Abstract
Two structurally homologous guanosine triphosphatase (GTPase) domains interact directly during signal recognition particle (SRP)–mediated cotranslational targeting of proteins to the membrane. The 2.05 angstrom structure of a complex of the NG GTPase domains of Ffh and FtsY reveals a remarkably symmetric heterodimer sequestering a composite active site that contains two bound nucleotides. The structure explains the coordinate activation of the two GTPases. Conformational changes coupled to formation of their extensive interface may function allosterically to signal formation of the targeting complex to the signal-sequence binding site and the translocon. We propose that the complex represents a molecular “latch” and that its disengagement is regulated by completion of assembly of the GTPase active site.
Ffh, the prokaryotic homolog of SRP54, and FtsY, the homolog of its receptor SRα, are GTPase components of the signal recognition targeting pathway that interact directly during cotranslational targeting of proteins to the membrane (1–3). They function at sequential steps, Ffh binding to the hydrophobic signal sequence as it emerges from the ribosome and FtsY interacting with Ffh to effect release of the signal peptide to the membrane translocon. Subsequently, coordinate stimulation of the guanosine triphosphate (GTP) hydrolysis activity of the two proteins leads, ultimately, to disengagement of the targeting complex (4–6 ). The SRP GTPases exhibit distinct properties relative to other members of the GTPase superfamily, including relatively low nucleotide affinity and rapid nucleotide exchange (7, 8). The interactions of the SRP at the membrane evolve through several stages, and GTP binding and GTP hydrolysis play different roles in the engagement and disengagement of the complex (9–12); thus, GTP binding is required, but in the presence of a nonhydrolyzable GTP analog the proteins can enable targeting of a single chain but cannot recycle (12). Biochemical studies of the eukaryotic SRP consistent with “empty-site” behavior before formation of the targeting complex (11), mutational studies of prokarytic SRP suggesting uncoupling of GTPase function from receptor interaction (9), and a kinked binding mode for the GTP analog GMPPNP in its complex with Thermus aquaticus Ffh (10), together support the existence of a “primed” binding mode that functions to load nucleotide without activation until assembly of the productive complex (10). These data suggest that the SRP GTPases may function by a logic distinct from that of the classic GTPase “switch.”
We have determined the structure of the GTP-dependent heterodimeric complex of the core GTPases of Ffh and FtsY that are integral to SRP-mediated targeting. These structurally homologous SRP GTPase subunits are termed NG, because they comprise an α-helical N domain that packs against a G-domain fold similar to other GTPases (10, 13–17 ). The GTPase subunits are modular; in Ffh the NG domain occurs at the N terminus of the polypeptide, and in FtsY, at the C terminus. Like other GTPases, the sequences of the SRP GTPases are characterized by four conserved motifs, I to IV, that reflect residues directly involved in nucleotide binding and hydrolysis. Several additional sequence motifs, termed ALLEADV, DARGG, and DGQ (table S1), are distinctive to the SRP GTP-ases. The G domain includes an insertion box subdomain (IBD), distal to the N domain interface, that contains GTPase sequence motif II and had been thought to provide the site of interaction in the targeting complex (13). We report the 2.05 Å resolution x-ray structure of the engagement complex of the NG domains of Ffh and FtsY from T. aquaticus, stabilized with the nonhydrolyzable GTP analog, β-γ methylene-guanosine 5′-triphosphate (GMPPCP) (18, 19). The structure was determined by molecular replacement using the G domain of T. aquaticus Ffh as a search model (20), followed by autotracing with ARP/wARP (21). The resulting electron density map (fig. S1) provided an unambiguous identification of the two polypeptides present in the asymmetric unit. The crystallized proteins retain both GTP hydrolysis and reciprocal GTPase activating protein (GAP) activities (fig. S2).
The NG domains of Ffh and FtsY associate longitudinally as a symmetric heterodimeric complex, with the N, G, and IBD regions of each protein interacting primarily with their respective counterparts (Fig. 1). The pattern of contacts of each protein across the interface, their main-chain and side-chain configurations, and their ligand-binding interactions, are remarkably similar. The G domains can be superimposed by a rotation of 179° with a root mean square Cα deviation of only 1.10 Å. Two GMPPCP molecules are buried within a composite active site chamber formed at the center of the complex by the direct apposition of the two nucleotide binding sites (fig. S3). The GTP analogs are integral to the formation of the heterodimer interface. Here, we refer primarily to structure of the Ffh NG; when we refer to FtsY, we note the corresponding residue in the Ffh sequence in parentheses. With one important exception, the interactions of equivalent residues in FtsY NG are identical to those in Ffh.
Fig. 1.

Overall structure of the heterodimeric Ffh/FtsY NG domain complex. (A) Ribbon representation viewed perpendicular to the dimer axis, which is vertical in the figure. The N domains (blue) and C-terminal helices (golden) of the two proteins are at the top, and their IBD domains (purple) are at the bottom. The two active sites are brought into direct apposition to form an active site chamber at the center of the G domains (gray), where the buried GMPPCP ligands are shown. The motif I P-loops of the two proteins pack adjacent to each other (*). The structure is highly symmetric, with the exception of the smaller N domain of FtsY, and all secondary structure elements adopt the same orientation in both proteins. (B) The structure viewed along the two-fold axis further highlights the symmetry of the complex. The viewpoint is toward the IBD, at the bottom of the diagram in (A).
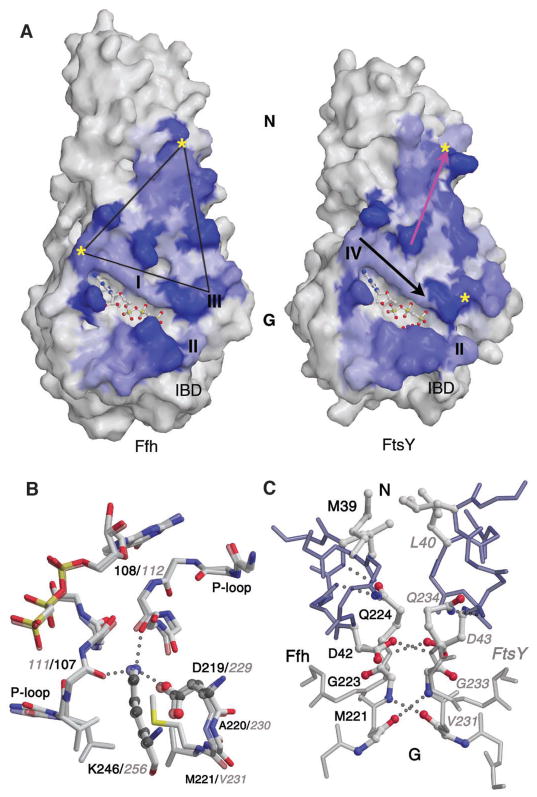
The contact between the two proteins is extensive, burying ~1800 Å2 accessible surface area, and it comprises three distinct interfaces (Fig. 2A). The first is a large area that we term the latch, which extends a triangular footprint from the N domain ~30 Å toward the shared active site. A second interface comprises helix/helix interactions local to the IBD that includes residues of the catalytic ensemble (Fig. 1B). Between them are the two nucleotides bound within the active site chamber. The two motif I P-loops, which hydrogen bond to the phosphates of the bound nucleotides, are separated by only ~4 Å and are stabilized by an aspartate/lysine framework that defines the edge of the latch adjacent to the active site (Fig. 2B). The buried salt bridge between Asp219 and Lys246 [and, in an approximately antiparallel configuration, between Asp229(219) and Lys256(246)] positions the lysine side chain to form hydrogen bonds to both P-loops, thus symmetrically bridging the heterodimer interface. The lysine side chain of the framework arises from the sequence motif IV common to all GTPases (22); the aspartate arises from a sequence motif in the loop preceding helix α3 that is conserved in SRP GTPases but previously had no known function (the DGQ motif, Asp219 to Gln224 in T. aquaticus Ffh NG) (table S1). Residues of the DGQ motif also make critical contributions to the network of symmetric main-chain and side-chain hydrogen-bond interactions that extends from the active site toward the intramolecular N/G domain interface. There, hydrophobic residues of the AL-LEADV motif of the N domain that are solvent-exposed in the monomeric proteins (23) generate a symmetric hydrophobic interface between the N domains of Ffh and FtsY (Fig. 2C).
Fig. 2.
An extensive interaction surface. (A) The molecular surfaces of the Ffh monomer (left) and the FtsY monomer (right) are shown, shaded by the change in accessible surface area at each residue between the monomer and in the heterodimer. The blue areas define the protein-protein contact. The GTP binding motifs I to IV are indicated, and the Mg2+ nucleotide ligands are shown in ball and stick representation. A symmetric triangular contact region above the active site cavity is termed the latch. The IBD regions of the two proteins contact one another below the active site cleft. The packing orientation in the complex can be visualized by rotating the monomers to overlay the yellow asterisks. Arrows on the surface of the FtsY monomer highlight the orientation of the Asp/Lys framework (black) and the latch interface (pink) presented in the following panels. (B) The framework formed by Asp229(219) of the DGQ motif (see table S1) and Lys256(246) of motif IV from both monomers is shown superimposed to emphasize the symmetry between Ffh and FtsY in the complex. This symmetric interaction lies approximately along the diagonal ridge located above the active site clefts in(A). The lysine hydrogen bonds to both P-loops, thus bridging the interface. In all figures, residues from FtsY are labeled in gray italics font and from Ffh in black font. (C) The symmetric latch interface between the N and G domains, corresponding to the close loop contacts seen above the adjacent P-loops in Fig. 1A. The conserved hydrophobic residues of the ALLEADV motifs of the N domains (top) and the symmetric glycine pair of the DGQ motifs of the G domains (bottom) are shown along with the pair of bridging aspartate and glutamine residues.
Formation of the interaction surface spanning the N and G domains depends on a relative rigid-body motion of the N and G domains that, in Ffh, translates the distal loops of the N domain by ~12 Å from their position in the nucleotide primed structure (Fig. 3A). Flexibility of the intramolecular N/G interface is well established (24, 25); however, the orientation of the N domain adopted in the heterodimer is well outside the range of conformations previously observed. The DARGG loop, which mediates an interaction between the nucleotide binding site and the N domain (24 ), is reoriented to interact across the heterodimer interface, thus uncoupling the positions of the two domains (fig. S4). Movement of the N domain repositions several conserved hydrophobic residues, including the alanines of the ALLEADV motif, Leu247 of motif IV, and Gly253 of the DARGG loop, to create a new packing interaction for the amphipathic C-terminal helix (fig S4). Consequently, the helix reorients, symmetrically in the two proteins (Fig. 1B), into a new packing configuration. In Ffh, this reorientation can be seen as a 34° hinge motion that translates the C terminus ~10 Å toward the heterodimer interface (Fig. 3A), displacing the N terminus of the NG domain. Similar rearrangement occurs in FtsY, as evidenced by comparison with the structure of Escherichia coli FtsY (15) and by the strict association of proteolysis at Ala21(20) with formation of the heterodimeric complex (18).
Fig. 3.
Conformational changes generate the heterodimer interface. (A) The structure of the Ffh NG domain with GMPPNP bound (1JPJ.pdb) (in lighter colors) is superimposed with its structure in the complex. The N domain moves as a rigid body toward helix α3 of the G domain; this shift, in turn, is coupled to conformational rearrangement in the DGQ motif at the N terminus of α3, enabling formation of the extensive heterodimeric contact there. Helix α4 moves with the N domain, accommodated by an ~2.9 Å translation of the remainder of helix α3. Note the concurrent reorientation of the C-terminal helix. (B) G-domain conformational changes associated with complex formation are limited to the loops of conserved sequence motifs. The magnitude of the shifts are mapped so that the largest shifts (~6.5 Å) are the darkest shaded regions. (C) Reorientation of motifs II and III upon complex formation. The left panel shows the Ffh NG GMPPNP structure, the right panel Ffh NG in the complex. The side chain of motif III residue Leu192 moves to insert into a pocket across the heterodimer interface, between the guanine base and Gly259(249) that follows motif IV. Movement of this leucine and the accompanying rearrangement of the motif III backbone allows the P-loop to open sufficiently to accommodate the nucleotide in an extended conformation (10). Motif II residues Asp135 and Arg138 move into the catalytic chamber. The same configuration is observed in FtsY.
Concerted rearrangement of four conserved sequence motifs of the G domain, at loops of the β-α GTPase fold, accompanies the motion of the N domain and completes formation of the latch interface surface (Fig. 3B). Conformational changes of the P-loop and motif III allow the nucleotide to shift from a kinked binding conformation, observed in the structure of the GMPPNP-bound Ffh (10), to the extended binding conformation characteristic of other GTPase structures that is adopted in the heterodimeric complex. Key residues in the transition include the conserved Leu192 of motif III, which is solvent-exposed in the monomeric protein (13). This residue shifts ~6 Å, removing a steric constraint that prevents the P-loop from accommodating the γ-PO4 in an extended conformation (10) and placing the side chain into a hydrophobic pocket across the heterodimer interface that is created between the nucleotide bound to FtsY and the conserved Gly259(249) following motif IV (Fig. 3C). The opening of the motif I P-loop, an ~1.6 Å shift at Gln107 that accommodates the γ-PO4, in turn effects the hydrogen bonding interactions that mediate the aspartate/lysine framework (Fig. 2B). These interactions involve the residues of the DGQ motif, which itself rearranges to a conformation that allows the hydrogen bonding and packing interactions at the center of the heterodimer interface (Fig. 2C). This is coupled to a slight rotation of adjacent helix α3, and the displacement of the DARGG loop and helix α4 that accommodates movement of the N domain relative to the G domain (Fig. 3A). Remarkably, the two GTP analogs sequestered within the resulting catalytic chamber interact directly, forming a reciprocating pair of short hydrogen bonds between the ribose O3′ hydroxyl of one and the γ-phosphate oxygen of the other (Fig. 4A). This coordination of the protein structural changes with the position of the γ-PO4, itself involved in a reciprocal hydrogen bond at the center of the heterodimer interface, provides a mechanism by which the GTP- and GDP-bound structural states of the SRP GTPases can discriminate formation and dissociation of the targeting complex.
Fig. 4.
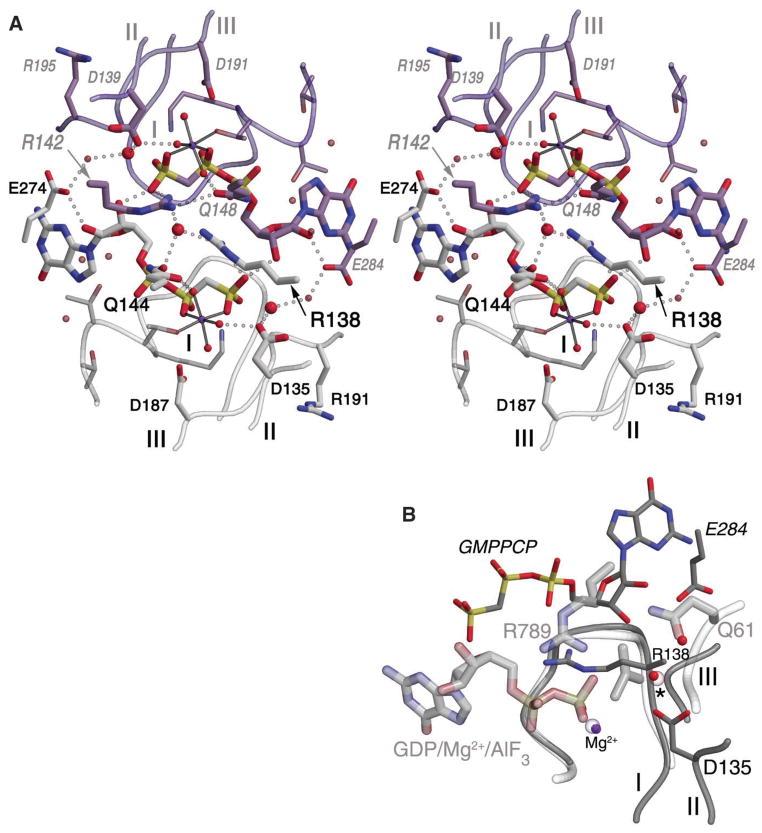
A composite active site. (A) A stereo image shows the nucleotides interacting directly and symmetrically across the complex interface within the active site chamber. Residues from Ffh are shown in gray, from FtsY in purple. Ffh motif II residue Arg138 extends from the right, and Arg142(138) (FtsY) from the left. A water molecule is located asymmetrically between them (large central red sphere). Each GMPPCP ribose O3′ hydroxyl forms a short hydrogen bond with the γ-phosphate oxygen of the other across the active site chamber. The putative catalytic water molecule at each active center is shown as a large red sphere, hydrogen bonded to Asp135 [and Asp139(135)] and hydrogen bonded to another water molecule that interacts across the dimer interface with invariant Glu284(274). Two other invariant arginines, Arg191 and Arg195(191), point away from the active site chamber but may be reoriented by further conformational rearrangement of motif III. (B) Superimposition of the Ffh NG with Ras/RasGAP in complex with GDP/Mg/AlF3 (1WQ1.pdb). Active site ligands, loops, and side chains from the Ras/RasGAP complex are shown with thicker lines and labeled in gray. Only the nucleotide bound to FtsY is shown for the heterodimer; the other superimposes well with the Ras/RasGAP ligands, as shown by the position of the magnesium ion from Ffh (small purple sphere center) and the putative catalytic water (small red sphere, *). An additional active site water (above right), which hydrogen bonds Wat13 and Glu284, is nearly superimposed with the Ras motif III Gln61 OE2. Gly190 of Ffh motif III is only 2 Å from the AlF3 bound in the Ras complex, suggesting that motif III undergoes a further conformational change to accommodate the transition-state structure of the Ffh/FtsY heterodimer.
The conserved residues of motif II that are likely to mediate hydrolysis of GTP are directed into the catalytic chamber by a conformational change local to the motif that is not tightly coordinated to formation of the complex. Similar backbone configurations of the motif have been observed in structures of the apo FtsY and a Mg2+GDP-bound complex of Ffh (14, 15). As a result of the rearrangement (Fig. 3C), residues that form the two-fold symmetric packing interaction of the IBD are shifted into place, and the side chains of Asp135, Arg138, and Gln144 become arrayed along one face of the bound nucleotide pair (Fig. 4A). It is likely that these residues, from both Ffh and FtsY, provide much of the catalytic machinery in the heterodimeric complex. Thus, the aspartate side chain, positioned by hydrogen bonding a coordinating water of the magnesium ion, in turn positions a candidate nucleophilic water 3.3 Å from the γ-phosphate. That water, Wat13 [and Wat507(13) at the FtsY active center], is stabilized by an additional hydrogen bond to a neighboring water molecule that interacts with the side chain of Glu284(274), contributed across the interface (Fig. 4A). The glutamine side chain, Gln144, is directed into the active site from the α1a helix of the IBD and forms hydrogen bonds to one of the coordinating waters of the magnesium ion and to an α-phosphate oxygen of the bound nucleotide. The arginine side chain, perhaps the most important player, lies between the two bound nucleotides and is positioned by hydrogen bonds to the γ-phosphate of GMPPCP bound to Ffh and to a buried water, Wat86. It is also poised 3.3 Å from the α-phosphate of the nucleotide bound to FtsY (Fig. 4A).
The positioning of the two motif II arginines, Arg138 and Arg142(138), within the catalytic chamber is clearly consistent with their playing the role of “arginine finger,” possibly in both cis and trans configurations relative to the two nucleotides. Comparison with the structure of the Ras/RasGAP/GDP/AlF3 complex (26 ) reveals that, although displaced around the phosphate chain, the relation of Arg138 to the β-γ phosphate linkage is almost identical to that of the arginine finger supplied by RasGAP (Fig. 4B). Interestingly, it is with these side chains that the striking symmetry that holds throughout the heterodimeric structure breaks down most notably—the position of the corresponding Arg142(138) of FtsY is markedly different (Fig. 4A), turned away from the chamber center and forming an additional hydrogen bond with Gln148(144). This asymmetry—which is enforced by the water, Wat86, also asymmetric, bound between the two arginine side chains—suggests that the arrangement of the arginines may alternate within the chamber, perhaps contributing sequentially to hydrolysis of each nucleotide.
The structure of the GMPPCP-stabilized complex represents, at best, a ground state of the GTP hydrolysis reaction, and we can infer from comparison with changes observed between the structures of other GTPases (26, 27) that additional conformational change accompanies progression to the transition state of GTP hydrolysis. The SRP GTPases must evolve through distinct structural states in the targeting complex, including initial engagement, transfer of the bound peptide, GTP hydrolysis, and disengagement (10–12, 28–32). That the interactions between the catalytic regions provided by the IBD are, in this structure, much less extensive than in the “latch” region is consistent with the different functional regions having different roles in the kinetics of complex formation, transfer, and release. Furthermore, several conserved residues likely to also contribute to catalysis adopt conformations that are not consistent with either catalytic or interface function. These residues, which can be considered to be in a “pending” state, include the invariant arginine of motif III, Arg191 [FtsY Arg195(191)], which shifts away from the catalytic site (Fig. 3C), and conserved motif I residue, Gln107 [Asn111(107) in FtsY] (fig. S5). Both are associated with a water-filled passage between the catalytic center and bulk solvent that is inconsistent with desolvation of the nucleophilic water observed in structures of GTPase/GAP complexes (26 ). In addition, molecular modeling suggests that motif III Gly190 must also shift to accommodate a transition-state structure (Fig. 4B). Finally, the α2 helix, which includes a number of highly conserved residues and which corresponds to the switch 2 region of GTPases that commonly provides the site of interaction between GTPases and their activating proteins (33), undergoes little change in the GMPPCP-stabilized heterodimer. We interpret these observations to suggest that this heterodimeric complex is in a latched, but not catalytic, state. Interestingly, the invariant residues Glu284(274)—which interacts directly with the bound nucleotide in its own active site and also across the heterodimer interface—and Arg191 are situated so that a relatively small rearrangement of motif III and helix α2 would allow the formation of a salt bridge between them (Fig. 4A). Assembly of a catalytic conformation of motif III and helix α2, perhaps by interaction with the M domain of Ffh, perhaps by interaction with other components of the targeting complex, provides a mechanism by which disengagement of the heterodimer may be regulated after signal peptide transfer.
That each of the disparate sequence motifs of the SRP GTPases contributes to an integrated set of interactions across most of the protein surface is consistent with previous mutagenesis studies and explains the behavior of DARGG motif mutants of the E. coli SRP that strongly inhibited SRP-SR interaction (9); these can now be understood in terms of the extensive structural interface of the heterodimer latch. Mutational and cross-linking studies have demonstrated interaction between signal sequence recognition and the function of the GTPase domain (9, 23, 34, 35). Particularly interesting are mutations of the ALLEADV motif that affect signal sequence binding (23); its intimate association with the heterodimer interface suggests that formation of the targeting complex may modulate a signal peptide-binding activity, perhaps one correlated with the rearrangement of the C-terminal helix implied in this structure. The tight coupling between the N and G domains in the heterodimeric complex, which symmetrically shifts the C-terminal helices and displaces the N termini, provides an allosteric mechanism by which interaction between the two GTPases may be communicated at this stage to the accessory domains of the SRP and its receptor. The N domain can therefore be seen to be a sensor, not of nucleotide occupancy (13, 24), but of formation of the GTP-dependent targeting complex.
Finally, the observation that the SRP GTPases behave as reciprocal GTPase activating proteins (5) can now be understood to be a consequence of the formation of a shared catalytic chamber between them. Indeed, the reciprocal hydrogen bonding between the bound nucleotides may itself be catalytically important. However, the structure of the complex also demonstrates how the initial engagement of the two proteins can function as a latch, in that a number of structural elements, including the bound nucleotides, contribute to an intricate interface that is unlikely to dissociate until two subsequent steps, signal peptide transfer followed by nucleotide hydrolysis, occur. This kind of mechanism for the SRP GTPases is consistent with a process requiring assembly of multiple components, and it can be distinguished from one in which the GTPases act along a signaling pathway. Extending the metaphor, the GTP molecules themselves can be imagined as “explosive bolts” in that they are integral to the interface that holds the proteins together, and so promote transfer of the translating ribosomal cargo, but that they also provide, by their hydrolysis, the “explosion” that disengages the components of the targeting complex (fig. S6). This conception of the role of GTP is somewhat distinct from the classic GTPase switch model and provides insight into the logic of the SRP GTPases that may be relevant to understanding other GTPases that function in the assembly of cellular components.
Supplementary Material
Acknowledgments
We thank C. W. Carter, S. McGovern, U. D. Ramirez, and S. R. Sprang for critical reading of this manuscript, and Panchika Prangkio for her contribution. This work was supported by grant GM58500 from the NIH. Portions of this work were performed at the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center, Sector 5, and BioCARS, Sector 14, of the Advanced Photon Source (APS) at Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. DND-CAT was supported by DuPont, Dow, the state of Illinois, and the NSF. Use of the BioCARS was supported by NIH, National Center for Research Resources, under grant number RR07707. Support from the R. H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility is acknowledged. Coordinates and structure factors have been deposited with the Protein Data Bank and assigned accession code 1OKK.
Footnotes
Note added in proof: A structure of a similar complex of the SRP GTPases in a different crystal form was independently determined and is reported by Egea et al. (36).
References and Notes
- 1.Keenan RJ, Freymann DM, Stroud RM, Walter P. Annu Rev Biochem. 2001;70:755. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein HD, et al. Nature. 1989;340:482. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 3.Römisch K, et al. Nature. 1989;340:478. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 4.Miller JD, Wilhelm H, Gierasch L, Gilmore R, Walter P. Nature. 1993;366:351. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- 5.Powers T, Walter P. Science. 1995;269:1422. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 6.Valent QA, et al. EMBO J. 1998;17:2504. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moser C, Mol O, Goody RS, Sinning I. Proc Natl Acad Sci USA. 1997;94:11339. doi: 10.1073/pnas.94.21.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Biochemistry. 2001;40:15224. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y, et al. EMBO J. 2001;20:6724. doi: 10.1093/emboj/20.23.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padmanabhan S, Freymann DM. Structure. 2001;9:859. doi: 10.1016/s0969-2126(01)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rapiejko PJ, Gilmore R. Cell. 1997;89:703. doi: 10.1016/s0092-8674(00)80253-6. [DOI] [PubMed] [Google Scholar]
- 12.Song W, Raden D, Mandon EC, Gilmore R. Cell. 2000;100:333. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- 13.Freymann DM, Keenan RJ, Stroud RM, Walter P. Nature. 1997;385:361. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 14.Focia PJ, Alam H, Lu T, Ramirez UD, Freymann DM. Proteins. 2004;54:222. doi: 10.1002/prot.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montoya G, Svensson C, Luirink J, Sinning I. Nature. 1997;385:365. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 16.Montoya G, Kaat K, Moll R, Schäfer G, Sinning I. Structure. 2000;8:515. doi: 10.1016/s0969-2126(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 17.Vetter IR, Wittinghofer A. Science. 2001;294:1299. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 18.Shepotinovskaya IV, Freymann DM. Biochim Biophys Acta. 2002;1597:107. doi: 10.1016/s0167-4838(02)00287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepotinovskaya IV, Focia PJ, Freymann DM. Acta Crystallogr. 2003;D59:1834. doi: 10.1107/s0907444903016573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Materials and methods are available as supporting material on Science Online.
- 21.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. Acta Crystallogr. 1997;D53:448. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 22.Sequence motifs are defined for T. aquaticus Ffh and FtsY in table S1.
- 23.Newitt JA, Bernstein HD. Eur J Biochem. 1997;245:720. doi: 10.1111/j.1432-1033.1997.00720.x. [DOI] [PubMed] [Google Scholar]
- 24.Freymann DM, Keenan RJ, Stroud RM, Walter P. Nat Struct Biol. 1999;6:793. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez UD, et al. J Mol Biol. 2002;320:783. doi: 10.1016/s0022-2836(02)00476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffzek K, et al. Science. 1997;277:333. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 27.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Cell. 1997;89:251. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 28.Rapiejko PJ, Gilmore R. Mol Biol Cell. 1994;5:887. doi: 10.1091/mbc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connolly T, Rapiejko PJ, Gillmore R. Science. 1991;252:1171. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 30.de Leeuw E, et al. EMBO J. 2000;19:531. doi: 10.1093/emboj/19.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millman JS, Qi HY, Vulcu F, Bernstein HD, Andrews DW. J Biol Chem. 2001;276:25982. doi: 10.1074/jbc.M011331200. [DOI] [PubMed] [Google Scholar]
- 32.Miller JD, Bernstein HD, Walter P. Nature. 1994;367:657. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 33.Sprang SR. Annu Rev Biochem. 1997;66:639. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 34.Lütcke H, High S, Römisch K, Ashford AJ, Dobberstein B. EMBO J. 1992;11:1543. doi: 10.1002/j.1460-2075.1992.tb05199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cleverley RM, Gierasch LM. J Biol Chem. 2002;277:46763. doi: 10.1074/jbc.M207427200. [DOI] [PubMed] [Google Scholar]
- 36.Egea PF, et al. Nature. 2004;427:215. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.