Abstract
Protein-RNA interactions play an important role in many biological processes. The ability to predict the molecular structures of protein-RNA complexes from docking would be valuable for understanding the underlying chemical mechanisms. We have developed a novel non-redundant benchmark dataset for protein-RNA docking and scoring. The diverse dataset of 72 targets consists of 52 unbound-unbound test complexes, and 20 unbound-bound test complexes. Here, unbound-unbound complexes refer to cases in which both binding partners of the co-crystallized complex are either in apo form or in a conformation taken from a different protein-RNA complex, whereas unbound-bound complexes are cases in which only one of the two binding partners has another experimentally determined conformation. The dataset is classified into three categories according to the interface RMSD and the percentage of native contacts in the unbound structures: 49 easy, 16 medium, and 7 difficult targets. The bound and unbound cases of the benchmark dataset are expected to benefit the development and improvement of docking and scoring algorithms for the docking community. All the easy-to-view structures are freely available to the public at http://zoulab.dalton.missouri.edu/RNAbenchmark/.
Keywords: Benchmarking, protein-RNA interactions, molecular docking, scoring function, molecular recognition
1 INTRODUCTION
Due to the cost and technical difficulty of experimental structure determination, molecular docking has become an important computational tool for studying biomolecular recognition (1–15). Over the past few decades, a variety of docking algorithms have been developed. Meanwhile, it is commonly believed that selection of structures for benchmarking is important to the development of docking algorithms and scoring functions (e.g., refs (16–22)) because of two reasons. First, benchmark datasets can be used for validation of docking algorithms and scoring functions. Second, comparative assessments of different docking and scoring algorithms on the same benchmark datasets may provide valuable insights into how to improve the existing algorithms and how to develop novel methods (22–24).
A good set of structures for benchmarking binding mode predictions should possess three features. First, a benchmark dataset should consist of diverse targets in order to test the robustness of docking/scoring algorithms. Second, only experimentally determined structures should be selected for benchmarking so as to prevent introduction of computational errors. Finally, the benchmark structures should include both the bound and unbound structures of the binding partners so as to reflect conformational changes upon binding.
Several good benchmark datasets have been developed for protein-protein docking and protein-DNA docking (17–19, 25–28). The docking community urgently needs novel datasets with diverse targets to be assembled for benchmarking protein-RNA docking algorithms, because of the critical role played by protein-RNA interactions in many biological processes such as protein synthesis, DNA replication, regulation of gene expression and defense against pathogens (29–37).
With the increasing number of experimentally determined structures of RNAs and protein-RNA complexes deposited in the Protein Data Bank (PDB) (38), development of a benchmark dataset for protein-RNA docking has become feasible. In the present study, we have developed a large benchmark dataset which consists of 72 diverse targets for protein-RNA docking from the PDB, referred to as RNABenchmark 1.0. Each target in the benchmark dataset includes both the co-crystalized partners and their corresponding unbound structures so as to reflect the conformational changes upon binding. The benchmark dataset will be beneficial to those in the docking community who are studying protein-RNA interactions.
2 MATERIALS AND METHODS
We have developed a non-redundant benchmark dataset of 72 protein-RNA targets from the PDB. Similary to other benchmark datasets in the macromolecular docking field (17, 19, 25–27), each target in our benchmark dataset contains the bound structures and at least one unbound structure(s) for unbound docking. Here, we follow the definitions of bound and unbound structures in the protein docking field (25, 26). Namely, if two structures belong to the binding partners in an experimentally determined complex, they are defined as the bound structures of this complex; otherwise if a structure is in free form or belongs to a binding partner in another complex, it is defined as an unbound structure. The bound structures in the benchmark are used as a “reference” to check the conformational changes between the bound and unbound structures as well as the accuracy of the predicted binding modes from unbound docking. As a reference, the bound structures need to be as accurate as possible. Therefore, we have restricted the bound structures to be crystal structures that are often thought to be relatively more accurate. However, we have removed this restriction when searching for the corresponding unbound structures that are used for testing how successful a docking/scoring algorithm can handle the conformational changes between the bound and unbound structures due to the changes of physiological and/or experimental (e.g. X-ray, NMR, etc.) conditions — a major purpose of a macromolecular docking benchmark dataset.
Specifically, we queried all the X-ray crystal structures with resolution better than 4.0 Å to identify those PDB entries that contain at least one protein and one RNA chain but no DNA chains. As of April 12, 2011, the search yielded a total of 859 entries. These PDB entries were manually examined and the adequate protein-RNA complexes were kept. Here, an adequate protein-RNA complex is defined as a structure that meets all of the following criteria. First, both the protein and the RNA should belong to the same biological unit. Second, the number of the residues in the protein should lie between 20 and 1000, and the number of the residues in the RNA should range from 20 to 200. Third, there should be no more than six chains in the protein or RNA, respectively. Lastly, the complexes that contain only backbone atoms in the protein or in the RNA should be excluded. 313 structures of protein-RNA complexes met the inclusion criteria.
The selected complexes were then clustered according to their sequence similarities in order to remove the redundancy. If any chain in the protein of a complex has at least 30% sequence identity with a chain in the protein from another complex, or if any chain in the RNA of a complex has at least 70% sequence identity with a chain in the RNA from another complex, the two complexes were grouped into the same cluster. We set a higher sequence identity cutoff for RNA because similar percentages of homology result in much larger differences in RNA structures than in protein structures (39). According to the clustering criterion, the 313 complexes were then grouped into 87 clusters. The crystal structure with the best resolution in each cluster was selected as the cluster’s representative, resulting in 87 bound structures.
To obtain the corresponding unbound structures, we searched all the sequences in the PDB against each chain in the above 87 pairs of bound structures using BLAST (40). If a protein or RNA structure in the PDB had more than 90% sequence identity to the bound structure and the alignment covered more than 90% of the shorter sequence, the structure was considered as a candidate for the unbound structure. If there are multiple unbound candidates for a bound structure, the unbound structure was selected according to the following priorities: highest sequence identity, highest-resolution crystal structure unless only NMR structures were available, and closest length. For each NMR structure which consists of an ensemble of structures, the first model was selected as a representative of the unbound structure. Only those targets for which there was an unbound structure for the protein or the RNA were kept, reducing the target number to 72. It should be noted that most of the unbound structures are other bound structures with different docking partners or in a different condition due to the limited number of free protein or RNA conformations in the PDB. These 72 targets form our benchmark dataset of bound and unbound structures for the assessment of protein-RNA docking.
3 RESULTS
Table 1 lists the 72 targets in our benchmark dataset for protein-RNA docking. More information can be found in the table provided at our website (http://zoulab.dalton.missouri.edu/RNAbenchmark/). For convenience, each target is named by the PDB entry of the complex for the bound structures. To make the benchmark dataset easy to use, the unbound structures of the proteins and the RNAs were superimposed onto their respective bound structures using Chimera (41), which can be viewed with the Jmol program. Jmol is an open-source Java viewer for chemical structures in 3D (http://jmol.sourceforge.net/). By using the interactive Jmol viewer, users can easily examine and compare the bound and unbound structures in both ribbon and atomic modes. More interactive features will be added in the next release. For each target, following the sequence alignment, a residue number mapping between the bound and unbound structures was obtained for the protein and the RNA, respectively. Based on the residue mapping, a second set of mapped bound and unbound structures was created by removing the mis-matched residues in the alignment from the original structure files. This set of mapped bound and unbound structures will be useful for docking evaluations because the bound and the unbound structures have the same number of residues in the same order. Thus, every target of the benchmark dataset consists of a pair of complexed bound structures and their unbound structure(s) from the PDB for the protein and/or the RNA, their mapped bound and unbound structures, and two files on residue number mappings for the protein and RNA, respectively. All the binding interfaces of the bound and unbound structures were manually checked and no gaps were found that would significantly affect the binding between the protein and the RNA. Unusual amino acids or nucleic acid residues in the bound and unbound structures are also specified in the table listed at the website (http://zoulab.dalton.missouri.edu/RNAbenchmark/) for the convenience of docking preparation.
Table 1.
Benchmarking structures for protein-RNA docking.
| Complex of bound structures a |
Unbound structure(s)b |
Typec | Irmsd (Å) |
fnat | ΔSASAd (Å2) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| PDBID | Protein | RNA | Protein | Prmsd | RNA | Rrmsd | ||||
| ‘Easy’ (49) | ||||||||||
| 1C0A_A:B | Aspartyl tRNA synthetase | Aspartyl tRNA | 1IL2_A | 0.945 | 1EFW_C | 1.770 | PBU/RBU | 0.972 | 0.861 | 4175 |
| 1DFU_P:MN | Ribosomal protein l25 | 5S rRNA fragment | 3OFQ_V | 0.784 | 1FEU_CB | 4.548 | PBU/RBU | 1.001 | 0.696 | 1688 |
| 1E8O_CD:E | Signal recognition particle protein | 7SL RNA | 1E8O_AB | 0.930 | 1RY1_E | 0.001 | PBU/RB | 0.654 | 0.900 | 1434 |
| 1F7Y_A:B | 30S ribosomal protein S15 | 16S ribosomal RNA fragment | 2VQE_O | 0.803 | 1DK1_B | 0.275 | PBU/RBU | 0.589 | 0.963 | 2427 |
| 1FFY_A:T | Isoleucyl-tRNA synthetase | Isoleucyl-tRNA | 1QU3_A | 1.367 | 1QU2_T | 0.000 | PBU/RB | 0.840 | 0.879 | 4971 |
| 1G1X_FH:IJ | 30S ribosomal protein S6, S18 | 16S ribosomal RNA fragment | 2VQE_FR | 0.557 | 1G1X_DE | 1.678 | PBU/RBU | 1.059 | 0.879 | 2282 |
| 1GAX_B:D | Valyl-tRNA synthetase | tRNA(Val) | 1GAX_A | 0.411 | 1IVS_C | 0.547 | PBU/RBU | 0.426 | 0.904 | 5193 |
| 1H4S_AB:T | Prolyl-tRNA synthetase | tRNApro(cgg) | 1HC7_AB | 1.385 | 1H4Q_T | 0.188 | PBU/RBU | 0.840 | 0.773 | 2481 |
| 1HQ1_A:B | signal recognition particle protein | 4.5S RNA domain IV | 3LQX_A | 0.470 | 1DUL_B | 0.127 | PBU/RBU | 0.371 | 0.957 | 1364 |
| 1J1U_A:B | Tyrosyl-tRNA synthetase | tRNA(Tyr) | 1U7D_A | 1.272 | PBU/RB | 1.014 | 0.833 | 1113 | ||
| 1JBS_A:C | Restrictocin | Sarcin/Ricin domain RNA analog | 1JBR_A | 0.628 | 1JBT_C | 0.520 | PBU/RBU | 0.637 | 0.844 | 1267 |
| 1JID_A:B | Signal recognition particle protein | Helix 6 of human srp RNA | 3KTV_B | 0.637 | 1L1W_A | 1.967 | PBU/RBU | 1.062 | 0.907 | 1362 |
| 1K8W_A:B | tRNA Pseudouridine Synthase B | T Stem-Loop RNA | 1R3F_A | 1.924 | 1ZL3_B | 0.229 | PBU/RBU | 1.398 | 0.800 | 2607 |
| 1KOG_CD:K | Threonyl-tRNA synthetase | Threonyl-tRNA synthetase mRNA | 1EVL_AB | 0.658 | 1KOG_I | 0.958 | PBU/RBU | 0.775 | 0.827 | 2709 |
| 1LNG_A:B | Signal recognition particle protein | 7S.S srp RNA | 3NDB_A | 0.624 | 2V3C_M | 2.005 | PBU/RBU | 0.952 | 0.868 | 2367 |
| 1MMS_A:C | Ribosomal protein L11 | 23S ribosomal RNA fragment | 2JQ7_A | 1.798 | 1OLN_C | 0.000 | PBU/RB | 1.254 | 0.791 | 2455 |
| 1N78_B:D | Glutamyl-tRNA synthetase | tRNA(Glu) | 1J09_A | 2.126 | 2DXI_C | 0.905 | PBU/RBU | 1.883 | 0.824 | 4570 |
| 1Q2R_C:F | Queuine tRNA-ribosyltransferase | a stem-loop RNA substrate | 1R5Y_A | 0.731 | 1Q2S_E | 0.487 | PBU/RBU | 1.051 | 0.770 | 2677 |
| 1QTQ_A:B | Glutaminyl-tRNA synthetase | tRNA Gln II | 1GTR_A | 0.403 | 1QRS_B | 0.600 | PBU/RBU | 0.381 | 0.937 | 5183 |
| 1R3E_A:CDE | tRNA pseudouridine synthase B | a stemCloop RNA | 1ZE2_A | 0.925 | PBU/RB | 0.618 | 0.942 | 3276 | ||
| 1S03_H:A | 30S ribosomal protein S8 | spc Operon mRNA | 3OFO_H | 1.165 | 1S03_B | 1.631 | PBU/RBU | 0.988 | 0.830 | 1743 |
| 1SJ3_P:R | Small nuclear ribonucleoprotein A | Precursor form of the Hepatitis Delta virus ribozyme | 1M5O_C | 0.385 | 1VC7_B | 0.366 | PBU/RBU | 0.383 | 0.898 | 1867 |
| 1T0K_B:CD | 60S ribosomal protein L30 | mRNA | 3O58_Z | 1.389 | PBU/RB | 0.901 | 0.758 | 1008 | ||
| 1YVP_B:EF | 60-kDa SS-A/Ro ribonucleoprotein | Y RNA sequence, first strand, second strand | 1YVR_A | 1.437 | 1YVP_CD | 0.311 | PBU/RBU | 1.514 | 0.875 | 1538 |
| 2AKE_A:B | Tryptophanyl-tRNA synthetase | transfer RNA-Trp | 2DR2_A | 0.273 | 2AZX_C | 3.304 | PBU/RBU | 0.692 | 0.976 | 1122 |
| 2ANR_A:B | Neuro-oncological ventral antigen 1 | RNA aptamer hairpins | 2ANN_B | 0.549 | PB/RBU | 0.252 | 0.921 | 1219 | ||
| 2AZ0_AB:CD | B2 protein | double-stranded RNA (dsRNA) | 2B9Z_AB | 1.391 | 2AZ2_CD | 0.466 | PBU/RBU | 1.122 | 0.721 | 1969 |
| 2BH2_A:C | 23S rRNA (uracil-5-)-methyltransferase RumA | 23S ribosomal RNA fragment | 1UWV_A | 1.413 | 2BH2_D | 1.047 | PBU/RBU | 1.359 | 0.738 | 4043 |
| 2CSX_B:D | Methionyl-tRNA synthetase | tRNA(Met) | 2CSX_A | 0.335 | 2CT8_C | 1.026 | PBU/RBU | 0.477 | 0.855 | 2249 |
| 2CZJ_E:F | SsrA-binding protein | tmRNA | 1WJX_A | 1.488 | 2CZJ_B | 0.566 | PBU/RBU | 1.145 | 0.821 | 2424 |
| 2DU3_A:D | O-phosphoseryl-tRNA synthetase | tRNA | 2DU5_A | 0.390 | 2DU4_C | 0.625 | PBU/RBU | 0.423 | 0.875 | 1405 |
| 2FK6_A:R | Ribonuclease Z | tRNA(Thr) | 1Y44_A | 0.730 | PBU/RB | 0.939 | 0.950 | 1531 | ||
| 2GJW_AB:EFH | tRNA-splicing endonuclease | a bulge-helix-bulge RNA | 1R0V_AB | 1.646 | PBU/RB | 1.547 | 0.804 | 2889 | ||
| 2QUX_DE:F | Coat protein | a viral RNA | 2QUD_AB | 0.751 | 2QUX_C | 0.483 | PBU/RBU | 0.615 | 0.840 | 1673 |
| 2RFK_A:DE | Probable tRNA pseudouridine synthase B | Guide RNA 1, Guide RNA 2 | 3LWR_A | 0.972 | 3HJY_CD | 2.580 | PBU/RBU | 1.092 | 0.813 | 3068 |
| 2XDB_A:G | a protein toxin (ToxN) | a specific RNA antitoxin (ToxI) | 2XD0_A | 0.404 | PBU/RB | 0.319 | 0.947 | 1883 | ||
| 2ZM5_A:C | tRNA delta(2)-isopentenylpyrophosphate transferase | tRNA(Phe) | 3FOZ_A | 0.546 | 2ZXU_C | 0.185 | PBU/RBU | 0.335 | 0.971 | 3935 |
| 2ZNI_AB:C | Pyrrolysyl-tRNA synthetase | Bacterial tRNA | 2ZNJ_AB | 1.458 | 2ZNI_D | 1.066 | PBU/RBU | 1.219 | 0.798 | 3887 |
| 2ZUE_A:B | Arginyl-tRNA synthetase | tRNA-Arg | 2ZUF_B | 0.270 | PB/RBU | 0.127 | 0.992 | 4592 | ||
| 3CIY_A:CD | Toll-like receptor 3 | double-stranded RNA | 3CIG_A | 1.196 | PBU/RB | 1.367 | 0.833 | 2184 | ||
| 3DD2_H:B | Thrombin heavy chain | an RNA aptamer | 1GJ5_H | 0.394 | PBU/RB | 0.275 | 0.854 | 1508 | ||
| 3EPH_A:E | tRNA isopentenyltransferase | tRNA | 3EPK_A | 0.248 | 3EPJ_E | 0.409 | PBU/RBU | 0.229 | 0.967 | 4815 |
| 3FOZ_A:C | tRNA delta(2)- isopentenylpyrophosphate transferase | tRNA(Phe) | 2ZXU_A | 0.530 | 2ZM5_C | 0.711 | PBU/RBU | 0.351 | 0.928 | 4053 |
| 3HHZ_O:R | Nucleocapsid protein | viral genomic RNA (vRNA) | 3PTX_A | 1.568 | 2GIC_R | 0.971 | PBU/RBU | 0.867 | 0.763 | 2164 |
| 3LRR_A:CD | Probable ATP-dependent RNA helicase DDX58 | double-stranded RNA | 3LRN_A | 0.401 | PBU/RB | 0.348 | 1.000 | 664 | ||
| 3LWR_ABC:DE | Pseudouridine synthase Cbf5, Ribosome biogenesis protein Nop10, 50S ribosomal protein L7Ae | H/ACA RNA | 3LWP_ABC | 0.259 | 3HJW_DE | 0.794 | PBU/RBU | 0.394 | 0.917 | 5190 |
| 3MOJ_B:A | ATP-dependent RNA helicase dbpA | 23S ribosomal RNA fragment | 2G0C_A | 1.029 | PBU/RB | 0.916 | 0.774 | 1758 | ||
| 3OL9_M:NO | Polymerase | Positive-strand RNA | 3OL6_A | 0.561 | 3OLB_BC | 0.390 | PBU/RBU | 0.376 | 0.965 | 3665 |
| 3OVB_A:C | CCA-Adding Enzyme | tRNA | 3OV7_A | 0.328 | 3OUY_C | 0.472 | PBU/RBU | 0.352 | 0.924 | 2754 |
| ‘Medium’ (16): | ||||||||||
| 1F7U_A:B | Arginyl-tRNA synthetase | tRNA(Arg) | 1BS2_A | 3.393 | 1F7V_B | 0.876 | PBU/RBU | 2.573 | 0.800 | 5139 |
| 1IL2_A:C | Aspartyl-tRNA synthetase | Aspartyl transfer RNA | 1EQR_A | 1.813 | 1ASY_R | 1.545 | PBU/RBU | 1.696 | 0.671 | 4086 |
| 1R9F_A:BC | Core protein P19 | small interfering RNA | 3CZ3_EF | 4.464 | PB/RBU | 2.096 | 0.711 | 1723 | ||
| 1RC7_A:BCDE | Ribonuclease III | double-stranded RNA (dsRNA) | 1YYO_A | 3.397 | 1DI2_CDEF | 1.079 | PBU/RBU | 3.781 | 0.462 | 1866 |
| 1SER_AB:T | Seryl-tRNA synthetase | tRNAser | 1SES_AB | 1.936 | PBU/RB | 2.411 | 0.419 | 2259 | ||
| 1UN6_C:E | Transcription factor IIIA | 5S ribosomal RNA fragment | 2HGH_A | 2.337 | 1UN6_F | 2.571 | PBU/RBU | 2.181 | 0.659 | 1750 |
| 2BTE_D:E | Aminoacyl-tRNA synthetase | tRNA(Leu) transcript with anticodon cag | 1H3N_A | 3.900 | 2BYT_B | 1.854 | PBU/RBU | 2.758 | 0.679 | 3575 |
| 2FMT_A:C | Methionyl-tRNA fMet formyltransferase | Formyl-methionyl-tRNAfMet2 | 1FMT_A | 1.168 | 3CW5_A | 3.358 | PBU/RBU | 2.462 | 0.418 | 2941 |
| 2NUG_AB:CDEF | Ribonuclease III | double-stranded RNA | 2NUF_AB | 3.512 | PBU/RB | 3.047 | 0.608 | 5932 | ||
| 2UWM_A:C | Selenocysteine-specific elongation factor | SECIS mRNA | 1LVA_A | 7.787 | 1WSU_E | 0.617 | PBU/RBU | 3.190 | 0.654 | 932 |
| 2VPL_A:B | 50S ribosomal protein L1 | Fragment of mRNA for L1-operon containing regulator L1-binding site | 2OV7_A | 1.249 | 1U63_B | 3.678 | PBU/RBU | 1.703 | 0.506 | 2341 |
| 2ZKO_AB:CD | Non-structural protein 1 | double-stranded RNA (dsRNA) | 2Z0A_AB | 2728 | 2ZI0_CD | 4.294 | PBU/RBU | 3.874 | 0.522 | 2466 |
| 2ZZM_A:B | Uncharacterized protein MJ0883 | tRNA(Leu) | 2ZZN_A | 1.857 | PBU/RB | 1.581 | 0.733 | 4491 | ||
| 3ADD_A:C | L-seryl-tRNA(Sec) kinase | Selenocysteine tRNA | 3ADC_A | 2.625 | 3ADB_C | 0.781 | PBU/RBU | 2.327 | 0.500 | 2942 |
| 3FTF_A:CD | Dimethyladenosine transferase | 16S rRNA fragment | 3FTD_A | 2.410 | 3FTE_CD | 2.327 | PBU/RBU | 2.460 | 0.783 | 1693 |
| 3HL2_C:E | O-phosphoseryl-tRNA(Sec) selenium transferase | tRNASec | 3HL2_A | 0.214 | 3A3A_A | 4.008 | PBU/RBU | 2.598 | 0.421 | 975 |
| Difficult (7): | ||||||||||
| 1H3E_A:B | tyrosyl-tRNA synthetase | Wild-type tRNAtyr(Gua) | 1H3F_A | 9.444 | PBU/RB | 11.454 | 0.143 | 2224 | ||
| 100A_B:D | Nuclear factor NF-kappa-B p105 subunit | RNA aptamer | 1NFK_A | 6.193 | 2JWV_A | 5.444 | PBU/RBU | 5.564 | 0.354 | 1742 |
| 1U0B_B:A | Cysteinyl-tRNA synthetase | Cysteinyl tRNA | 1LI5_A | 1.001 | 1B23_R | 7.303 | PBU/RBU | 4.257 | 0.347 | 4558 |
| 2HW8_A:B | 50S ribosomal protein L1 | mRNA | 1AD2_A | 6.727 | 1ZHO_B | 1.518 | PBU/RBU | 5.305 | 0.595 | 2334 |
| 2IPY_A:C | Iron-responsive element-binding protein 1 | Ferritin IRE RNA | 2B3Y_A | 11.737 | 2IPY_D | 0.606 | PBU/RBU | 8.601 | 0.317 | 2857 |
| 2R8S_HL:R | Fab heavy chain, Fab light chain | p4-p6 RNA ribozyme domain | 3IVK_AB | 5.609 | 1HR2_A | 4.345 | PBU/RBU | 3.585 | 0.393 | 2510 |
| 2V3C_C:M | Signal recognition 54 kDa protein | 7S.SSRP RNA | 3NDB_B | 13.863 | 1LNG_B | 2.005 | PBU/RBU | 13.256 | 0.165 | 2876 |
The chain IDs of the interacting protein and RNA in a complex is separated by “:”, in which the former chain stands for the protein and the later chain is for the RNA.
“Prmsd” (“Rrmsd”) stands for the global RMSD (Å) of the protein (RNA) between its bound and unbound structures after optimal superimposition. The column is left blank when there is no unbound structure. In such a case, the bound structure can be used for unbound docking in the benchmark dataset.
“PBU/RBU” stands for the subset of the targets in which both the protein (P) and the RNA (R) are represented in bound and unbound conformations, and “PB/RBU” for the subset in which the protein is found only in bound conformations, and the RNA is present in bound and unbound conformations. Similarly with “PBU/RB”.
ΔSASA stands for the change of solvent access surface areas (SASA) of the protein and the RNA upon complex formation, in which SASA is calculated by the program NACCESS (44)
The 72 targets are grouped into three categories, ‘easy’, ‘medium’ and ‘difficult’ cases. The categories are classified based on two parameters, Irmsd and fnat (Table 2). Irmsd is the root mean square deviation (RMSD) of the interface region between the bound and the unbound structures of a target after optimal superimposition. The interface is defined as those residues of the bound structures having at least one atom that is within 10 Å from the other partner. The superimposition was based on one backbone atom for each residue, i.e., Cα atoms for the protein and C4′ atoms for the RNA (42). The fnat parameter of a complex is defined as the fraction of the native contacts in the unbound structures, namely, the ratio of the number of native residue-residue contacts in the superimposed unbound structures to the number of residue contacts in the native bound structures. A pair of residues from different partners are defined as contact residues if they are within 5 Å of each other (43). According to the criteria, the benchmark dataset contains 49 ‘easy’ targets, 16 ‘medium’ targets, and 7 ‘difficult’ targets (Table 1).
Table 2.
Criteria to categorize the targets using Irmsd and fnat.
| Category | Criterion |
|---|---|
| Easy | (Irmsd ≤ 1.5 Å ) OR (fnat ≥ 0.8) |
| Medium | (1.5 Å < Irmsd ≤ 4.0 Å) AND (0.4 ≤ fnat < 0.8) |
| Difficult | (Irmsd > 4.0 Å) OR (fnat < 0.4) |
It should be noted that ideally the difficulty-based categorization of the targets should be classified according to the docking results such as the number of hits in the top predictions. However, such docking results often depend on the docking algorithm and docking parameters in use, which could result in inconsistency in categorization by different research groups. Therefore, in the present work, we rely on the two parameters that are commonly used by the docking community, Irmsd and fnat, to classify the targets in the benchmark dataset (17).
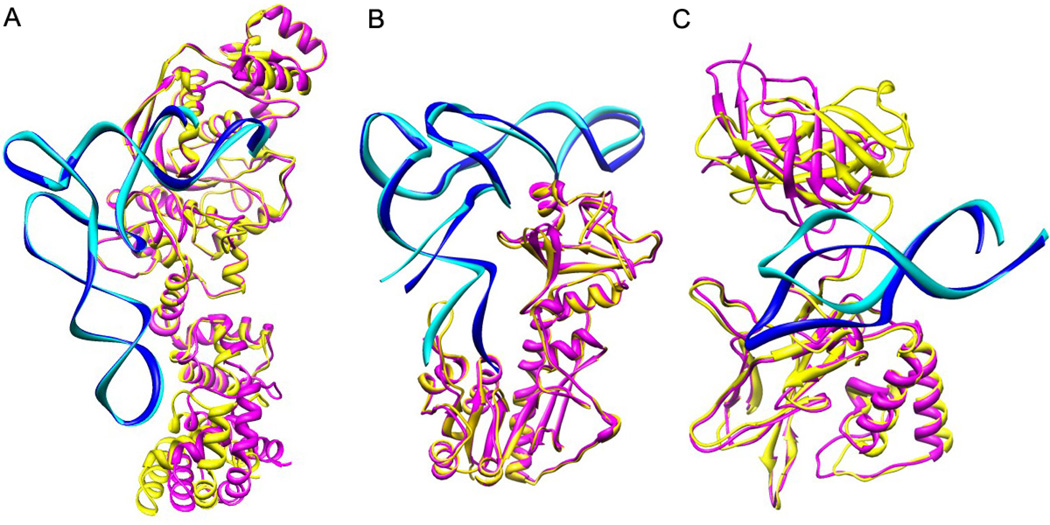
The classification by Irmsd and fnat is a reflection of the conformational changes between the unbound and bound structures, particularly the conformational changes at the binding interface. Normally, the ‘easy’ targets have small conformational changes and thus keep a large percentage of the native contacts (Figure 1A). These targets are good for validating the performance of a semi-rigid docking algorithm in which protein flexibility is considered implicitly. The easy targets can also be used to examine the efficiency of rigid-body sampling — the first step for all docking algorithms. The ‘medium’ targets often involve significant conformational changes from the unbound to bound states (Figure 1B). Therefore, docking the ‘medium’ targets may require explicit consideration of protein flexibility during sampling. Otherwise, the correct binding modes would not be ranked in the top predictions. For ‘difficult’ targets, there are often global conformational changes such as large domain movements via hinges between the unbound and bound structures (Figure 1C). In some extreme cases, the binding site may be even blocked in the unbound structures due to the large conformational changes. For example, in Table 1, Target 2HW8 has the binding interface partially blocked in its unbound structure. Moreover, the binding interface of Target 2IPY is fully blocked in its unbound structure. Therefore, when docking the difficult targets, protein flexibility must be considered. The correct binding mode may be completely missed if the large conformational change is not explicitly considered during sampling.
Figure 1.
Comparison of the bound and unbound structures of three targets, in which the bound/unbound conformations of the protein are colored in yellow/magenta and the bound/unbound conformations of the RNA are colored in blue/cyan. (A) ‘Easy’ target 1N78 (Irmsd = 1:883 Å, fnat = 0:824). (B) ‘Medium’ target 2FMT (Irmsd = 2:462 Å, fnat = 0:418). (C) ‘Difficult’ target 1OOA (Irmsd = 5:564 Å, fnat = 0:354).
An important feature for individual unbound structures is the overall conformational change between the bound and unbound structures. We have calculated the global RMSD between the bound and the bound structures after optimal superimposition based on one backbone atom per residue, which are listed in Table 1. It can be seen from the table that overall the unbound structures tend to have smaller global RMSDs for easy targets, and larger global conformational changes for difficult targets (e.g. 13.86 °A for the protein of 2V3C), as what is expected.
It is also notable from Table 1 that for a few targets only one unbound structure can be found from PDB for one of the two binding partners; the other binding partner has no available unbound structure. A second feature of Table 1 is that a few unbound RNA structures such as 1E8O have a very small global RMSD due to the fact that both the bound and unbound structure were solved by the same group. To take this phenomenon into account, we have introduced a new concept — the “unbound conformation”, that is, an unbound structure with a global RMSD greater than 0:1 Å; otherwise, the unbound structure is defined as a “bound conformation”. According to the availability of unbound conformations for the protein and the RNA, the benchmark dataset of 72 targets are divided into three categories, ‘PBU/RBU’, ‘PBU/RB’ and ’PB/RBU’, where “PBU/RBU” stands for those targets in which both the Protein (P) and the RNA (R) have the bound (B) and unbound (U) conformations, “PB/RBU” for the targets in which the protein is only found in the bound conformation, while the RNA is present in both the bound and the unbound conformations, and “PBU/RB” has a similar definition. Following the classification, the benchmark dataset consists of 52 ‘PBU/RBU’ targets, 17 ‘PBU/RB’ targets, and 3 ‘PB/RBU’ targets. Considering the conformational changes between the unbound (U) and the bound (B) conformations, it is expected that the ‘easy’ category should contain the largest number of ‘PBU/RB’ or ‘PB/RBU’ targets, and the ‘difficult’ category should have the largest percentage for ‘PBU/RBU’ targets. This is indeed the case, as shown in Table 1.
To measure the size of a binding interface for each target, we also calculated the change of the solvent accessible surface areas (SASA) of the protein and the RNA upon binding. ΔSASA is defined as (SASA of the protein + SASA of the RNA – SASA of the complex). Here, the SASA was calculated with the NACCESS program (44), in which the probe radius was set to 1.4 Å.
4 DISCUSSIONS
Compared to the field of protein docking, the RNA-docking field is relatively young with only a small number of published examples. This phenomenon may be attributed to three reasons. First, it is challenging to predict the three-dimensional (3D) structure of an RNA from its sequence, which has limited the application of computationally predicted RNA 3D structures to molecular docking. Unlike proteins whose sequences are conserved among homologues, RNA molecules show conservation in secondary and tertiary structures but not in primary sequences. In addition to the native structure which corresponds to the global minimum, there exist many metastable conformations which correspond to the local minima on the free energy landscape of RNA folding. It is therefore challenging to predict RNA 3D structures from sequences by homology modeling, as shown in Target 33 of the CAPRI experiment (45, 46). Second, compared to experimentally-determined protein structures or protein-protein complex structures, there are very limited RNA structures or protein-RNA complex structures in the PDB that can be used for the development, validation and improvement of RNA docking algorithms. Therefore, a well-prepared benchmark dataset of protein-RNA complexes is urgently needed. Lastly, it is challenging to account for conformational changes in proteins and particularly in RNA molecules upon binding, a reason for us to provide both the bound and unbound structures in our dataset.
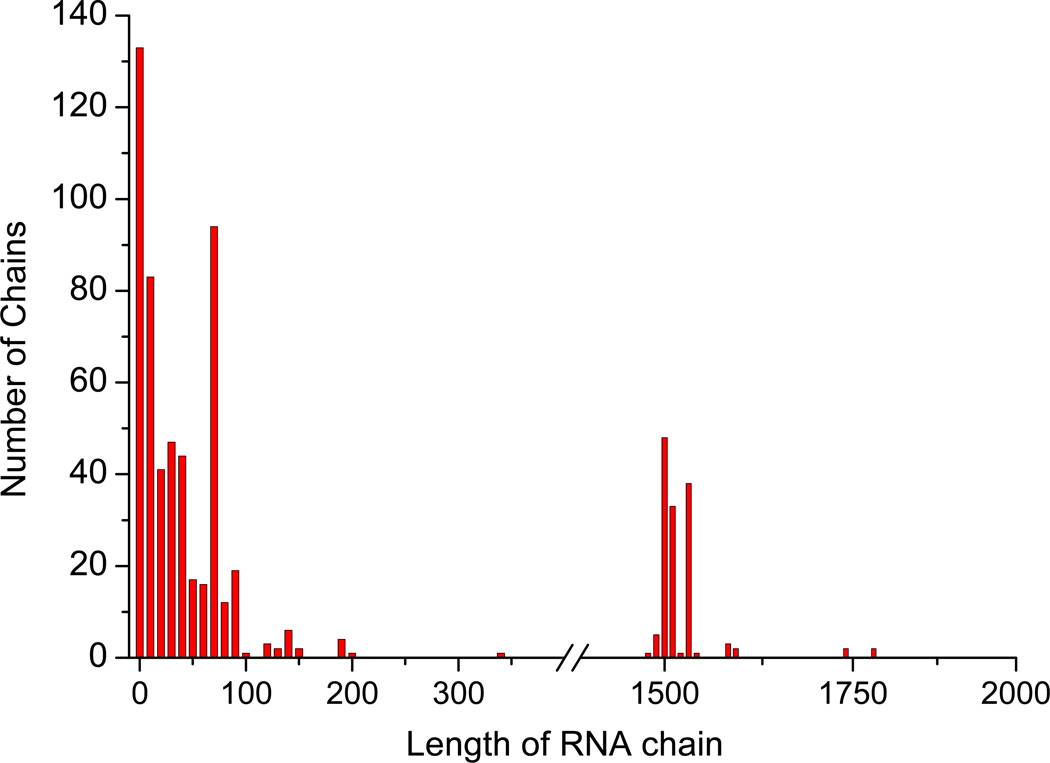
During the development of our benchmark dataset, we have limited the size of the RNA to 20 ~ 200 nucleotides because of the following reasons. If an RNA chain is too short, it cannot fold into a stable 3D structure, or it is normally part of a larger RNA. If an RNA chain is too long, say more than 1000 nucleotides, it may be too challenging for the existing RNA structure prediction algorithms to predict a reliable 3D structure and its conformational change that can be used for docking calculations. As shown in Figure 2, for the 859 protein-RNA complexes initially extracted from the PDB (see the Materials and Methods section), the lengths of their RNA chains are mainly distributed in two regions. The first region ranges from 1 to 200 nucleotides and consists of different types of RNA molecules. The other region is between 1400 and 1800 nucleotides, which correspond to ribosomal RNAs (rRNA).
Figure 2.
The distribution of the lengths of the RNA chains in the 859 protein-RNA complexes obtained from our initial PDB query. See the text for detail.
Despite the rich source of ribosomes in the PDB, these complexes are not included in the present release of the RNA benchmark because most of the existing docking algorithms are designed for two-body docking (17, 19, 25–27). A target in a benchmark dataset normally contains only one biologically important binding interface that is formed by two binding bodies, e.g. two protein structures for protein docking benchmarks, or one protein structure and one RNA structure for protein-RNA docking benchmark in this case. In contrast, a ribosome complex usually consists of a large RNA subunit that has more than 1000 nucleotides and multiple protein chains which are embedded in the RNA. For example, the ribosome 1JJ2 includes one rRNA chain of 2922 nucleic acids and 28 protein chains. These protein chains/structures form multiple separate protein-RNA interfaces with the rRNA. The complexity involved in such multi-body binding problems is beyond the scope of present docking algorithms.
Therefore, based on the distribution of the RNA sizes shown in Figure 2, we have restricted the maximum size for the RNA molecules to 200 nucleotides in the current benchmark dataset. However, this restriction does not exclude protein-rRNA interactions from the benchmark dataset. As shown in Table 1, there are quite a few targets on protein-rRNA fragment interactions that may serve as good examples for investigation of their binding mechanisms. Given the biological importance of ribosomes, we will include ribosomes as a special category in the next version of our protein-RNA docking benchmark dataset. The ribosomal structures will be useful for the development and assessment of multi-body docking algorithms, and may also serve as a benchmark for application of traditional two-body docking algorithms to ribosome research.
Theoretically speaking, we shall not limit the number of the chains in each binding partner so as to collect as many protein-RNA interfaces as possible in our benchmark dataset. However, more chains in a binding partner also mean much less possibility in finding the corresponding unbound structure with the same number of chains from the PDB, leading to fewer effective targets in the dataset. Considering the fact that some RNA molecules may break up into several chains in experimental conditions and that some protein structures might exist as an oligomer of multiple identical chains (e.g. a hexamer of six chains), we have limited the number of the chains in the protein or RNA to be no more than six when constructing the present benchmark dataset, which keeps sufficient number of effective cases in the dataset without leaving out those important oligomers that have multiple chains.
Furthermore, a benchmark dataset should be diverse to represent different types of proteins and RNA molecules. In the present study, we have used sequence as an index for diversity, a commonly-used index by other benchmark datasets (19, 25). However, as aforementioned, unlike proteins, RNA molecules are conserved in secondary and tertiary structures but not in sequences. Therefore, we have used a stricter clustering method to diversify our selection of the protein-RNA complexes. Namely, two complexes are grouped into the same cluster if the two proteins have higher than 30% sequence identity or if the two RNA molecules have higher than 70% sequence identity. It is noted that the present sequence cutoff for RNA (70%) is lower than the cutoff used in the literature (19). To consider the structural diversity of RNA molecules, secondary and tertiary structures would be a better clustering index than sequences, which will be addressed in the future when the benchmark dataset is updated.
To measure the induced fit and conformational adaptation upon binding, we have calculated the RMSD between the bound and unbound structures. Despite the RMSD metric is widely used for benchmark datasets by the docking community (17–19, 25–28), it should be noted that RMSD is a crude, global measurement of conformational changes. For RNA structures, other metrics such as the consideration of specific interactions like non-Watson-Crick base pairing would provide more informative measures on the similarity of RNA structures. The reliability in predicting non-Watson-Crick base pairs (47) directly determine the accuracy of the predicted RNA structures and conformational changes, which is important for RNA-protein docking.
For the calculation of interface RMSDs in the present study, for simplicity, each residue is represented by one backbone atom, i.e., Cα for the protein and C4′ for the RNA. It should be noted that unlike proteins for which each residue is commonly represented by the Cα backbone atom in reduced models, RNA molecules have different reduced representations for each nucleotide, such as the use of P and C4′, respectively (19,42). An advantage of using C4′ over P is that DNA and RNA molecules normally contain C4′ atoms in each nucleotide but may miss P atoms in the terminal residues in some PDB files such as 1YVP. However, different representations will not result in significant differences in the measured RMSD values.
5 CONCLUSION
We have constructed a benchmark dataset for protein-RNA docking, which consists of 52 unbound/unbound cases and 20 unbound/bound cases. All the bound and unbound structures in the benchmark dataset are extracted from experimentally determined structures in the PDB, reflecting real conformational changes of the proteins and RNAs upon binding. The diverse bound and unbound structures may serve as a benchmark to assess the performance of docking and scoring algorithms on protein-RNA interactions. All the structures in the benchmark dataset listed in Table 1 are freely available at http://zoulab.dalton.missouri.edu/RNAbenchmark/. As a public resource of the RNA docking community, the benchmark dataset will be updated annually with the increasing number of protein-RNA complexes deposited in the PDB.
ACKNOWLEDGMENTS
We thank Sam Grinter for proofreading the manuscript. XZ is supported by NIH grant R21GM088517 and NSF CAREER Award 0953839. The computations were performed on the HPC resources at the University of Missouri Bioinformatics Consortium (UMBC).
References
- 1.Wodak SJ, Janin J. J. Mol. Biol. 1978;124:323–342. doi: 10.1016/0022-2836(78)90302-9. [DOI] [PubMed] [Google Scholar]
- 2.Muegge I, Rarey M. Rev. Comput. Chem. 2001;17:1–60. [Google Scholar]
- 3.Shoichet BK, McGovern SL, Weih B, Irwin JJ. Curr. Opin. Chem. Biol. 2002;6:439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- 4.Smith GR, Sternberg MJ. Curr. Opin. Struct. Biol. 2002;12:28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 5.Halperin I, Ma B, Wolfson H, Nussinov R. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 6.Brooijmans N, Kuntz ID. Annu. Rev. Biophys. Biomol. Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 7.Schneidman-Duhovny D, Nussinov R, Wolfson HJ. Curr. Med. Chem. 2004;11:91–107. doi: 10.2174/0929867043456223. [DOI] [PubMed] [Google Scholar]
- 8.Kitchen DB, Decornez H, Furr JR, Bajorath J. Nat. Rev. Drug Discov. 2004;3:935–948. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 9.Gray JJ. Curr. Opin. Struct. Biol. 2006;16:183–193. doi: 10.1016/j.sbi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Bonvin AM. Curr. Opin. Struct. Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Sousa SF, Fernandes PA, Ramos M. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 12.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. Proteins. 2008;73:271–289. doi: 10.1002/prot.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolb P, Ferreira RS, Irwin JJ, Shoichet BK. Curr. Opin. Biotech. 2009;20:429–436. doi: 10.1016/j.copbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S-Y, Zou X. Int. J. Mol. Sci. 2010;11:3016–3034. doi: 10.3390/ijms11083016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S-Y, Grinter SZ, Zou X. Phys. Chem. Chem. Phys. 2010;12:12899–12908. doi: 10.1039/c0cp00151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janin J, Henrick K, Moult J, Ten Eyck L, Sternberg MJE, Vajda S, Vasker I, Wodak SJ. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 17.Hwang H, Vreven T, Janin J, Weng Z. Proteins. 2010;78:3111–3114. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kastritis P, Moal I, Hwang H, Weng Z, Bates P, Bonvin A, Janin J. Protein Sci. 2011;20:482–491. doi: 10.1002/pro.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Dijk M, Bonvin AM. Nucleic Acids Res. 2008;36:e88. doi: 10.1093/nar/gkn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, Douguet D, Tovchigrechko A, Vakser IA. Proteins. 2007;69:845–851. doi: 10.1002/prot.21714. [DOI] [PubMed] [Google Scholar]
- 21.Irwin JJ, Shoichet BK. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunbar JB, Jr, Smith RD, Yang CY, Ung PM, Lexa KW, Khazanov NA, Stuckey JA, Wang S, Carlson HA. J. Chem. Inf. Model. 2011;51:2036–2046. doi: 10.1021/ci200082t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S-Y, Zou X. J. Chem. Inf. Model. 2011;51:2107–2114. doi: 10.1021/ci200080g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang S-Y, Zou X. J. Chem. Inf. Model. 2011;51:2097–2106. doi: 10.1021/ci2000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen R, Mintseris J, Janin J, Weng Z. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 26.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z. Proteins. 2005;60:214–216. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 27.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijk M, Bonvin AM. Nucleic Acids Res. 2010;38:5634–5647. doi: 10.1093/nar/gkq222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabian MR, Sonenberg N, Filipowicz W. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 30.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Licatalosi DD, Darnell RB. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorkovic ZJ. Trends Plant Sci. 2009;14:229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Lukong KE, Chang KW, Khandjian EW, Richard S. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Lunde BM, Moore C, Varani G. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansfield KD, Keene JD. Biol. Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittal N, Roy N, Babu MM, Janga SC. Proc. Natl Acad. Sci. USA. 2009;106:20300–20305. doi: 10.1073/pnas.0906940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammad MM, Donti TR, Sebastian Yakisich J, Smith AG, Kapler GM. EMBO J. 2007;26:5048–5060. doi: 10.1038/sj.emboj.7601919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capriotti E, Marti-Renom MA. Curr. Bioinform. 2008;3:32–45. [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Brandman R, Brandman Y, Pande VS. PLoS One. 2012;7:e29377. doi: 10.1371/journal.pone.0029377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Méndez R, Leplae R, Lensink MF, Wodak SJ. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 44.Hubbard SJ, Thornton JM. NACCESS Computer Program. London: Department of Biochemistry and Molecular Biology, University College; 1993. [Google Scholar]
- 45.Lensink MF, Wodak SJ. Proteins. 2010;78:3073–3084. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 46.Huang SY, Zou X. Proteins. 2010;78:3096–3103. doi: 10.1002/prot.22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leontis NB, Stombaugh J, Westhof E. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]