and mechanical methods to study intact cells or to analyze the intracellular contents of lysed, single cells. Microchambers were used to trap up to 340 single-cell pairs while tracking intercellular signaling.184 In another study, a multiplexed platform integrated cell culture and stimulation with downstream analysis, including live cell fluorescent microscopy and flow cytometry, to profile toll-like receptor 4 (TLR4) signaling in single lipopolysaccharide-stimulated macrophages.185 Additionally, well-controlled fluid flow has transported single cells through microchannels for density measurements.186 Another device measured the deformability of red blood cells as they passed through a constriction and distinguished healthy cells from those parasitized by Plasmodium falciparum based on stiffness.187 Other devices use on-chip lysis for analysis of intracellular contents.125,132,188 One device captured and isolated 100 cells in reversibly sealed microchambers for incubation, washing, labeling, and eventual lysis.189 This platform was used to monitor single-cell NADPH/NADH dynamics and the toxic impact of the alkaloid camptothecin.
Advanced microfluidic systems are using quantitative single-cell measurements to reveal biologically important heterogeneity in cell populations. More than ten types of secreted proteins from single immune effector cells were measured using 1040 3-nL microchambers, demonstrating functional heterogeneity in phenotypically similar cytotoxic T-cells.190 Digital microfluidic devices have proven especially useful in single-cell analysis due their programmability and inherent compartmentalization of small volumes. One multipurpose platform consisting of 95 separate storage chambers sorted, isolated, and processed single cells by alternating droplet immobilization by flow-controlled wetting.191 This chip was used to genotype single bacteria from multiple environments ranging from deep-sea sediments to the human oral cavity. Another digital device incorporated a rolling-circle-enhanced enzyme activity detection (REEAD) assay to detect rare, aberrant single cells from a wild-type population.192 Through the continued advancement of μTAS technology, single-cell assays will be more readily available to researchers, reducing high experimental costs and long assay times and revealing results obscured by ensemble measurements of heterogeneous cell populations.
Manipulating and Sorting Individual Cells
Precise control of small volumes makes microfluidic technologies ideal for manipulating individual cells for fusion, transfection, staining, or sorting. Cell pairing is a critical step in fusing genetically dissimilar cell types to create hybridomas. Recent microscale systems employed dielectrophoresis to selectively mate pairs of cells for subsequent fusion.193 In addition to cell pairing, multiple strategies have been described for sorting, capturing, and collecting rare cells. While traditional size- and deformability-based separations have been plagued by clogging, a novel device overcame these issues by incorporating a series of microstructure funnels to flow cells irreversibly, even under oscillatory flow conditions.194 Another strategy employed dielectrophoresis to isolate C2C12 myoblasts from differentiating myotubes.195 A centrifuge-on-a-chip harnessed fluid vortices to combine cell sorting with subsequent sample processing, including concentration, staining, and wash steps on-chip.196
Exceptionally rare cell types, such as circulating tumor cells (CTCs), require further refinement of cell sorting methods. Because of mounting epidemiological and molecular evidence that CTC numbers predict cancer metastasis and patient outcome, CTC enrichment and recovery is a growing area of research. While macroscale immunomagnetic collection of CTCs via anti-epithelial cell adhesion molecule (EpCAM) labeling remains the gold standard, several competing microscale platforms have been developed. These platforms have extended the capabilities of macroscale collection to improve enrichment rates and recover viable CTCs. For example, magnetically-labeled CTCs were detected by a micro-Hall detector with parallel sensor arrays at higher sensitivity than currently possible with clinical standards.20 A robust, single-channel device for immunomagnetic collection of CTCs by anti-EpCAM binding captured 86-90% of cancer cells spiked into whole blood at flow rates of 10 mL/h.197 This device functioned on the same principle as macro-scale implementations such as CellSearch™ by Veridex, but reduced reagent consumption by 25% and improved capture efficiency. Moving away from immunomagnetic capture, “virtual aliquots” were produced as plugs of whole blood containing a target cell were identified by fluorescence and separated from continuous flow with high recovery rates (93%).198 The limitations of anti-EpCAM capture have also been demonstrated; a microfluidic device composed of functionalized micropillars increased capture rates over CellSearch™ up to 400-fold using an antibody to a prostate-specific membrane antigen in samples from 25 prostate cancer patients in a clinical setting.199 Most immunomagnetic and surface capture methods irreversibly bind tumor cells, preventing their recovery and reducing cell viability. By anchoring capture antibodies to an alginate biopolymer, researchers enabled rapid and efficient release of captured CTCs by incubation with alginate lysase.200 To move CTC devices from the research laboratory to the clinic, future devices should offer efficient capture, on-demand release, and sensitive analysis of these rare cells.
CLINIC
Successful devices for clinical applications must perform as well or better than current assays, adhere to stringent production and operation requirements, and have a medically-useful dynamic range with high sensitivity and specificity. When these criteria are met, microfluidic technology offers several advantages over conventional assays. Parallelization, automation, and multiplexing reduce the total read time for time-sensitive clinical assays. Reduced reagent consumption and automated sample preparation minimize assay costs and bring quality clinical assays to resource-poor settings. Recent research efforts have focused on platforms with one or more of the following functions: handling of soluble analytes, analysis of intact or lysed cells, and integration of assays for point-of-care devices.
Soluble Analytes
Microfluidic platforms have advanced analysis of soluble analytes in human fluid samples, including blood, urine, saliva, sputum, and tears. In particular, automated pre-processing of clinical samples has enabled efficient analyses. For example, recent methods have separated analytes from whole human blood using microfiltration,201 cationic isotachophoresis,202 or paper-based203 flow prior to subsequent analysis. Fluid control was the central focus of a device used to count CD4+ lymphocytes for HIV staging from a fingerstick.204 The device featured an antibody-impregnated hydrogel layer that controlled antibody release via swelling once blood was applied. Two devices integrated sample separation with on-chip analysis of cardiac biomarkers.203,205 Microdevices have also quantified other important serum components, including sugar moiety alterations on bloodstream proteins (useful in stratifying patients with esophageal cancer)206 and cobalt ions (contained in the vitamin B12 complex).207
Because of their ability to filter and separate analytes, paper microfluidic devices are well-suited to separations of human-derived fluids. Filtration can be achieved either horizontally203,208 or vertically,209,210 and applied to blood,203,209 urine,208 or sputum.210 For example, one device added a second stage to a conventional lateral flow assay to form a “two1dimensional paper network” that enhanced the sensitivity of an immunoassay-based pregnancy test.208 While these devices were composed of paper, a more traditionally-fabricated digital device reconstituted and manipulated paper-based samples for dried blood spot analysis using off-chip MS.116 Devices like these integrate multiple sample preparation steps on-chip with high precision and control.
While most reports on sample preparation focused on blood, other fluid samples are routinely analyzed in clinical labs. One group used microchip electrophoresis to separate and quantify three markers of kidney stone formation in urine in a single 10-min run.211 While blood and urine sample volumes are generally milliliter-scale, microfluidic platforms also offer the ability to assay soluble analytes in lower-volume samples such as saliva, sputum, and tears. A simulated sputum sample was analyzed using a foldable paper-based device,210 and one group used on-chip alkaline electrophoresis to analyze protein biomarkers in tear samples from autoimmune patients in just 5 s.212 These samples were microliter-scale, demonstrating the utility of microfluidic platforms for low-volume samples that could not be accurately assessed by conventional methods.
Cellular Analysis
Sensitive, specific cell-based assays on-chip have been applied to intact or lysed human cells and to detection of pathogenic microorganisms in clinical samples. In some devices, the readout was based on the flow characteristics of cells in the sample. One study, for example, used microfluidic flow to study the deformability of cells obtained from chest wall fluid to screen for cancer and achieved sensitivity and specificity of 91% and 86%, respectively (Figure 7a).213 In another example, researchers decreased sample oxygen content to induce sickling of erythrocytes and simulate the vaso-occlusions of sickle cell disease.214 Occlusions decreased the flow of sample despite a constant driving pressure, and this readout was used to identify and stratify patients with sickle cell disease. A related device examined occlusions due to platelet aggregation, a key pathological event in heart attack and stroke,215 and innovative hematocrit measurements were based on progressive packing of erythrocytes in serpentine channels (Figure 7b).205
Figure 7.
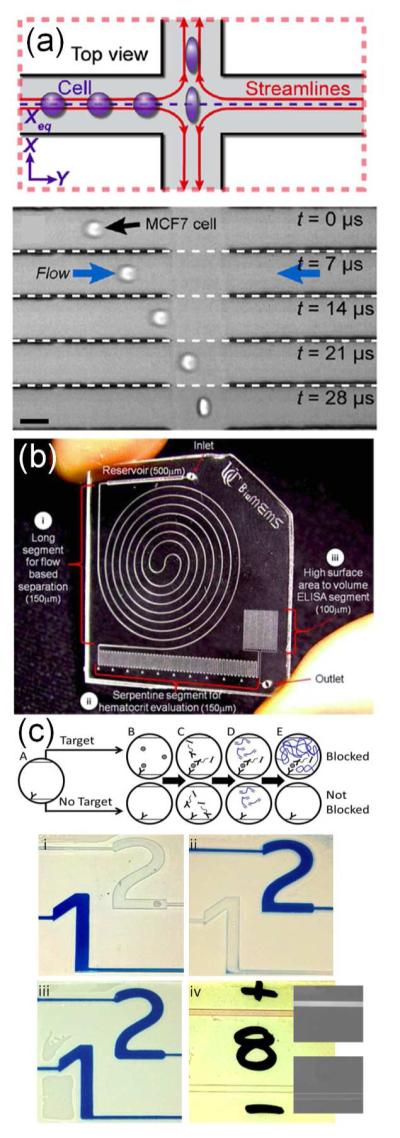
Clinically-relevant microfluidic devices for (a, b) whole cell analyses and (c) easy readout. (a) Hydrodynamic pressure applied to cells from the chest wall of patients allowed identification of malignancy by monitoring the mechanical deformability of single cells at a rate of 2000 cells/s. Reproduced from ref 213. Copyright 2012, National Academy of Sciences, USA. (b) A microfluidic device for ELISA-based measurements from whole blood. The device separated plasma from red blood cells for subsequent immunosensing. The serpentine channels also allowed for quantification of serum hematocrit, based on the length of the packed red blood cells. Reproduced from ref 205 with permission of The Royal Society of Chemistry. (c) Antigen-responsive microfluidic valves for easily interpreted results. Introduction of an antigen into an antibody-packed column led to polymerization and blockage of subsequent flow, which was used to visualize test results. Reproduced from ref 228 with permission of The Royal Society of Chemistry.
Some clinical applications require recovery of intact, viable cells after an assay or post-treatment. Storage of viable oocytes benefitted from the precise fluid handling for controlled delivery of cryo-protective agents (CPAs).216 The device controlled loading of CPAs with user-defined concentration profiles in as little as 15 min, which reduced both the osmotic damage and the toxicity of the CPAs to the oocytes. Another device monitored glucose metabolism by delivering a fluorogenic substrate to up to 10 live mouse embryos.217 The reaction was monitored without using DNA-damaging UV excitation or electrically-driven flow, and the resulting data could be used to select for human embryos most likely to implant successfully. Another group purified hematopoietic stem cells from harvested bone marrow for regenerative medicine using a PDMS filter that achieved greater than 90% enrichment and post-separation viability.218
Microorganisms
A number of clinical assays detect medically-relevant microorganisms, and recent microfluidic platforms have improved on larger-scale detection approaches. When utilized on-chip, these methods have detected viruses, such as respiratory viruses,219,220 hepatitis B and C,134,221 and HIV,221 as well as bacteria, most notably M. tuberculosis (Figure 8a),136 methicillin-resistant Staphylococcus aureus (MRSA),222 and Treponema pallidum, which causes syphilis.223 Because of the specificity of genetic and protein biomolecules, PCR and immunoassays represent the mainstay of microbiological detection for microfluidic platforms. For example, SlipChips enabled multivolume digital RT-PCR to quantify viral loads of HIV and hepatitis C virus.221 Immune- and PCR-based approaches were also combined to collect and detect, respectively, H1N1 influenza viruses from a throat swab.219 Another approach used nanostructured microelectrodes coated with sequence-specific peptide nucleic acid probes to detect genetic material from pathogens electrocatalytically.222 To inform treatment decisions, a recent study evaluated antibiotic sensitivity in a bacterial population in 100 min using a “microviscometer.”224 Monitoring the rotational period of magnetic beads in a bacterial suspension was used to quantify bacterial proliferation in the presence of various antibiotic concentrations. Ongoing efforts are needed to increase integration to minimize sample preparation and the use of expensive lab equipment, in order to translate these devices to the bedside, the clinic, or resource-poor areas.
Figure 8.
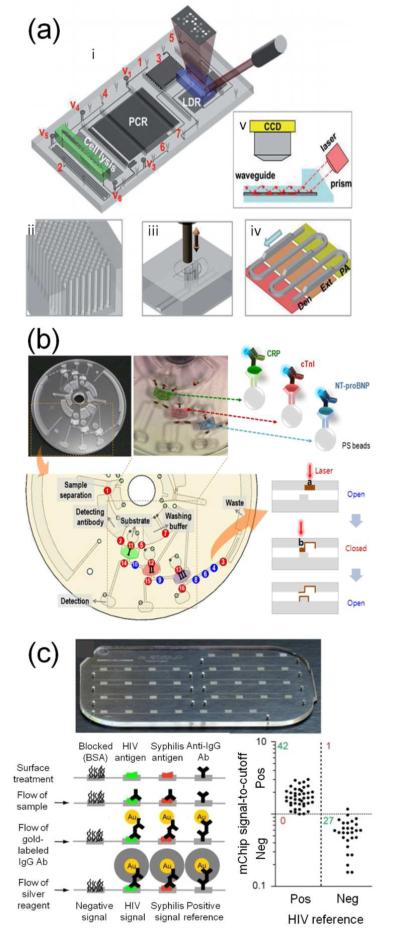
(a) Nucleic acid amplification and (b, c) immunosensing-based microfluidic devices for clinical use. (a) Integrated platform for detecting drug resistance genotypes in Mycobacterium tuberculosis. The system incorporated a cell lysis region, PCR amplification and LDR-based detection with integrated optics. Reproduced from ref 136 with permission of The Royal Society of Chemistry. (b) Immunosensing ELISA performed on a lab-on-a-disc to measure levels of human C-reactive protein, cardiac troponin I, or N-terminal prohormone of brain natriuretic peptide. Reproduced from ref 226. Copyright 2012 American Chemical Society. (c) Immunosensing device for detecting infectious microorganisms (HIV and Treponema pallidum, which causes syphilis). This device was field-tested in resource-poor areas of Rwanda. Reproduced from ref 223. Adapted by permission from Macmillan Publishers Ltd: Nature Medicine, copyright 2012.
Point-of-Care
Devices that achieve short sample-to-answer processing times, require little outside equipment, and provide an easily-interpreted readout are especially suitable as point-of-care (POC) platforms. While these features are difficult to achieve, many groups have advanced the field by integrating multiple steps onto a single chip. For example, a tunable acoustic device used multiple frequencies to complete an entire immunoassay using a single transducer and a lens-free detection system.225 Similarly, a “lab-on-a-disc” integrated sample loading, mixing, and incubation by spinning the disc at different speeds, followed by multiplexed analysis of three protein biomarkers (Figure 8b).226
POC devices that operate with minimal external equipment and provide clear readout are especially useful in resource-poor regions, such as rural areas or the developing world. For example, the “Squeeze-chip,” which generated precise flow patterns using only finger pumping by the operator, removed the need for outside pumps.227 Devices that provided clear readout without expert interpretation of results or complex analytical equipment included a device in which antibody analytes induced polymerization of a blocking material in indicator channels (Figure 7c).228 Similarly, an RT-PCR-based device indicated influenza virus detection within 1 min of operation using fluorophore-labeled primers that could be immobilized on an absorbent pad and visualized.229 Paper microfluidic devices are especially useful because they store well and are low cost, and four examples paired these advantages with simple, colorimetric readouts appropriate for use by technicians with limited training.203,208,209,230 An excellent POC example, a microfluidic ELISA called the “mChip,” was developed through collaboration of chemists, engineers, clinicians, and public health workers (Figure 8c).223 mChips were made inexpensively ($0.10/device) and rapidly (40 s) on a large scale, featured discrete reagent delivery using manual syringe pumping, and achieved quantitative detection of HIV or syphilis bacterial infection in approximately 5 min. Interpretation was performed qualitatively or quantitatively using a cell-phone-like device to measure optical density. Most importantly, the device was validated with samples from patients during extensive field-testing and optimization in Rwanda.
Microfluidic devices intended for clinical applications have much to gain from collaborations between physicians, clinical chemists and engineers for two reasons. First, clinicians have ready access to patient samples, the analysis of which is the true gold standard for a new clinical device. We chose many of the devices in this section because they were validated using patient samples from both diseased and normal subjects. Second, the experiences of medical professionals help highlight critical gaps in medical diagnostic technology. For example, several devices already exist that measure glucose in human serum with sufficient analytical power, but a new platform that lowers detection limits for cardiac biomarkers would improve early detection of heart attacks. Establishing a dialogue with clinicians makes certain that new devices meet a medical need.
FIELD
Speed and Portability
The same advances in speed and portability that benefit point-of-care analyses also make non-clinical analytical technologies field-ready. In particular, the development of portable detection systems and sample preparation methods facilitates successful transition of μTAS from the lab to the field.
Integration
Speed and portability benefit from fully integrated systems, which often contain complex electrical, mechanical, and optical components. Advanced integration simplifies devices, making them more user-friendly, and often increases information content. Recent advances in this area have included a water-activated, self-heating cartridge for a nucleic acid amplification chip231 and a microfabricated thermal modulator for sample transfer in 2D gas chromatography.232 Advances in self-powered devices included integrated batteries capable of powering electrochromic detection,233 an on-chip fuel cell that supplied electrical and pumping power (Figure 9a),234 and galvanic cells that powered an LED when sample was added to a paper-based device.235 Another autonomous system was a lab-on-a-robot, which contained a temperature-controlled microfluidic device integrated with capillary electrophoresis that could be operated by long-range remote control on an all-terrain vehicle.236 In other cases, integrated components increased the information content produced by lab-on-a-chip devices. For example, a recent device for environmental samples integrated a 3D mixer, an LED array, photodetector, and accompanying electronics for colorimetric detection at seven wavelengths, allowing determination of phenols, chromium (VI), and nitrite.237 While integrated sample pre-treatment is still needed to prepare this device for field applications, the automation and robustness of the device make it appropriate for environmental automatic alert stations.
Figure 9.
These highly portable devices are suitable for use in the field. (a) A self-powered microfluidic device that included a methanol fuel cell. Reproduced from ref 234 with permission of The Royal Society of Chemistry. (b) An on-chip distiller for measuring SO2 concentrations in wine at the production or bottling site. Reproduced from ref 243 with permission of The Royal Society of Chemistry. (c) An integrated immunoassay device for monitoring algal cyanotoxins in natural waters. Reproduced from ref 76 with permission of The Royal Society of Chemistry.
Sample Preparation
Streamlining sample preparation is a key challenge in any analysis, and field applications must accommodate diverse samples and matrices, including gases, aerosols, heterogeneous solids, such as foodstuffs and soil, and biological cells. Integrated sample preparation is often necessary for field-ready devices and has the additional advantage of increasing analysis speed. One notable example of on-chip sample clean-up was a microfluidic gradient elution moving boundary electrophoretic (GEMBE) device capable of removing dirt from aqueous environmental samples.238 Recent devices for microfluidic liquid-liquid extractions included a centrifugal chip with automated pneumatic recirculation239 and a honey-comb patterned silicon microchannel capable of extracting sulfides from both water and oil.240 For gas phase or airborne analytes, devices may transfer the sample to an aqueous phase. For example, a microfluidic aerosol-to-hydrosol sampler converted analytes from a gas to a liquid phase and was coupled to silicon nanowire sensors for near real-time detection of airborne viruses.241 Another device utilized a 3D gas/liquid laminar flow to extract and concentrate ammonia from air and operated continuously for three weeks in a cleanroom at nearly 100% extraction efficiency.242 Another on-chip system distilled gaseous sulfur dioxide (SO2) from wine; this device will allow wine producers to measure SO2on site, rather than shipping samples to laboratories (Figure 9b).243 Other devices accepted real-world samples with minimal processing, such as a device to measure methanol content in red wine with an integrated microreactor and detection channels designed to fit into a standard UV spectrophotometer.244
Environmental Safety and Public Health
Low cost, portable, and high-speed microfluidic systems contribute to many environmental safety and public health applications. Recent advances are providing rapid, on-demand testing for hazardous contaminates in our water and air.
Water safety
Water analysis has a long history of field-compatible methods, and microfluidic devices are extending and adding to existing techniques for on-site water testing. Water analysis is conducted for public safety reasons as well as to monitor changes in naturally occurring species for environmental studies. Many groups have made improvements in the cost and portability of devices that measure chemical pollutants in water, especially for applications in countries with increasing industrialization. Multiplexed assays based on paper microfluidics230 and microchip electrophoresis245 have been demonstrated for rapid, portable heavy metal detection in water samples. However, neither of these devices consistently yielded limits of detection below the World Health Organization guidelines for metals of interest.246 In order to be useful for public safety testing, the detection limits of these devices must be decreased. A recent device for nitrate and nitrite testing exemplified the characteristics required for practical application of microfluidic water analysis devices; this system had an environmentally-relevant dynamic range, measured samples every 10 min, and was continuously deployed in a river for 26 days.247 These benchmarks of sensitive, robust operation should be goals for all researchers developing field-ready systems.
Detecting microorganisms in water for environmental and public safety applications is similar to chemical detection but often includes the additional challenge of species specificity. Devices to identify pathogenic, waterborne bacteria included an immunoassay with electrochemical impedance spectroscopy detection,248 a multiplexed nanoparticle-based DNA barcode immunoassay with electrophoresis,249 a compact, modular device for PCR and array-based detection,250 and a fluorescence in situ hybridization (FISH) assay coupled to a microfluidic flow cytometer.251 This latter detection strategy was used to identify bacteria at a contaminated environmental site in Hanford, WA. Both non-pathogenic and pathogenic algae were also targets for microfluidic detection. Measuring changes in the type of algae species can be an effective way to monitor the environmental impact of climate change. For this reason, identification of five algae species was conducted using a machine-learning algorithm to classify the intensity distribution of laser light from a microfabricated waveguide.252 In addition, several devices were designed to assay algal biochemicals for public safety reasons. A microfluidic immunoassay measured three algal toxins in 2/3 the time of conventional ELISA and with 1000x lower reagent volumes (Figure 9c).76 Another device used a microchannel integrated with an LED, filter set, and photodiode to measure the fluorescence of chlorophyll in algae as an indicator for algal stress in the presence of chemical pollutants.253
Security, Defense, and Public Safety
Portable, automated microfluidic systems have obvious advantages for security, defense, and workplace safety applications. A recent device successfully detected biological and chemical warfare agents, including anthrax, ricin, and explosives. The eight-channel device used a commercial detector with disposable cartridges containing antibody-coated waveguides to monitor binding of anthrax spores, F. tularensis cells, Vaccinia virus, and ricin toxin.254 Antibody-coated field effect transistors (FETs) provided another sensitive non-optical detection technique for lab-on-a-chip applications. On-chip FET sensors have been integrated in microchannels241 and a digital device;255 both platforms were applied to virus detection on a seconds to minutes timescale. Hazardous aerosols, particulates, and gases are additional public health and workplace safety issues that can be addressed by microfluidics. Two recent devices assayed particulate matter for oxidative activity256 and metal content.257 The first apparatus included a particle-to-liquid sampling system and used a colorimetric reaction with oxidized dithiothreitol to determine oxidative activity.256 The second device dissolved metal particulates, such as those from machining or transportation services, and detected μg-levels using filter paper-based fluidics to decrease turn-around time from sampling to measurement.257 Other instruments with workplace safety applications detected potentially hazardous gases using chemiresistors.258,259 In one example, chemiresistive sensing was paired with microfluidic GC separation for in-home screening for trichloroethylene (TCE) 259 a toxic gas that can seep into homes from groundwater. The portable system detected TCE levels below the Environmental Protection Agency regional screening limit and identified sources of contamination in real-world environments.
In most public safety applications, rapid results are critical; however, many binding-based detection methods integrate signal over time, which can limit analysis speed. For example, several of the devices discussed here require 10-60 min to obtain signal-to-noise ratios sufficient for reliable detection.76,248,249,254 While this time frame is acceptable for some applications, in emergency situations dangerous chemicals and infectious pathogens must be detected as rapidly as possible. Consequently, the signal-to-noise ratio as a function of time must be improved for many of these systems to establish their utility in real-life situations.
Challenging Environments
Some field environments present unique technical challenges, especially for analyses conducted at sea or in outer space. Measuring chemical or biological species in the ocean requires robust, sensitive, and high-throughput devices to achieve high spatial resolution on moving vehicles. A submersible microfluidic platform performed displacement-based immunoaffinity assays for explosive compounds from bombs planted by terrorists or left by former military operations (Figure 10a).260 This device detected trinitrotoluene (TNT) 60 times faster and with two orders of magnitude more sensitivity than conventional techniques. Other devices targeted biologically-relevant analytes. For example, a device for the colorimetric detection of ocean nutrients utilized a multiplexed stop-flow system to perform measurements with high spatial resolution, allowing it to be deployed on moving vehicles, such as buoys or ocean gliders.261 Another device for an underwater bioluminescence assay of microbial activity was successfully operated at a shallow hot spring in Okinawa (Figure 10b).262 The microfluidic device, electronics and photomultiplier tube were contained in a pressurized vessel and waste was collected rather than dispensed into the environment. In another system, acoustic radiation was used to collect and trap microorganisms in a microfluidic device at a depth of 1200 meters.263 These submersible microfluidic devices have displayed impressive robustness and accuracy in the ocean – one of the most analytically challenging environments on the planet – where they are surrounded by high salt concentrations, high pressures, fluctuating temperatures, and abundant biological activity.
Figure 10.
μTAS have been designed for deployment in highly challenging environments, including (a) the ocean, (b) hot springs, and (c) outer space. (a) A submersible microfluidic device for detecting explosive compounds in the ocean. Reprinted from ref 260. Copyright 2011 American Chemical Society. (b) A pressurized vessel containing a photomultiplier tube, electronics, and a microfluidic device for measuring microbial activity in a shallow hot spring. Reproduced from ref 262 with permission of The Royal Society of Chemistry. (c) A microfluidic device for voltage clamp studies of oocytes during parabolic flight. Reproduced from ref 265 with permission of The Royal Society of Chemistry.
Another challenging environment for analysis is outer space, where devices must be compact and completely autonomous for months on end. Highly integrated and automated instrumentation enables the use of microfluidic devices on unmanned space flights. A recent system executed completely automated fluorescent-labeling, electrophoretic separation, and detection of amino acids on a single chip for the first time for eventual application in astrobiology studies on Mars and elsewhere.264 Microfluidic devices have also been developed to study the effects of gravitational fields265 and ionizing radiation266 on biological processes. For example, microfluidic patch clamping experiments during parabolic flight showed varying sodium channel activity under hyper- and microgravity (Figure 10c).265 In addition to affecting biological specimens, microgravity changes fluidic behavior in μTAS. In microgravity, cells do not settle and convective fluid flow is minimal. These considerations affected a study of Bacillus subtilis germination conducted in low earth orbit.266 A deeper understanding of the effects of microgravity on fluidic operations coupled with a further increase in robustness will make μTAS valuable research tools for extraterrestrial exploration.
CONCLUSIONS AND OUTLOOK
The wide range of laboratory, clinical, and field applications discussed here highlights the versatility of μTAS. While miniaturized analysis is not well-suited to every application, exceptional device performance can be achieved by thoughtfully matching the strengths of microscale systems to specific applications. New research areas (such as the novel physical and biological applications noted here) demonstrate that microfluidics has much to contribute beyond the well-established area of microscale separations.
In the research laboratory, microfluidic systems continue to reduce reagent consumption, decrease analysis time, and automate analyses in traditional applications like biochemical assays. These benefits also contribute to on-chip syntheses of molecular and supramolecular entities. Although synthetic applications are not directly relevant to analysis, the development of microreactors on-chip represents a unique opportunity for microfluidics researchers to contribute to interdisciplinary efforts that may have downstream analytical relevance, for example, contrast agents for clinical imaging. The characteristic attributes of microdevices have also enhanced systems for cell culture, manipulation, and analysis. As biological applications of microfluidics mature, devices provide more sophisticated cell culture platforms and probe biological systems across a broad range of complexities from cells to tissues to organisms.
A true lab-on-a-chip should not require an external, macroscale laboratory to support its operation. Consequently, μTAS designed for clinical and field applications must be robust and portable. To convince clinicians, field biologists, environmental scientists, and public safety officers to adopt μTAS in their work, researchers must analyze real samples in real-world scenarios and non-laboratory settings. To accomplish this, researchers must often address challenges beyond chemical analysis and find engineering solutions for portable power sources, intuitive readout, and remote operation. As interdisciplinary teams advance microfluidic technology, μTAS will contribute to new analytical methods in the research lab and beyond.
ACKNOWLEDGMENT
The authors thank Jessica Park Lindsey for preparing the table of contents graphic. This work was supported by grants from the National Institutes of Biomedical Imaging and Bioengineering, R01EB012549 (NLA); National Cancer Institute, F31CA171631 (AJD) and F32CA162574 (ATM); National Institute of General Medical Sciences: Minority Opportunities in Research grant K12GM000678 (MLK) and training grant T32GM007040 (DMO).
ABBREVIATIONS
- ATR-FTIR
attenuated total reflection Fourier-transform infrared
- CTC
circulating tumor cell
- CPA
cryo-protective agent
- ELISA
enzyme-linked immunosorbent assay
- EGF
epidermal growth factor
- EpCAM
epithelial cell adhesion molecule
- ECM
extracellular matrix
- FET
field effect transistor
- FISH
fluorescence in situ hybridization
- GEMBE
gradient elution moving boundary electrophoresis
- HUVEC
human umbilical vein endothelial cell
- MRI
magnetic resonance imaging
- MRSA
methicillin-resistant Staphylococcus aureus
- μTAS
micro total analysis systems
- POC
point-of-care
- PDMS
polydimethylsiloxane
- PCR
polymerase chain reaction
- PMMA
polymethylmethacrylate
- PTFE
polytetrafluoroethylene
- PET
positron emission topography
- qPCR
quantitative polymerase chain reaction
- RT-PCR
reverse-transcriptase polymerase chain reaction
- REEAD
rolling-circle-enhanced enzyme activity detection
- SPR
surface plasmon resonance
- TLR4
toll-like receptor 4
- TCE
trichloroethylene
- TNT
trinitrotoluene
Biographies
Michelle L. Kovarik studied chemistry at Saint Louis University and went on to obtain her Ph.D. in analytical chemistry from Indiana University-Bloomington in 2009. She is currently a SPIRE postdoctoral scholar at the University of North Carolina-Chapel Hill. Her research interests focus on biological and biochemical applications of micro- and nanofluidic devices.
Douglas M. Ornoff received his B.Sc. in Chemistry from the University of North Carolina-Chapel Hill in 2007, concentrating in biological and analytical chemistry. After a postbaccalaureate fellowship at NIEHS studying virology and host-pathogen interactions, he entered into a combined M.D./Ph.D. program at the University of North Carolina School of Medicine and is currently a Ph.D. candidate in pharmacology. His graduate research is at the interface of microscale analysis and pulmonary physiology. His research interests lie in applying bioanalytical chemistry and microfluidics to fundamental problems in respiratory cell physiology.
Adam T. Melvin studied chemical engineering and chemistry at the University of Arizona and went on to obtain his Ph.D. in chemical engineering at North Carolina State University in 2010 under the direction of Jason Haugh. He is currently an NIH postdoctoral fellow at the University of North Carolina-Chapel Hill in the Department of Chemistry. His previous research focused on the live cell imaging of PI3K dynamics during fibroblast chemotaxis, while his current work involves the bioengineering of novel substrates targeting the ubiquitin proteasome system.
Nicholas C. Dobes received his B.S. in Chemistry in 2009 from Benedictine University. He is currently a graduate student in the Department of Chemistry at the University of North Carolina-Chapel Hill. A former recipient of Parent Project Muscular Dystrophy’s Weisman Fellowship, his current research involves the development and modification of microsystems for the culture, isolation and analysis of primary muscle stem cells.
Yuli Wang received his Ph.D. in Materials Sciences in 2003 from the University of California at Irvine. Since then he has worked with Nancy Allbritton in developing new lab-on-a-chip technologies to support biologists in addressing important biomedical problems. He is currently a research associate in the Department of Chemistry at the University of North Carolina-Chapel Hill. Besides his academic research, he is interested in entrepreneurship and has co-founded two startup companies: Intellego Corporation and Cell Microsystems. His current research involves developing platforms for studying intestinal and airway epithelial cells and tissues.
Alexandra J. Dickinson received her B. A. in Chemistry and History at Swarthmore College in 2009. Currently, she is an NIH predoctoral fellow studying analytical chemistry at the University of North Carolina-Chapel Hill. Her research focuses on improving the throughput of single-cell analysis using capillary electrophoresis.
Philip C. Gach received his B.S. in Biochemistry in 2007 from Indiana University. He obtained his Ph.D. in analytical chemistry from the University of North Carolina-Chapel Hill where he currently works as a postdoctoral scholar. His current research involves developing platforms for sorting single adherent cells.
Pavak K. Shah received his B.S. in Biomedical Engineering from the North Carolina State University in 2009 concentrating in Bioinstrumentation. His current research focuses on the development of automated platforms for the study of complex phenotypes in single cells,
Nancy L. Allbritton received her Ph.D. in Medical Physics/Medical Engineering from the Massachusetts Institute of Technology, and her M.D. from the Johns Hopkins University. Upon completion of a postdoctoral fellowship in cell biology at Stanford University, she joined the faculty of the University of California at Irvine. She joined the University of North Carolina-Chapel Hill (UNC) as the Debreczeny Distinguished Professor in the Department of Chemistry in 2007 followed by appointment as Professor and Chair of the Department of Biomedical Engineering in the School of Medicine at UNC and the College of Engineering at North Carolina State University in 2009.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- (1).Hitzbleck M, Gervais L, Delamarche E. Lab Chip. 2011;11:2673–2679. doi: 10.1039/c1lc20282k. [DOI] [PubMed] [Google Scholar]
- (2).Vulto P, Podszun S, Meyer P, Hermann C, Manz A, Urban GA. Lab Chip. 2011;11:1596–1602. doi: 10.1039/c0lc00643b. [DOI] [PubMed] [Google Scholar]
- (3).Liu CC, Thompson JA, Bau HH. Lab Chip. 2011;11:1688–1693. doi: 10.1039/c1lc20089e. [DOI] [PubMed] [Google Scholar]
- (4).Luecha J, Hsiao A, Brodsky S, Liu G, Kokini J. Lab Chip. 2011;11:3419–3425. doi: 10.1039/c1lc20726a. [DOI] [PubMed] [Google Scholar]
- (5).Liu H, Crooks R. J. Am. Chem. Soc. 2011;133:17564–17566. doi: 10.1021/ja2071779. [DOI] [PubMed] [Google Scholar]
- (6).Jamal M, Zarafshar AM, Gracias DH. Nat. Commun. 2011:2. doi: 10.1038/ncomms1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Mussard W, Kebir N, Kriegel I, Esteve M, Semetey V. Angew. Chem. Int. Edit. 2011;50:10871–10874. doi: 10.1002/anie.201101029. [DOI] [PubMed] [Google Scholar]
- (8).Tropmann A, Tanguy L, Koltay P, Zengerle R, Riegger L. Langmuir. 2012;28:8292–8295. doi: 10.1021/la301283m. [DOI] [PubMed] [Google Scholar]
- (9).Vila-Planas J, Fernandez-Rosas E, Ibarlucea B, Demming S, Nogues C, Plaza J, Dominguez C, Buttgenbach S, Llobera A. Nat. Protoc. 2011;6:1642–1655. doi: 10.1038/nprot.2011.383. [DOI] [PubMed] [Google Scholar]
- (10).Nakazato H, Kawaguchi H, Iwabuchi A, Hane K. Lab Chip. 2012;12:3419–3425. doi: 10.1039/c2lc40178a. [DOI] [PubMed] [Google Scholar]
- (11).Lin S, Crozier K. Lab Chip. 2011;11:4047–4051. doi: 10.1039/c1lc20574a. [DOI] [PubMed] [Google Scholar]
- (12).Krishnan M, Erickson D. Lab Chip. 2012;12:613–621. doi: 10.1039/c1lc20891h. [DOI] [PubMed] [Google Scholar]
- (13).Fei P, He Z, Zheng CH, Chen T, Men YF, Huang YY. Lab Chip. 2011;11:2835–2841. doi: 10.1039/c1lc20425d. [DOI] [PubMed] [Google Scholar]
- (14).Lee W, Luo YH, Zhu QR, Fan XD. Opt. Express. 2011;19:19668–19674. doi: 10.1364/OE.19.019668. [DOI] [PubMed] [Google Scholar]
- (15).Shilton RJ, Glass NR, Chan P, Yeo LY, Friend JR. Appl. Phys. Lett. 2011:98. [Google Scholar]
- (16).Glass N, Shilton R, Chan P, Friend J, Yeo L. Small. 2012;8:1881–1888. doi: 10.1002/smll.201102282. [DOI] [PubMed] [Google Scholar]
- (17).Ding X, Lin SC, Kiraly B, Yue H, Li S, Chiang IK, Shi J, Benkovic SJ, Huang T. J. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11105–11109. doi: 10.1073/pnas.1209288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nie BQ, Xing SY, Brandt JD, Pan TR. Lab Chip. 2012;12:1110–1118. doi: 10.1039/c2lc21168h. [DOI] [PubMed] [Google Scholar]
- (19).Belardi J, Schorr N, Prucker O, Ruhe J. Adv. Funct. Mater. 2011;21:3314–3320. [Google Scholar]
- (20).Issadore D. Sci. Transl. Med. 2012:4. doi: 10.1126/scitranslmed.3003747. DOI: 10.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ryan H, Song S, Zass A, Korvink J, Utz M. Anal. Chem. 2012;84:3696–3702. doi: 10.1021/ac300204z. [DOI] [PubMed] [Google Scholar]
- (22).Gabrielsson EO, Tybrandt K, Berggren M. Lab Chip. 2012;12:2507–2513. doi: 10.1039/c2lc40093f. [DOI] [PubMed] [Google Scholar]
- (23).Cheng S, Wu Z. Adv. Funct. Mater. 2011;21:2282–2290. [Google Scholar]
- (24).Orth A, Schonbrun E, Crozier KB. Lab Chip. 2011;11:3810–3815. doi: 10.1039/c1lc20114j. [DOI] [PubMed] [Google Scholar]
- (25).Kundu P, Bhattacharyya TK, Das SJ. Micromech. Microeng. 2012;22:15. [Google Scholar]
- (26).Shepherd RF, Ilievski F, Choi W, Morin SA, Stokes AA, Mazzeo AD, Chen X, Wang M, Whitesides GM. Proc. Natl. Acad. Sci. U.S.A. 2011 doi: 10.1073/pnas.1116564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Jensen K, Lee J, Bohr T, Bruus H, Holbrook N, Zwieniecki M. RSC Interface. 2011;8:1155–1165. doi: 10.1098/rsif.2010.0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nguyen D, Leho Y, Esser-Kahn A. Lab Chip. 2012;12:1246–1250. doi: 10.1039/c2lc00033d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Morin SA, Shepherd RF, Kwok SW, Stokes AA, Nemiroski A, Whitesides GM. Science. 2012;337:828–832. doi: 10.1126/science.1222149. [DOI] [PubMed] [Google Scholar]
- (30).Samuel R, Sant HJ, Jiao F, Johnson CR, Gale BK. J. Micromech. Microeng. 2011:21. doi: 10.1088/0960-1317/21/9/095027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Luu L, Roman PA, Mathews SA, Ramella1Roman JC. Biomed. Opt. Express. 2012;3:1350–1364. doi: 10.1364/BOE.3.001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gunda NSK, Bera B, Karadimitriou NK, Mitra SK, Hassanizadeh SM. Lab Chip. 2011;11:3785–3792. doi: 10.1039/c1lc20556k. [DOI] [PubMed] [Google Scholar]
- (33).Singh R, Olson M. Environ. Sci. Technol. 2011;45:8780–8787. doi: 10.1021/es201706w. [DOI] [PubMed] [Google Scholar]
- (34).Pearce CI, Wilkins MJ, Zhang CY, Heald SM, Fredrickson JK, Zachara JM. Environ. Sci. Technol. 2012;46:7992–8000. doi: 10.1021/es301050h. [DOI] [PubMed] [Google Scholar]
- (35).Krebs T, Schroen K, Boom R. Lab Chip. 2012;12:1060–1070. doi: 10.1039/c2lc20930f. [DOI] [PubMed] [Google Scholar]
- (36).Abolhasani M, Singh M, Kumacheva E, Gunther A. Lab Chip. 2012;12:1611–1618. doi: 10.1039/c2lc21043f. [DOI] [PubMed] [Google Scholar]
- (37).Schultz KM, Furst EM. Lab Chip. 2011;11:3802–3809. doi: 10.1039/c1lc20376b. [DOI] [PubMed] [Google Scholar]
- (38).Chen C, Sarkar A, Song Y, Miller M, Kim S, Griffith L, Lauffenburger D, Han J. J. Am. Chem. Soc. 2011;133:10368–10371. doi: 10.1021/ja2036628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yun JY, Jambovane S, Kim S-K, Cho S-H, Duin EC, Hong JW. Anal. Chem. 2011;83:6148–6153. doi: 10.1021/ac201177g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).SooHoo J, Herr J, Ramsey J, Walker G. Anal. Chem. 2012;84:2195–2201. doi: 10.1021/ac202461h. [DOI] [PubMed] [Google Scholar]
- (41).Rob T, Liuni P, Gill P, Zhu S, Balachandran N, Berti P, Wilson D. Anal. Chem. 2012;84:3771–3779. doi: 10.1021/ac300365u. [DOI] [PubMed] [Google Scholar]
- (42).Jegou A, Niedermayer T, Orban J, Didry D, Lipowsky R, Carlier M, Romet-Lemonne G. PLoS Biol. 2011;9:e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Jiang Y, Douglas NR, Conley NR, Miller EJ, Frydman J, Moerner WE. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16962–16967. doi: 10.1073/pnas.1112244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Goldsmith RH, Tabares LC, Kostrz D, Dennison C, Aartsma TJ, Canters GW, Moerner WE. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17269–17274. doi: 10.1073/pnas.1113572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ainla A, Jeffries G, Brune R, Orwar O, Jesorka A. Lab Chip. 2012;12:1255–1261. doi: 10.1039/c2lc20906c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhou Y, Pang Y, Huang Y. Anal. Chem. 2012;84:2576–2584. doi: 10.1021/ac203469v. [DOI] [PubMed] [Google Scholar]
- (47).Momotenko D, Cortes-Salazar F, Lesch A, Wittstock G, Girault H. Anal. Chem. 2011;83:5275–5282. doi: 10.1021/ac2006729. [DOI] [PubMed] [Google Scholar]
- (48).Chung AJ, Cordovez B, Jasuja N, Lee DJ, Huang XT, Erickson D. Microfluid. Nanofluid. 2012;13:345–352. [Google Scholar]
- (49).Staykova M, Holmes DP, Read C, Stone HA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9084–9088. doi: 10.1073/pnas.1102358108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Richmond DL, Schmid EM, Martens S, Stachowiak JC, Liska N, Fletcher DA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9431–9436. doi: 10.1073/pnas.1016410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Xu X, et al. Nat. Biotechnol. 2011;29:735–U131. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Hirose Y, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4263–4268. doi: 10.1073/pnas.1117560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Johnson BE, Glauser DA, Dan-Glauser ES, Halling DB, Aldrich RW, Goodman MB. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20784–20789. doi: 10.1073/pnas.1116795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Illing PT, et al. Nature. 2012;486:554–558. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- (55).Wei P, Wong WW, Park JS, Corcoran EE, Peisajovich SG, Onuffer JJ, Weiss A, Lim WA. Nature. 2012;488:384–388. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).White R, Quake S, Curr KJ. Virol. Methods. 2012;179:45–50. doi: 10.1016/j.jviromet.2011.09.017. [DOI] [PubMed] [Google Scholar]
- (57).Walter K, et al. Clin. Cancer Res. 2012;18:2360–2373. doi: 10.1158/1078-0432.CCR-11-2635-T. [DOI] [PubMed] [Google Scholar]
- (58).Windbergs M, Weitz D. Small. 2011;7:3011–3015. doi: 10.1002/smll.201100520. [DOI] [PubMed] [Google Scholar]
- (59).Miller OJ, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:378–383. doi: 10.1073/pnas.1113324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Dimov IK, Kijanka G, Park Y, Ducree J, Kang T, Lee LP. Lab Chip. 2011;11:2701–2710. doi: 10.1039/c1lc20105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Lim K, Park J, Rhee S, Yoon T. Cytom. Part A. 2012;81A:691–697. doi: 10.1002/cyto.a.22079. [DOI] [PubMed] [Google Scholar]
- (62).Mao SF, Gao D, Liu W, Wei HB, Lin JM. Lab Chip. 2012;12:219–226. doi: 10.1039/c1lc20678h. [DOI] [PubMed] [Google Scholar]
- (63).Kim J, Taylor D, Agrawal N, Wang H, Kim H, Han A, Rege K, Jayaraman A. Lab Chip. 2012;12:1813–1822. doi: 10.1039/c2lc21202a. [DOI] [PubMed] [Google Scholar]
- (64).Churski K, Kaminski TS, Jakiela S, Kamysz W, Baranska-Rybak W, Weibel DB, Garstecki P. Lab Chip. 2012;12:1629–1637. doi: 10.1039/c2lc21284f. [DOI] [PubMed] [Google Scholar]
- (65).Vega N, Allison K, Khalil A, Collins J. Nat. Chem. Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung C.-k., Pourmand N, Austin RH. Science. 2011;333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- (67).Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. Science. 2012;335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Cao J, Kursten D, Schneider S, Knauer A, Gunther P, Kohler J. Lab Chip. 2012;12:474–484. doi: 10.1039/c1lc20584f. [DOI] [PubMed] [Google Scholar]
- (69).Shen J, Zhou Y, Lu T, Peng J, Lin Z, Huang L, Pang Y, Yu L, Huang Y. Lab Chip. 2012;12:317–324. doi: 10.1039/c1lc20845d. [DOI] [PubMed] [Google Scholar]
- (70).He M, Novak J, Julian B, Herr A. J. Am. Chem. Soc. 2011;133:19610–19613. doi: 10.1021/ja207963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Zheng CH, Wang JW, Pang YH, Wang JB, Li WB, Ge ZG, Huang YY. Lab Chip. 2012;12:2487–2490. doi: 10.1039/c2lc40145b. [DOI] [PubMed] [Google Scholar]
- (72).Lovchik RD, Kaigala GV, Georgiadis M, Delamarche E. Lab Chip. 2012;12:1040–1043. doi: 10.1039/c2lc21016a. [DOI] [PubMed] [Google Scholar]
- (73).Mai J, Sommer GJ, Hatch AV. Anal. Chem. 2012;84:3538–3545. doi: 10.1021/ac203076p. [DOI] [PubMed] [Google Scholar]
- (74).Gaster RS, Xu L, Han S-J, Wilson RJ, Hall D. a., Osterfeld SJ, Yu H, Wang SX. Nat. Nanotechnol. 2011;6:314–320. doi: 10.1038/nnano.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Henley W, Dennis P, Ramsey J. Anal. Chem. 2012;84:1776–1780. doi: 10.1021/ac202445g. [DOI] [PubMed] [Google Scholar]
- (76).Zhang J, Liu S, Yang P, Sui G. Lab Chip. 2011;11:3516–3522. doi: 10.1039/c1lc20516a. [DOI] [PubMed] [Google Scholar]
- (77).Hahn Y, Park J. Lab Chip. 2011;11:2045–2048. doi: 10.1039/c0lc00569j. [DOI] [PubMed] [Google Scholar]
- (78).Kim J, Jensen EC, Megens M, Boser B, Mathies RA. Lab Chip. 2011;11:3106–3112. doi: 10.1039/c1lc20407f. [DOI] [PubMed] [Google Scholar]
- (79).Dykstra P, Roy V, Byrd C, Bentley W, Ghodssi R. Anal. Chem. 2011;83:5920–5927. doi: 10.1021/ac200835s. [DOI] [PubMed] [Google Scholar]
- (80).Lynn N, Tobet S, Henry C, Dandy D. Anal. Chem. 2012;84:1360–1366. doi: 10.1021/ac202314n. [DOI] [PubMed] [Google Scholar]
- (81).Ghosh T, Mastrangelo C. Analyst. 2012;137:2381–2385. doi: 10.1039/c2an35045a. [DOI] [PubMed] [Google Scholar]
- (82).Hughes AJ, Lin RKC, Peehl DM, Herr AE. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5972–5977. doi: 10.1073/pnas.1108617109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Fallah-Araghi A, Baret J, Ryckelynck M, Griffiths A. Lab Chip. 2012;12:882–891. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- (84).Wang J, et al. Proc. Natl. Acad. Sci.U.S.A. 2011;108:6909–6914. doi: 10.1073/pnas.1014753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Fraikin J, Teesalu T, McKenney C, Ruoslahti E, Cleland A. Nat. Nanotechnol. 2011;6:308–313. doi: 10.1038/nnano.2011.24. [DOI] [PubMed] [Google Scholar]
- (86).Debs BE, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11570–11575. doi: 10.1073/pnas.1204514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Wu A, Kawahara T, Rapicavoli N, van Riggelen J, Shroff E, Xu L, Felsher D, Chang H, Quake S. Lab Chip. 2012;12:2190–2198. doi: 10.1039/c2lc21290k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).McKenna B, Evans J, Cheung M, Ehrlich D. Nat. Methods. 2011;8:401–403. doi: 10.1038/nmeth.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Fidalgo LM, Maerkl SJ. Lab Chip. 2011;11:1612–1619. doi: 10.1039/c0lc00537a. [DOI] [PubMed] [Google Scholar]
- (90).Siuti P, Retterer S, Doktycz M. Lab Chip. 2011;11:3523–3529. doi: 10.1039/c1lc20462a. [DOI] [PubMed] [Google Scholar]
- (91).Greener J, Tumarkin E, Debono M, Kwan C, Abolhasani M, Guenther A, Kumacheva E. Analyst. 2012;137:444–450. doi: 10.1039/c1an15940b. [DOI] [PubMed] [Google Scholar]
- (92).Park C, Maurya R, Lee J, Kim D. Lab Chip. 2011;11:1941–1945. doi: 10.1039/c1lc20071b. [DOI] [PubMed] [Google Scholar]
- (93).Barikbin Z, Rahman M, Khan S. Small. 2012;8:2152–2157. doi: 10.1002/smll.201102748. [DOI] [PubMed] [Google Scholar]
- (94).Theberge A, Mayot E, El Harrak A, Kleinschmidt F, Huck W, Griffiths A. Lab Chip. 2012;12:1320–1326. doi: 10.1039/c2lc21019c. [DOI] [PubMed] [Google Scholar]
- (95).Lee JS, Lee SH, Kim JH, Park CB. Lab Chip. 2011;11:2309–2311. doi: 10.1039/c1lc20303g. [DOI] [PubMed] [Google Scholar]
- (96).Zhang J, Coulston RJ, Jones ST, Geng J, Scherman OA, Abell C. Science. 2012;335:690–694. doi: 10.1126/science.1215416. [DOI] [PubMed] [Google Scholar]
- (97).Wang C, Sinton D, Moffitt M. J. Am. Chem. Soc. 2011;133:18853–18864. doi: 10.1021/ja2067252. [DOI] [PubMed] [Google Scholar]
- (98).Roig Y, Marre S, Cardinal T, Aymonier C. Angew. Chem. Int. Edit. 2011;50:12071–12074. doi: 10.1002/anie.201106201. [DOI] [PubMed] [Google Scholar]
- (99).Ji X-H, Cheng W, Guo F, Liu W, Guo S-S, He Z-K, Zhao X-Z. Lab Chip. 2011;11:2561–2568. doi: 10.1039/c1lc20150f. [DOI] [PubMed] [Google Scholar]
- (100).Jiang K, Xue C, Arya C, Shao C, George E, DeVoe D, Raghavan S. Small. 2011;7:2470–2476. doi: 10.1002/smll.201100514. [DOI] [PubMed] [Google Scholar]
- (101).Kim J, Li Z, Park I. Lab Chip. 2011;11:1946–1951. doi: 10.1039/c1lc20079h. [DOI] [PubMed] [Google Scholar]
- (102).Witters D, Vergauwe N, Ameloot R, Vermeir S, De Vos D, Puers R, Sels B, Lammertyn J. Adv. Mater. 2012;24:1316–1320. doi: 10.1002/adma.201104922. [DOI] [PubMed] [Google Scholar]
- (103).Kinahan M, Filippidi E, Koster S, Hu X, Evans H, Pfohl T, Kaplan D, Wong J. Biomacromolecules. 2011;12:1504–1511. doi: 10.1021/bm1014624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Laza S, Polo M, Neves A, Cingolani R, Camposeo A, Pisignano D. Adv. Mater. 2012;24:1304–1308. doi: 10.1002/adma.201103357. [DOI] [PubMed] [Google Scholar]
- (105).Thiele J, Windbergs M, Abate AR, Trebbin M, Shum HC, Forster S, Weitz DA. Lab Chip. 2011;11:2362–2368. doi: 10.1039/c1lc20298g. [DOI] [PubMed] [Google Scholar]
- (106).Zhang Q, Xu L, Zhou Y, Wang J, Chen J. Ind. Eng. Chem. Res. 2011;50:13805–13812. [Google Scholar]
- (107).Kitazoe K, Wang J, Kaji N, Okamoto Y, Tokeshi M, Kogure K, Harashima H, Baba Y. Lab Chip. 2011;11:3256–3262. doi: 10.1039/c1lc20392d. [DOI] [PubMed] [Google Scholar]
- (108).Chen D, et al. J. Am. Chem. Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- (109).Tran T, Nguyen C, Kim D, Lee Y, Huh K. Lab Chip. 2012;12:589–594. doi: 10.1039/c1lc20769e. [DOI] [PubMed] [Google Scholar]
- (110).Martz T, Sheeran P, Bardin D, Lee A, Dayton P. Ultrasound Med. Biol. 2011;37:1952–1957. doi: 10.1016/j.ultrasmedbio.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Keng PY, et al. Proc. Natl. Acad. Sci. U.S.A. 2011 [Google Scholar]
- (112).Liu K, et al. Mol. Imaging. 2011;10:1681176, 161–167. [Google Scholar]
- (113).Huang S, Zeng S, He Z, Lin B. Lab Chip. 2011;11:3407–3410. doi: 10.1039/c1lc20537d. [DOI] [PubMed] [Google Scholar]
- (114).Diguet A, Li H, Queyriaux N, Chen Y, Baigl D. Lab Chip. 2011;11:2666–2669. doi: 10.1039/c1lc20328b. [DOI] [PubMed] [Google Scholar]
- (115).Xu J, Chen R, Wang Y, Luo G. Lab Chip. 2012;12:2029–2036. doi: 10.1039/c2lc21193a. [DOI] [PubMed] [Google Scholar]
- (116).Jebrail M, Yang H, Mudrik J, Lafreniere N, McRoberts C, Al-Dirbashi O, Fisher L, Chakraborty P, Wheeler A. Lab Chip. 2011;11:3218–3224. doi: 10.1039/c1lc20524b. [DOI] [PubMed] [Google Scholar]
- (117).Luk V, Fiddes L, Luk V, Kumacheva E, Wheeler A. Proteomics. 2012;12:1310–1318. doi: 10.1002/pmic.201100608. [DOI] [PubMed] [Google Scholar]
- (118).Srigunapalan S, Eydelnant I, Simmons C, Wheeler A. Lab Chip. 2012;12:369–375. doi: 10.1039/c1lc20844f. [DOI] [PubMed] [Google Scholar]
- (119).Aijian AP, Chatterjee D, Garrell RL. Lab Chip. 2012;12:2552–2559. doi: 10.1039/c2lc21135a. [DOI] [PubMed] [Google Scholar]
- (120).Hadwen B, Broder G, Morganti D, Jacobs A, Brown C, Hector J, Kubota Y, Morgan H. Lab Chip. 2012;12:3305–3313. doi: 10.1039/c2lc40273d. [DOI] [PubMed] [Google Scholar]
- (121).Travagliati M, De Simoni G, Lazzarini C, Piazza V, Beltram F, Cecchini M. Lab Chip. 2012;12:2621–2624. doi: 10.1039/c2lc40396j. [DOI] [PubMed] [Google Scholar]
- (122).Sinz D, Darhuber A. Lab Chip. 2012;12:705–707. doi: 10.1039/c2lc21082g. [DOI] [PubMed] [Google Scholar]
- (123).Lee S, Lee S, Kang K. Appl. Phys. Lett. 2012:100. [Google Scholar]
- (124).Knowles TPJ, White DA, Abate AR, Agresti JJ, Cohen SIA, Sperling RA, De Genst EJ, Dobson CM, Weitz DA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14746–14751. doi: 10.1073/pnas.1105555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Martino C, Zagnoni M, Sandison M, Chanasakulniyom M, Pitt A, Cooper J. Anal. Chem. 2011;83:5361–5368. doi: 10.1021/ac200876q. [DOI] [PubMed] [Google Scholar]
- (126).Kodzius R, Xiao K, Wu J, Yi X, Gong X, Foulds I, Wen W. Sensor Actuat. B-Chem. 2012;161:349–358. [Google Scholar]
- (127).Heyries KA, Tropini C, Vaninsberghe M, Doolin C, Petriv OI, Singhal A, Leung K, Hughesman CB, Hansen CL. Nat. Methods. 2011;8:649–651. doi: 10.1038/nmeth.1640. [DOI] [PubMed] [Google Scholar]
- (128).Shen F, Davydova E, Du W, Kreutz J, Piepenburg O, Ismagilov R. Anal. Chem. 2011;83:3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Shen F, Sun B, Kreutz J, Davydova E, Du W, Reddy P, Joseph L, Ismagilov R. J. Am. Chem. Soc. 2011;133:17705–17712. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Pekin D, et al. Lab Chip. 2011;11:2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- (131).Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).White AK, VanInsberghe M, Petriv OI, Hamidi M, Sikorski D, Marra MA, Piret J, Aparicio S, Hansen CL. Proc. Natl. Acad. Sci. U.S.A. 2011 doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Mary P, Dauphinot L, Bois N, Potier M, Studer V, Tabeling P. Biomicrofluidics. 2011:5. doi: 10.1063/1.3596394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Zhi X, Liu Q, Zhang X, Zhang Y, Feng J, Cui D. Lab Chip. 2012;12:741–745. doi: 10.1039/c2lc20949g. [DOI] [PubMed] [Google Scholar]
- (135).Njoroge S, Witek M, Battle K, Immethun V, Hupert M, Soper S. Electrophoresis. 2011;32:3221–3232. doi: 10.1002/elps.201100274. [DOI] [PubMed] [Google Scholar]
- (136).Wang H, et al. Angew. Chem. Int. Ed. 2012;51:4349–4353. doi: 10.1002/anie.201200732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Martewicz S, Michielin F, Serena E, Zambon A, Mongillo M, Elvassore N. Integr. Biol. 2012;4:153–164. doi: 10.1039/c1ib00087j. [DOI] [PubMed] [Google Scholar]
- (138).Lo J, Wang Y, Blake A, Yu G, Harvat T, Jeon H, Oberholzer J, Eddington D. Anal. Chem. 2012;84:1987–1993. doi: 10.1021/ac2030909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Lee K, Boccazzi P, Sinskey A, Ram R. Lab Chip. 2011;11:1730–1739. doi: 10.1039/c1lc20019d. [DOI] [PubMed] [Google Scholar]
- (140).Eydelnant I, Uddayasankar U, Li B, Liao M, Wheeler A. Lab Chip. 2012;12:750–757. doi: 10.1039/c2lc21004e. [DOI] [PubMed] [Google Scholar]
- (141).Lecault V, et al. Nat. Methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- (142).Kim T, Cho Y. Lab Chip. 2011;11:1825–1830. doi: 10.1039/c1lc20234k. [DOI] [PubMed] [Google Scholar]
- (143).Lee K, Kim C, Yang J, Lee H, Ahn B, Xu L, Kang J, Oh K. Biomicrofluidics. 2012:6. doi: 10.1063/1.3687409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Vogel P, Halpin S, Martin R, Spence D. Anal. Chem. 2011;83:4296–4301. doi: 10.1021/ac2004746. [DOI] [PubMed] [Google Scholar]
- (145).Zhong M, Lee C, Croushore C, Sweedler J. Lab Chip. 2012;12:2037–2045. doi: 10.1039/c2lc21085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Xie Z, Zhang Y, Zou K, Brandman O, Luo C, Ouyang Q, Li H. Aging Cell. 2012;11:599–606. doi: 10.1111/j.1474-9726.2012.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Lee SS, Vizcarra IA, Huberts DHEW, Lee LP, Heinemann M. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4916–4920. doi: 10.1073/pnas.1113505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Wakamoto Y, Grosberg AY, Kussell E. Evolution. 2012;66:115–134. doi: 10.1111/j.1558-5646.2011.01418.x. [DOI] [PubMed] [Google Scholar]
- (149).Mondragón-Palomino O, Danino T, Selimkhanov J, Tsimring L, Hasty J. Science. 2011;333:1315–1319. doi: 10.1126/science.1205369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Pan J, Stephenson A, Kazamia E, Huck W, Dennis J, Smith A, Abell C. Integr. Biol. 2011;3:1043–1051. doi: 10.1039/c1ib00033k. [DOI] [PubMed] [Google Scholar]
- (151).Nadell CD, Bassler BL. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Meier B, Zielinski A, Weber C, Arcizet D, Youssef S, Franosch T, Rädler JO, Heinrich D. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11417–11422. doi: 10.1073/pnas.1014853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Scherber C, Aranyosi A, Kulemann B, Thayer S, Toner M, Iliopoulos O, Irimia D. Integr. Biol. 2012;4:259–269. doi: 10.1039/c2ib00106c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Primiceri E, Chiriaco MS, Dioguardi F, Monteduro AG, D’Amone E, Rinaldi R, Giannelli G, Maruccio G. Lab Chip. 2011;11:4081–4086. doi: 10.1039/c1lc20540d. [DOI] [PubMed] [Google Scholar]
- (155).Wu C, Asokan S, Berginski M, Haynes E, Sharpless N, Griffith J, Gomez S, Bear J. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Sun Y-S, Peng S-W, Cheng J-Y. Biomicrofluidics. 2012:6. doi: 10.1063/1.3671399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Brett M, DeFlorio R, Stone D, Eddington D. Lab Chip. 2012;12:3127–3134. doi: 10.1039/c2lc40398f. [DOI] [PubMed] [Google Scholar]
- (158).Polacheck WJ, Charest JL, Kamm RD. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11115–11120. doi: 10.1073/pnas.1103581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Zhang Q, Liu T, Qin J. Lab Chip. 2012;12:2837–2842. doi: 10.1039/c2lc00030j. [DOI] [PubMed] [Google Scholar]
- (160).Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Tong Z, Cheung L, Stebe K, Konstantopoulos K. Integr. Biol. 2012;4:847–856. doi: 10.1039/c2ib20036h. [DOI] [PubMed] [Google Scholar]
- (162).Xie Y, Zhang W, Wang L, Sun K, Sun Y, Jiang X. Lab Chip. 2011;11:2819–2822. doi: 10.1039/c0lc00562b. [DOI] [PubMed] [Google Scholar]
- (163).Przybyla LM, Voldman J. Proc. Natl. Acad. Sci. U.S.A. 2012;109:835–840. doi: 10.1073/pnas.1103100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Moledina F, Clarke G, Oskooei A, Onishi K, Günther A, Zandstra PW. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3264–3269. doi: 10.1073/pnas.1111478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Yeon J, Ryu H, Chung M, Hu Q, Jeon N. Lab Chip. 2012;12:2815–2822. doi: 10.1039/c2lc40131b. [DOI] [PubMed] [Google Scholar]
- (166).Zheng Y, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Forouzan O, Burns JM, Robichaux JL, Murfee WL, Shevkoplyas SS. Lab Chip. 2011;11:1924–1932. doi: 10.1039/c0lc00547a. [DOI] [PubMed] [Google Scholar]
- (168).Wood L, Ge R, Kamm R, Asada H. Integr. Biol. 2012;4:1081–1089. doi: 10.1039/c2ib20054f. [DOI] [PubMed] [Google Scholar]
- (169).Song JW, Munn LL. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15342–15347. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Miller JS, et al. Nature Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Grosberg A, Alford PW, McCain ML, Parker KK. Lab Chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Ma Z, Liu Q, Liu H, Yang H, Yun J, Eisenberg C, Borg T, Xu M, Gao B. Lab Chip. 2012;12:566–573. doi: 10.1039/c2lc20699d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Kim H, Huh D, Hamilton G, Ingber D. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- (174).Tavana H, Zamankhan P, Christensen P, Grotberg J, Takayama S. Biomed. Microdevices. 2011;13:731–742. doi: 10.1007/s10544-011-9543-5. [DOI] [PubMed] [Google Scholar]
- (175).Yetisen AK, Jiang L, Cooper JR, Qin Y, Palanivelu R, Zohar YJ. Micromech. Microeng. 2011:21. [Google Scholar]
- (176).Kumano I, Hosoda K, Suzuki H, Hirata K, Yomo T. Lab Chip. 2012;12:3451–3457. doi: 10.1039/c2lc40367f. [DOI] [PubMed] [Google Scholar]
- (177).Choudhury D, van Noort D, Iliescu C, Zheng B, Poon K, Korzh S, Korzh V, Yu H. Lab Chip. 2012;12:892–900. doi: 10.1039/c1lc20351g. [DOI] [PubMed] [Google Scholar]
- (178).Chung K, Zhan M, Srinivasan J, Sternberg P, Gong E, Schroeder F, Lu H. Lab Chip. 2011;11:3689–3697. doi: 10.1039/c1lc20400a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Hendricks M, Ha H, Maffey N, Zhang Y. Nature. 2012;487:99–103. doi: 10.1038/nature11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Albrecht DR, Bargmann CI. Nat. Methods. 2011;8:599–605. doi: 10.1038/nmeth.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (181).Carr J, Parashar A, Gibson R, Robertson A, Martin R, Pandey S. Lab Chip. 2011;11:2385–2396. doi: 10.1039/c1lc20170k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Liu P, Mao D, Martin R, Dong L. Lab Chip. 2012;12:3458–3466. doi: 10.1039/c2lc40459a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Lockery S, Hulme S, Roberts W, Robinson K, Laromaine A, Lindsay T, Whitesides G, Weeks J. Lab Chip. 2012;12:2211–2220. doi: 10.1039/c2lc00001f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Hong S, Pan Q, Lee L. Integr. Biol. 2012;4:374–380. doi: 10.1039/c2ib00166g. [DOI] [PubMed] [Google Scholar]
- (185).Wu M, Perroud T, Srivastava N, Branda C, Sale K, Carson B, Patel K, Branda S, Singh A. Lab Chip. 2012;12:2823–2831. doi: 10.1039/c2lc40344g. [DOI] [PubMed] [Google Scholar]
- (186).Grover WH, Bryan AK, Diez-Silva M, Suresh S, Higgins JM, Manalis SR. Proc. Natl. Acad. Sci. U.S.A. 2011;109:E1523–E1529. doi: 10.1073/pnas.1104651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Guo Q, Reiling S, Rohrbach P, Ma H. Lab Chip. 2012;12:1143–1150. doi: 10.1039/c2lc20857a. [DOI] [PubMed] [Google Scholar]
- (188).Wurm M, Zeng A. Lab Chip. 2012;12:1071–1077. doi: 10.1039/c2lc20918g. [DOI] [PubMed] [Google Scholar]
- (189).Eyer K, Kuhn P, Hanke C, Dittrich P. Lab Chip. 2012;12:765–772. doi: 10.1039/c2lc20876h. [DOI] [PubMed] [Google Scholar]
- (190).Ma C, et al. Nat. Med. 2011;17:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Leung K, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7665–7670. doi: 10.1073/pnas.1106752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Juul S, Ho Y, Koch J, Andersen F, Stougaard M, Leong K, Knudsen B. ACS Nano. 2011;5:8305–8310. doi: 10.1021/nn203012q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Kirschbaum M, Guernth-Marschner C, Cherre S, Pena A, Jaeger M, Kroczek R, Schnelle T, Muellerk T, Duschl C. Lab Chip. 2012;12:443–450. doi: 10.1039/c1lc20818g. [DOI] [PubMed] [Google Scholar]
- (194).McFaul S, Lin B, Ma H. Lab Chip. 2012;12:2369–2376. doi: 10.1039/c2lc21045b. [DOI] [PubMed] [Google Scholar]
- (195).Muratore M, Srsen V, Waterfall M, Downes A, Pethig R. Biomicrofluidics. 2012;6:034113. doi: 10.1063/1.4746252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Mach A, Kim J, Arshi A, Hur S, Di Carlo D. Lab Chip. 2011;11:2827–2834. doi: 10.1039/c1lc20330d. [DOI] [PubMed] [Google Scholar]
- (197).Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang X. Lab Chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Schiro P, Zhao M, Kuo J, Koehler K, Sabath D, Chiu D. Angew. Chem. Int. Edit. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (199).Kirby BJ, et al. PLoS One. 2012;7:e35976. doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Shah AM, et al. Anal. Chem. 2012;84:3682–3688. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Aran K, Fok A, Sasso LA, Kamdar N, Guan YL, Sun Q, Undar A, Zahn JD. Lab Chip. 2011;11:2858–2868. doi: 10.1039/c1lc20080a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Bottenus D, Hossan MR, Ouyang YX, Dong WJ, Dutta P, Ivory CF. Lab Chip. 2011;11:3793–3801. doi: 10.1039/c1lc20469f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Yang X, Forouzan O, Brown T, Shevkoplyas S. Lab Chip. 2012;12:274–280. doi: 10.1039/c1lc20803a. [DOI] [PubMed] [Google Scholar]
- (204).Beck M, Brockhuis S, van der Velde N, Breukers C, Greve J, Terstappen L. Lab Chip. 2012;12:167–173. doi: 10.1039/c1lc20565j. [DOI] [PubMed] [Google Scholar]
- (205).Browne AW, Ramasamy L, Cripe TP, Ahn CH. Lab Chip. 2011;11:2440–2446. doi: 10.1039/c1lc20144a. [DOI] [PubMed] [Google Scholar]
- (206).Mitra I, Zhuang Z, Zhang Y, Yu C-Y, Hammoud ZT, Tang H, Mechref Y, Jacobson SC. Anal. Chem. 2012;84:3621–3627. doi: 10.1021/ac203431s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Lok KS, Muttalib S, Lee PPF, Kwok YC, Nguyen NT. Lab Chip. 2012;12:2353–2361. doi: 10.1039/c2lc00037g. [DOI] [PubMed] [Google Scholar]
- (208).Fu E, Liang T, Houghtaling J, Ramachandran S, Ramsey SA, Lutz B, Yager P. Anal. Chem. 2011;83:7941–7946. doi: 10.1021/ac201950g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Vella S, Beattie P, Cademartiri R, Laromaine A, Martinez A, Phillips S, Mirica K, Whitesides G. Anal. Chem. 2012;84:2883–2891. doi: 10.1021/ac203434x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Govindarajan AV, Ramachandran S, Vigil GD, Yager P, Bohringer KF. Lab Chip. 2012;12:174–181. doi: 10.1039/c1lc20622b. [DOI] [PubMed] [Google Scholar]
- (211).Guo W, Fung Y. Electrophoresis. 2011;32:3437–3445. doi: 10.1002/elps.201100213. [DOI] [PubMed] [Google Scholar]
- (212).Karns K, Herr A. Anal. Chem. 2011;83:8115–8122. doi: 10.1021/ac202061v. [DOI] [PubMed] [Google Scholar]
- (213).Gossett DR, Tse HT, Lee SA, Ying Y, Lindgren AG, Yang OO, Rao J, Clark AT, Di Carlo D. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Wood DK, Soriano A, Mahadevan L, Higgins JM, Bhatia SN. Sci. Transl. Med. 2012;4:123ra126. doi: 10.1126/scitranslmed.3002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Li M, Ku DN, Forest CR. Lab Chip. 2012;12:1355–1362. doi: 10.1039/c2lc21145a. [DOI] [PubMed] [Google Scholar]
- (216).Heo Y, Lee H, Hassell B, Irimia D, Toth T, Elmoazzen H, Toner M. Lab Chip. 2011;11:3530–3537. doi: 10.1039/c1lc20377k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Heo Y, Cabrera L, Bormann C, Smith G, Takayama S. Lab Chip. 2012;12:2240–2246. doi: 10.1039/c2lc21050a. [DOI] [PubMed] [Google Scholar]
- (218).Schirhagl R, Fuereder I, Hall E, Medeiros B, Zare R. Lab Chip. 2011;11:3130–3135. doi: 10.1039/c1lc20353c. [DOI] [PubMed] [Google Scholar]
- (219).Ferguson B, Buchsbaum S, Wu T, Hsieh K, Xiao Y, Sun R, Soh H. J. Am. Chem. Soc. 2011;133:9129–9135. doi: 10.1021/ja203981w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Ritzi-Lehnert M, et al. Biomed. Microdevices. 2011;13:819–827. doi: 10.1007/s10544-011-9552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF. J. Am. Chem. Soc. 2011;133:17705–17712. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Lam B, Fang Z, Sargent E, Kelley S. Anal. Chem. 2012;84:21–25. doi: 10.1021/ac202599b. [DOI] [PubMed] [Google Scholar]
- (223).Chin CD, et al. Nat. Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- (224).Sinn I, Kinnunen P, Albertson T, McNaughton B, Newton D, Burns M, Kopelman R. Lab Chip. 2011;11:2604–2611. doi: 10.1039/c0lc00734j. [DOI] [PubMed] [Google Scholar]
- (225).Bourquin Y, Reboud J, Wilson R, Zhang Y, Cooper J. Lab Chip. 2011;11:2725–2730. doi: 10.1039/c1lc20320g. [DOI] [PubMed] [Google Scholar]
- (226).Park J, Sunkara V, Kim T, Hwang H, Cho Y. Anal. Chem. 2012;84:2133–2140. doi: 10.1021/ac203163u. [DOI] [PubMed] [Google Scholar]
- (227).Li WT, Chen T, Chen ZT, Fei P, Yu ZL, Pang YH, Huang YY. Lab Chip. 2012;12:1587–1590. doi: 10.1039/c2lc40125h. [DOI] [PubMed] [Google Scholar]
- (228).Berron B, May A, Zheng Z, Balasubramaniam V, Bowman C. Lab Chip. 2012;12:708–710. doi: 10.1039/c2lc21101g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Nagatani N, Yamanaka K, Ushijima H, Koketsu R, Sasaki T, Ikuta K, Saito M, Miyahara T, Tamiya E. Analyst. 2012;137:3422–3426. doi: 10.1039/c2an16294f. [DOI] [PubMed] [Google Scholar]
- (230).Hossain S, Brennan J. Anal. Chem. 2011;83:8772–8778. doi: 10.1021/ac202290d. [DOI] [PubMed] [Google Scholar]
- (231).Liu C, Mauk M, Hart R, Qiu X, Bau H. Lab Chip. 2011;11:2686–2692. doi: 10.1039/c1lc20345b. [DOI] [PubMed] [Google Scholar]
- (232).Serrano G, Paul D, Kim S-J, Kurabayashi K, Zellers ET. Anal. Chem. 2012;84:6973–6980. doi: 10.1021/ac300924b. [DOI] [PubMed] [Google Scholar]
- (233).Liu H, Crooks R. Anal. Chem. 2012;84:2528–2532. doi: 10.1021/ac203457h. [DOI] [PubMed] [Google Scholar]
- (234).Esquivel JP, Castellarnau M, Senn T, Lochel B, Samitier J, Sabate N. Lab Chip. 2012;12:74–79. doi: 10.1039/c1lc20426b. [DOI] [PubMed] [Google Scholar]
- (235).Thom NK, Yeung K, Pillion MB, Phillips ST. Lab Chip. 2012;12:1768–1770. doi: 10.1039/c2lc40126f. [DOI] [PubMed] [Google Scholar]
- (236).da Costa ET, Neves CA, Hotta GM, Vidal DTR, Barros MF, Ayon AA, Garcia CD, do Lago CL. Electrophoresis. 2012;33:2650–2659. doi: 10.1002/elps.201200273. [DOI] [PubMed] [Google Scholar]
- (237).da Rocha ZM, Martinez-Cisneros CS, Seabra AC, Valdes F, Gongora-Rubio MR, Alonso-Chamarro J. Lab Chip. 2012;12:109–117. doi: 10.1039/c1lc20747d. [DOI] [PubMed] [Google Scholar]
- (238).Strychalski EA, Henry AC, Ross D. Anal. Chem. 2011;83:6316–6322. doi: 10.1021/ac2011894. [DOI] [PubMed] [Google Scholar]
- (239).Kazarine A, Kong MCR, Templeton EJ, Salin ED. Anal. Chem. 2012;84:6939–6943. doi: 10.1021/ac301421k. [DOI] [PubMed] [Google Scholar]
- (240).Toda K, Ebisu Y, Hirota K, Ohira S-I. Anal. Chim. Acta. 2012;741:38–46. doi: 10.1016/j.aca.2012.06.036. [DOI] [PubMed] [Google Scholar]
- (241).Shen F, Tan M, Wang Z, Yao M, Xu Z, Wu Y, Wang J, Guo X, Zhu T. Environ. Sci. Technol. 2011;45:7473–7480. doi: 10.1021/es1043547. [DOI] [PubMed] [Google Scholar]
- (242).Hiki S, Mawatari K, Aota A, Saito M, Kitamori T. Anal. Chem. 2011;83:5017–5022. doi: 10.1021/ac200884z. [DOI] [PubMed] [Google Scholar]
- (243).Ju W, Fu L, Yang R, Lee C. Lab Chip. 2012;12:622–626. doi: 10.1039/c1lc20954j. [DOI] [PubMed] [Google Scholar]
- (244).Wang Y-N, Yang R-J, Ju W-J, Wu M-C, Fu L-M. Biomicrofluidics. 2012:6. doi: 10.1063/1.4746246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Liu B, Zhang Y, Mayer D, Krause H, Jin Q, Zhao J, Offenhausser A, Xu Y. Electrophoresis. 2012;33:1247–1250. doi: 10.1002/elps.201100626. [DOI] [PubMed] [Google Scholar]
- (246).World Health Organization . Guidelines for drinking-water quality. 4th ed. WHO Press; Geneva: 2011. pp. 307–442. [Google Scholar]
- (247).Beaton AD, Cardwell CL, Thomas RS, Sieben VJ, Legiret F-E, Waugh EM, Statham PJ, Mowlen MC, Morgan H. Environ. Sci. Technol. 2012;46:9548–9556. doi: 10.1021/es300419u. [DOI] [PubMed] [Google Scholar]
- (248).Li N, Brahmendra A, Veloso A, Prashar A, Cheng X, Hung V, Guyard C, Terebiznik M, Kerman K. Anal. Chem. 2012;84:3485–3488. doi: 10.1021/ac3003227. [DOI] [PubMed] [Google Scholar]
- (249).Jung J, Kim G, Seo T. Lab Chip. 2011;11:3465–3470. doi: 10.1039/c1lc20350a. [DOI] [PubMed] [Google Scholar]
- (250).Chen Y-W, Wang H, Hupert M, Witek M, Dharmasiri U, Pingle MR, Barany F, Soper SA. Lab Chip. 2012;12:3348–3355. doi: 10.1039/c2lc40805h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (251).Liu P, Meagher RJ, Light YK, Yilmaz S, Chakraborty R, Arkin AP, Hazen TC, Singh AK. Lab Chip. 2011;11:2673–2679. doi: 10.1039/c1lc20151d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (252).Schaap A, Rohrlack T, Bellouard Y. Lab Chip. 2012;12:1527–1532. doi: 10.1039/c2lc21091f. [DOI] [PubMed] [Google Scholar]
- (253).Lefevre F, Chalifour A, Yu L, Chodavarapu V, Juneau P, Izquierdo R. Lab Chip. 2012;12:787–793. doi: 10.1039/c2lc20998e. [DOI] [PubMed] [Google Scholar]
- (254).Bhatta D, Michel A, Villalba M, Emmerson G, Sparrow I, Perkins E, McDonnell M, Ely R, Cartwright G. Biosens. Bioelectron. 2011;30:78–86. doi: 10.1016/j.bios.2011.08.031. [DOI] [PubMed] [Google Scholar]
- (255).Choi K, Kim J-Y, Ahn J-H, Choi J-M, Im M, Choi Y-K. Lab Chip. 2012;12:1533–1539. doi: 10.1039/c2lc21203j. [DOI] [PubMed] [Google Scholar]
- (256).Sameenoi Y, Koehler K, Shapiro J, Boonsong K, Sun Y, Collett J, Volckens J, Henry C. J. Am. Chem. Soc. 2012;134:10562–10568. doi: 10.1021/ja3031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (257).Mentele M, Cunningham J, Koehler K, Volckens J, Henry C. Anal. Chem. 2012;84:4474–4480. doi: 10.1021/ac300309c. [DOI] [PubMed] [Google Scholar]
- (258).Hossein-Babaei F, Paknahad M, Ghafarinia V. Lab Chip. 2012;12:1874–1880. doi: 10.1039/c2lc00035k. [DOI] [PubMed] [Google Scholar]
- (259).Kim S, Burris D, Bryant-Genevier J, Gorder K, Dettenmaier E, Zellers E. Environ. Sci. Technol. 2012;46:6073–6080. doi: 10.1021/es204625w. [DOI] [PubMed] [Google Scholar]
- (260).Adams A, Charles P, Deschamps J, Kusterbeck A. Anal. Chem. 2011;83:8411–8419. doi: 10.1021/ac2009788. [DOI] [PubMed] [Google Scholar]
- (261).Ogilvie IRG, Sieben VJ, Mowlem MC, Morgan H. Anal. Chem. 2011;83:4814–4821. doi: 10.1021/ac200463y. [DOI] [PubMed] [Google Scholar]
- (262).Fukuba T, Aoki Y, Fukuzawa N, Yamamoto T, Kyo M, Fujii T. Lab Chip. 2011;11:3508–3515. doi: 10.1039/c1lc20523d. [DOI] [PubMed] [Google Scholar]
- (263).Jonsson J, Ogden S, Johansson L, Hjort K, Thornell G. Lab Chip. 2012;12:1619–1628. doi: 10.1039/c2lc00025c. [DOI] [PubMed] [Google Scholar]
- (264).Mora M, Greer F, Stockton A, Bryant S, Willis P. Anal. Chem. 2011;83:8636–8641. doi: 10.1021/ac202095k. [DOI] [PubMed] [Google Scholar]
- (265).Schaffhauser D, Andrini O, Ghezzi C, Forster I, Franco-Obregon A, Egli M, Dittrich P. Lab Chip. 2011;11:3471–3478. doi: 10.1039/c0lc00729c. [DOI] [PubMed] [Google Scholar]
- (266).Nicholson W, et al. Astrobiology. 2011;11:951–958. doi: 10.1089/ast.2011.0714. [DOI] [PubMed] [Google Scholar]