Abstract
MicroRNAs are master regulators of gene expression and control many biological pathways such as cell growth, differentiation and apoptosis. Deregulation of microRNA expression and activity results in a myriad of diseases including cancer. Recently, several reports have indicated that single nucleotide polymorphisms (SNPs) in microRNAs and microRNA-target sites impact microRNA biology and associate with cancer risk, treatment response and outcome. In this review we will describe these findings and discuss the possible future of utilizing these SNPs as diagnostic and prognostic markers in the clinic.
Keywords: MicroRNA, 3′ untranslated region, microRNA-target site, single nucleotide polymorphism, human disease, cancer, cancer risk, treatment response, personalized medicine
1. Introduction
Sequence analysis of the human diploid genome estimates that human populations are 99.5% identical at the DNA level (Levy et al., 2007; J. Wang et al., 2008). Therefore, factors leading to human diversity must arise from the remaining 0.5% of variable genetic information, comprised primarily of single nucleotide polymorphisms (SNPs). Approximately 10 million SNPs have been identified in the human genome, occurring at a frequency of approximately 1–3% (or 1 out every 100–300 nucleotides) in the normal population (Levy, et al., 2007; Sachidanandam et al., 2001; J. Wang, et al., 2008). SNPs can occur in coding and non-coding regions of the genome. While the vast majority of SNPs located in non-coding regions of the genome were believed to be silent, new evidence suggests that SNPs coincident with cis-regulatory elements play a critical role in defining human diversity and disease by regulating the nature and timing of gene expression (Buonocore et al., 2010; Dimas et al., 2009; Pastinen, Ge, & Hudson, 2006).
Cis-regulatory elements are sequence motifs in DNA and RNA that control gene expression (Pastinen, et al., 2006; Pastinen & Hudson, 2004). Cis-regulatory elements are often controlled by the concomitant expression of a requisite trans-acting factor. Transacting-factors function in response to stimuli and allow cells to fine tune gene expression and adapt to environmental or extracellular cues. Uncovering the relationship between cis-regulatory elements and the trans-acting factors that govern their expression is important to further our understanding of normal biological processes as well as disease.
2.1 MicroRNAs
MicroRNAs are a class of trans-acting RNAs found in eukaryotic organisms that bind to a cis-regulatory element in a target mRNA and regulate gene expression by inhibiting protein translation (Hobert, 2004). The first microRNAs discovered, lin-4 (R. C. Lee, Feinbaum, & Ambros, 1993; Wightman, Ha, & Ruvkun, 1993) and let-7 (Reinhart et al., 2000), were identified in C. elegans for their ability to control developmental timing and cell fate specification. The discovery that let-7 homologs displayed temporal expression in flies and mice indicated that microRNAs may have similar functions in higher order species (Pasquinelli et al., 2000). This prompted cloning efforts, which elucidated hundreds of new genes that encode for these trans-acting, small RNAs in worms, flies, mice and humans (Lagos-Quintana, Rauhut, Lendeckel, & Tuschl, 2001; Lagos-Quintana, Rauhut, Meyer, Borkhardt, & Tuschl, 2003; Lagos-Quintana et al., 2002; Lau, Lim, Weinstein, & Bartel, 2001).
MicroRNA genes are catalogued in the miRbase database (Griffiths-Jones, Grocock, van Dongen, Bateman, & Enright, 2006). According to the most recent release of miRbase, 21,264 precursor microRNAs and 25,141 mature microRNAs have been identified in 193 eukaryotic species as well as viruses (Kozomara & Griffiths-Jones, 2011). Of these, 1,600 precursor microRNAs and 2,042 mature microRNAs were cloned from human sources (Kozomara & Griffiths-Jones, 2011). MicroRNAs represent approximately 2% of the amount of protein-coding genes (Griffiths-Jones, 2004). MicroRNAs are believed to regulate up to 30% of all protein-coding genes (John et al., 2004; Krek et al., 2005; Lewis, Shih, Jones-Rhoades, Bartel, & Burge, 2003; Lim et al., 2005). As microRNA discovery extends to various cell, tissue and tumor types with the aide of deep-sequencing, the amount of annotated microRNAs will likely increase.
While many microRNAs display cell and tissue-specific expression patterns (Blower et al., 2007; Landgraf et al., 2007; Wienholds et al., 2005), elucidating the factors that govern microRNA expression in response to particular environmental cues and the specific mRNAs that are regulated in response to these cues remains a critical challenge to understanding how microRNAs function in human biology. Several recent studies have begun to uncover how extracellular stimuli such as growth factors (Seike et al., 2009; Suarez, Fernandez-Hernando, Pober, & Sessa, 2007), hormones (Klinge, 2009; Porkka et al., 2007), hypoxia (Kulshreshtha et al., 2007), DNA damage (Wagner-Ecker, Schwager, Wirkner, Abdollahi, & Huber, 2010; Weidhaas et al., 2007; Zhou et al., 2010) effect microRNA expression. Identifying the particular microRNAs and requisite mRNA targets that are sufficient to elicit a context-dependent, microRNA-mediated cellular response is critical, as they will likely provide useful diagnostic and prognostic biomarkers. Furthermore, uncovering how microRNA associated SNPs play a role in altering the normal biological processes in response to these cues is critical to understanding the molecular basis of how these variants play a role in disease onset and progression and will allow for the development of targeted therapeutics in the future.
MicroRNAs play a role in regulating many biological pathways including cell growth, differentiation and apoptosis (reviewed by (Esquela-Kerscher & Slack, 2006; He & Hannon, 2004) all of which are deregulated in cancer. MicroRNAs can function as both oncogenes and tumor suppressors (Croce, 2009; Hammond, 2006; B. Zhang, Pan, Cobb, & Anderson, 2007). Conditional deletion (He et al., 2007) or over-expression (Hayashita et al., 2005; Medina, Nolde, & Slack, 2010) of single microRNA genes is sufficient to drive tumorigenesis in mice. Consistent with these findings, it was found that 50% of all microRNA genes are in fragile regions of the genome that are frequently deleted, amplified and mis-expressed in human cancers (Calin & Croce, 2006; Calin et al., 2002; Calin et al., 2004). The role of SNPs in microRNAs and their binding sites are not surprisingly critical in cancer as well, as will be discussed in this review.
2.2 MicroRNA biogenesis
MicroRNA genes are located in the introns of protein-coding genes as well as in intergenic regions of the genome previously thought to be transcriptionally inactive (Saini, Griffiths-Jones, & Enright, 2007). About 45% of human microRNA genes are clustered together in groups of 2 or more and are individually generated from the polycistronic transcript (Saini, et al., 2007).
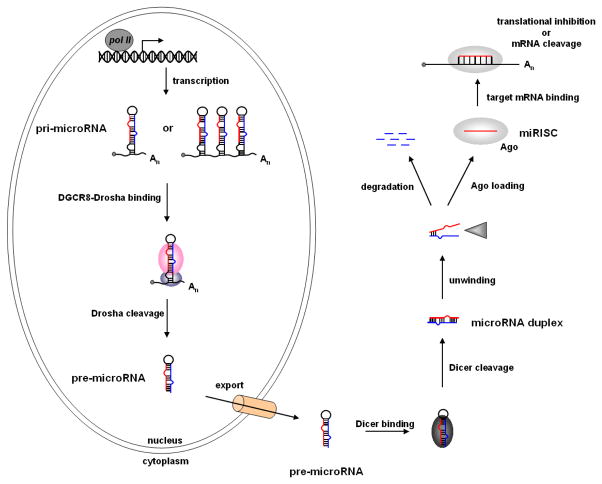
In mammalian systems, microRNAs are transcribed from the genome by RNA polymerase-II as a long primary transcript (or pri-microRNA) that is capped and polyadenylated (Cai, Hagedorn, & Cullen, 2004; Y. Lee et al., 2004). The pri-microRNA folds into a stem-loop structure and is bound by the double-strand RNA binding protein DGCR8 at the base of the stem (Han et al., 2004; Han et al., 2006). DGCR8 associates with the RNaseIII enzyme Drosha, which cleaves both strands of the pri-microRNA stem generating a shorter ~70 nucleotide stem-loop called the pre-microRNA (Gregory et al., 2004; Y. Lee et al., 2003) (Figure 1).
Figure 1.
The microRNA biogenesis pathway.
Exportin5 cooperatively binds the pre-microRNA hairpin and Ran-GTP and facilitates export of the RNA from the nucleus to the cytoplasm (Yi, Qin, Macara, & Cullen, 2003; Zeng & Cullen, 2004). In the cytoplasm the pre-microRNA is bound by the RNaseIII enzyme Dicer (Bernstein, Caudy, Hammond, & Hannon, 2001; Bernstein et al., 2003), which measures approximately 2-helical turns (22 nucleotides) up from the base and cleaves both strands of the stem generating a 22 nucleotide microRNA duplex (Macrae et al., 2006; H. Zhang, Kolb, Jaskiewicz, Westhof, & Filipowicz, 2004) (Figure 1).
The duplex is unwound by an RNA helicase (Chu & Rana, 2006; Salzman, Shubert-Coleman, & Furneaux, 2007) and the mature microRNA is loaded into 1 of 4 Argonuate proteins (Ago1–4) (Carmell, Xuan, Zhang, & Hannon, 2002; Farazi, Juranek, & Tuschl, 2008; Peters & Meister, 2007). The other strand of the duplex (or microRNA*) is often degraded. However in some cases, like miR-199 and miR-199* both strands of the microRNA duplex are loaded into an Ago protein (Czech et al., 2009; Okamura, Liu, & Lai, 2009). Ago is the heart of the microRNA-induced silencing complex (miRISC), which is guided by the microRNA to complementary elements in the 3′ UTR of a target mRNA (Carmell, et al., 2002; Peters & Meister, 2007). The miRISC negatively regulates gene expression by either mRNA cleavage or inhibiting translation (Valencia-Sanchez, Liu, Hannon, & Parker, 2006).
2.3 Determinants for microRNA target selection
To better understand how SNPs may be important in disrupting microRNA regulation of targets, it is important to understand the complexity of microRNA target selection. Target selection is based predominantly on the extent of Watson-Crick base pairing between the microRNA and mRNA and this is linked directly to the mechanism by which the mRNA is silenced. Nucleotides 2–7 (from the 5′ end of the microRNA), also called the microRNA ‘seed’, are a major determinant of mRNA target selection (Lewis, et al., 2003). Mutation(s) in either the seed or seed-complementary site inhibited microRNA activity and could be rescued with a compensatory mutation(s) highlighting the importance of seed sequence complementarity (Vaucheret, Vazquez, Crete, & Bartel, 2004; Vella, Choi, Lin, Reinert, & Slack, 2004). Complementarity in the seed region leads to translation repression (Pillai et al., 2005).
While Watson-Crick base pairing in the seed is absolutely critical for target recognition, there are other enhancing features that can strengthen microRNA target selection. Complementarity at position 8 of the microRNA, and the presence of an adenosine residue in the target mRNA opposite of nucleotide 1 enhance target recognition (Lewis, Burge, & Bartel, 2005). Additionally, more than 4 contiguous Watson-Crick base pairs between nucleotides 12–17 at the 3′ end of the microRNA also enhance target recognition (Grimson et al., 2007).
MicroRNA-directed cleavage occurs by hydrolysis of the phosphodiester backbone in the target mRNA opposite nucleotides 10 and 11, when there is complementarity between (at least) nucleotides 2–15 (Meister et al., 2004). While 1 or 2 single nucleotide mis-matches or G:U wobbles are tolerated, canonical Watson-Crick base pairing is absolutely critical between nucleotides 9–12 (Felice, Salzman, Shubert-Coleman, Jensen, & Furneaux, 2009; Martinez & Tuschl, 2004). However, it was recently demonstrated that centered pairing requiring 11–12 contiguous Watson-Crick base pairs, between nucleotides 4–15 of the microRNA, is also sufficient to direct target RNA cleavage (Shin et al.). While examples of microRNA-directed mRNA cleavage can occur in humans, computational analysis indicated that the amount of target sites predicted to fit this criteria are extremely rare (John, et al., 2004; Yekta, Shih, & Bartel, 2004).
3.1 SNPs in microRNAs and microRNA target sites
Because microRNA biogenesis and target selection is highly sequence dependent, germline sequence variants (such as SNPs) and posttranscriptional base modifications (such as ADAR editing) in either the microRNA or microRNA-target site can have profound effects on microRNA function. Interestingly, the first evidence that a microRNA-associated SNP could elicit gross morphologic defects was inherent to the initial discovery of the let-7 microRNA. The temperature sensitive let-7(n2853) mutation that results in C. elegans lethality is in fact a single nucleotide G>A point mutation at position 5 of the microRNA (Reinhart, et al., 2000). This mutation inhibits let-7 from targeting the lin-41 mRNA and results in reiteration of larval cell divisions in the adult worm (Reinhart, et al., 2000). While the let-7(n2853) mutation is chemically induced, it provides proof of principle evidence for this concept.
Sequencing analysis showed that microRNAs and microRNA target sites are highly conserved through evolution (Chen & Rajewsky, 2006). Furthermore, SNPs in microRNA genes are relatively rare (Saunders, Liang, & Li, 2007). These findings indicate that trans-acting microRNAs and the requisite cis-regulatory elements they regulate were under selective pressure during evolution. This suggests that the repertoire of SNPs that have been identified in microRNAs and microRNA target sites may represent a class of functional variants.
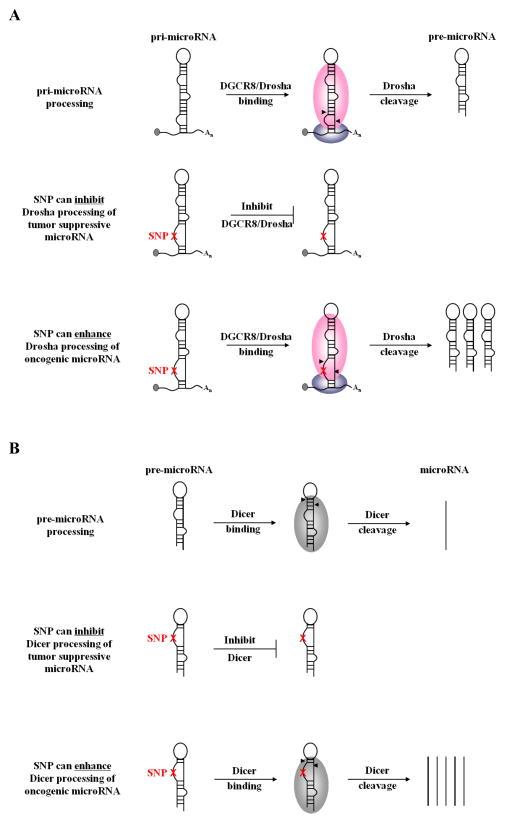
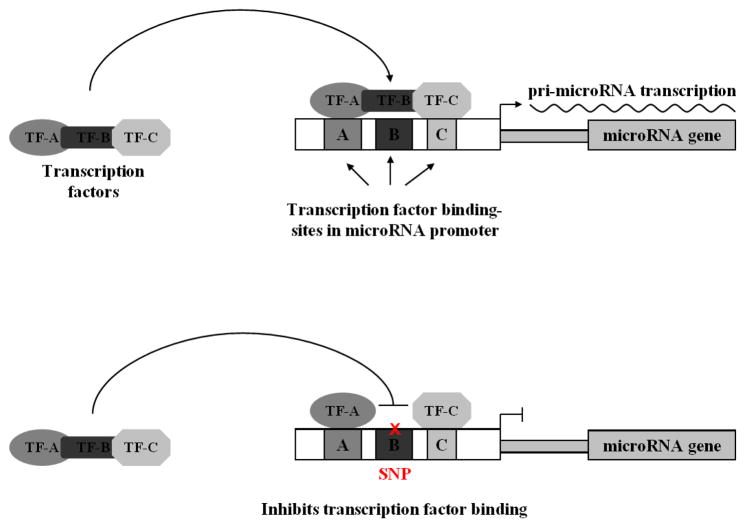
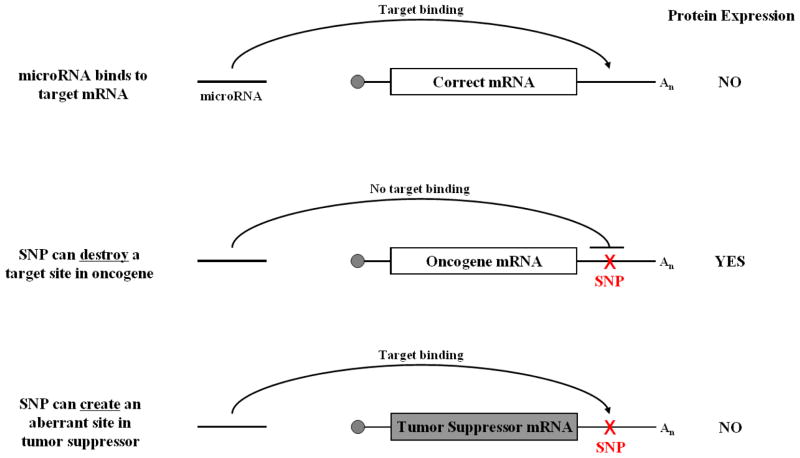
In theory, microRNA-associated SNPs can elicit cancer phenotypes by either creating a loss-of-function scenario whereby the expression, activity or targeting of a tumor suppressor microRNA is inhibited; or a gain-of-function scenario where by the expression, activity or targeting of an oncogenic microRNA is enhanced. SNPs can have director or indirect effects on microRNAs. Direct effects include SNPs in the pri-microRNA, pre-microRNA or mature microRNA that impair or enhance microRNA processing or function (Figure 3). Indirect effects include SNPs in microRNA promoters that affect transcription (Figure 2) and SNPs in an mRNA that create or destroy a target site (Figure 4). In the later half of this review we will discuss cancer-associated microRNA SNPs that affect microRNAs via three different mechanisms; microRNA transcription; microRNA precursor processing, and microRNA-mRNA binding.
Figure 3.
SNPs in pri-microRNA (A) or pre-microRNA (B) precursors can inhibit the processing of a tumor suppressor microRNA or can enhance the processing of an oncogenic microRNA.
Figure 2.
SNPs in microRNA promoters can prevent transcription factor binding and inhibit pri-microRNA transcription.
Figure 4.
SNPs in microRNA-target sites can either destroy an existing target site or create a target site in the wrong mRNA.
3.2 MicroRNA promoter SNPs
MicroRNA transcription is regulated by the same mechanisms that control protein-coding genes (Hobert, 2004; Marson et al., 2008). A number of well-characterized transcription factors control both mRNA and microRNA expression (He, He, Lim, et al., 2007; O’Donnell, Wentzel, Zeller, Dang, & Mendell, 2005). Transcription factors bind to conserved sequence motifs in the genome (typically) upstream of the gene for which they are controlling the transcription of (Orphanides, Lagrange, & Reinberg, 1996; Roeder, 1996). Because the recognition of a transcription factor to a particular DNA locus is sequence dependent (el-Deiry, Kern, Pietenpol, Kinzler, & Vogelstein, 1992; Maniatis, Goodbourn, & Fischer, 1987), variants in transcription factor binding sites could potentially alter microRNA expression (Figure 2). Computational analysis predicts that there are over 20,000 SNPs coincident with microRNA promoters in humans (Schmeier, Schaefer, MacPherson, & Bajic, 2011). However, the functional and phenotypic relevance of virtually all of these SNPs remains unclear.
The miR-34 family is transcriptionally upregulated following exposure to cytotoxic stress in a p53-dependent manner (He, He, Lowe, & Hannon, 2007; Hermeking, 2007). Mapping of the miR-34a and miR-34b/c promoters indicated that there are conserved p53 binding sites upstream of the miR-34 family that are required for transcription (Chang et al., 2007; He, He, Lim, et al., 2007). Loss of miR-34 function attenuates p53-mediated cell cycle arrest and apoptosis (He, He, Lim, et al., 2007; Raver-Shapira et al., 2007), which is congruent with increased cellular transformation and sensitivity to cytotoxic therapy (Kato et al., 2009). The rs4938723 T>C SNP located 423 nucleotides upstream of miR-34b/c is located in a transcription factor binding site and is predicted to attenuate GATA binding (Y. Xu et al., 2011). The rs4938723 SNP is associated with an increased risk of hepatocellular carcinoma in a case-control study of 501 Chinese individuals (Y. Xu, et al., 2011). While the precise mechanism in which the rs4938723 SNP associated with increased HCC risk is unknown, it phenocopies a TP53 loss-of-function mutation, and therefore likely inhibits expression of the miR-34 family resulting in enhanced cellular transformation.
3.3 SNPs in microRNA precursors
MicroRNA biogenesis proceeds through sequential processing steps mediated by the RNaseIII enzymes Drosha and Dicer (Kim, 2005) (Figure 1). These processing events rely heavily on proper folding of the precursor RNAs into a stem-loop structure (Han, et al., 2006). Variants in the pri-microRNA and/or pre-microRNA could alter secondary structure and inhibit or enhance pri-microRNA processing (Figure 3).
Patients with chronic lymphocytic leukemia (CLL) frequently have homozygous deletions at chromosome 13q13.4 (Calin, et al., 2002). This genomic locus encodes a polycistronic transcript from which, miR-15a and miR-16-1 are processed (Calin, et al., 2002; Lagos-Quintana, et al., 2001). These microRNAs are dynamically expressed during the cell cycle (Rissland, Hong, & Bartel, 2011) and target genes involved in regulating cell cycle progression (Linsley et al., 2007; Liu et al., 2008). They have also been shown to target genes involved in apoptosis and function as tumor suppressors (Bonci et al., 2008; Cimmino et al., 2005). In 2005, Carlo Croce’s group identified a homozygous C>T SNP coincident with the pri-miR-16-1 locus in two CLL patients with intact 13q13.4 (Calin et al., 2005). This SNP associated with decreased miR-16 expression in CLL cell lines derived from patients and inhibited pri-miR-16 processing in vitro (Calin, et al., 2005). Moreover, this SNP is associated with CLL-like disease in mice (Raveche et al., 2007), highlighting the importance of these microRNAs in tumorigenesis.
The miR-146 family, comprised of miR-146a and miR-146b, is transcriptionally activated in THP1 cells in response to LPS stimulation in an NF-κB-dependent manner (Taganov, Boldin, Chang, & Baltimore, 2006). NF-κB-mediated upregulation and ectopic expression of miR-146a/b inhibits migration and invasion of breast (Bhaumik et al., 2008; Hurst et al., 2009) and pancreatic (Li et al., 2010) cancer cell lines. Inhibition of migration and invasion attributed to targeting of IRAK-1 leading to subsequent inhibition of NF-κB by miR-146a in a negative feedback loop (Bhaumik, et al., 2008; Hurst, et al., 2009). A G>C SNP (rs2910164) in the pre-miR-146a was identified by associating with an increased risk of papillary thyroid carcinoma (Jazdzewski et al., 2008). Subsequent analysis showed association of the rs2910164 SNP with hepatocellular carcinoma (T. Xu et al., 2008), prostate cancer (B. Xu et al., 2010), and esophageal squamous cell carcinoma (Guo et al., 2010) in Han Chinese individuals. In a recent study the rs2910164 SNP was shown to associate with a decreased risk of bladder cancer and reduced risk of recurrence (M. Wang et al., 2012). Functional analysis established that the G>C alteration attenuates Drosha-mediated processing resulting in reduced miR-146a expression (Jazdzewski, et al., 2008). Interestingly, several reports have linked decreased miR-146a expression to androgen-independent prostate cancer (Lin, Chiang, Chang, & Ying, 2008; B. Xu et al.). The rs2910164 SNP is associated with hormone-refractory prostate cancer (B. Xu, et al., 2010). Therefore, it is plausible that miR-146a governs prostate cancer biology. However, in breast (Hurst, et al., 2009) and bladder (M. Wang, et al., 2012) cancer cell lines the rs2910164 SNP leads to upregulation of miR-146a expression. The discrepancy in data regarding the affect of the rs2910164 SNP on miR-146a expression highlights the ability of SNPs to dynamically alter microRNA expression in a tissue specific manner.
A homozygous T>C SNP (rs11614914) in pre-miR-196a-2 is associated with an increased risk of lung (Tian et al., 2009), breast (Hoffman et al., 2009) and gastric cancer (Peng et al., 2010) in Chinese populations, whereas, in Caucasian populations where the allele frequency is reversed, a C>T homozygous variant of the rs11614914 SNP associated with an increased risk of oesophageal cancer in non-smokers (Ye et al., 2008). While the T>C variant reduces the expression of miR-196a-2 (Hoffman, et al., 2009), the target genes sufficient to drive tumorigenesis are unknown and it is possible that this microRNA can function as both a tumor suppressor and oncogene. Homozygous rs11614914 T>C variants were associated with poor survival in patients diagnosed with non-small cell lung cancer indicating the importance of these SNPs as possible diagnostic markers for cancer prognosis (Hu et al., 2008). Taken together these observations indicate that SNPs in pre-microRNA regions can play a dynamic role in microRNA processing and cancer biology.
3.4 SNPs in microRNA target sites
Sequence complementarity is a major determinant for microRNA-target recognition (Bartel, 2009). Therefore, SNPs in mRNAs can alter microRNA binding by either creating a new site or destroying an existing target site (Figure 4). Computational analysis indicates that there are approximately 20,000 SNPs coincident with conserved human microRNA target sites with putative functionality (Chen & Rajewsky, 2006). Michel George’s group was the first to report that a 3′ UTR SNP could create an aberrant microRNA target site. They showed that a homozygous SNP in the 3′ UTR of myostatin caused muscular hypertrophy in Texel sheep (Clop et al., 2006). The G>A SNP creates an aberrant target site for miR-1 and miR-206, which are highly expressed in skeletal muscle and specifically target the variant allele (Clop, et al., 2006). Whereas, Matthew State’s group showed that a Tourette’s syndrome associated SNP in the 3′ UTR of SLITRK1 destroyed a target site for miR-189, and was the first evidence that a 3′ UTR SNP could inhibit microRNA binding (Abelson et al., 2005). Since these seminal observations, microRNA binding site SNPs have been identified in cancer and appear to function as biomarkers for disease risk, treatment response and outcome.
Let-7 is a tumor suppressor microRNA that regulates the expression of the KRAS (Johnson et al., 2005), MYC (Kumar, Lu, Mercer, Golub, & Jacks, 2007) and HMGA2 (Y. S. Lee & Dutta, 2007; Mayr, Hemann, & Bartel, 2007) oncogenes. Let-7 expression is frequently down regulated in many types of cancer and this is associated with poor prognosis in lung cancer (Karube et al., 2005; Takamizawa et al., 2004). A heterozygous T>G SNP (rs61764370) in the 3′ UTR of KRAS associates with an increased risk for non-small cell lung carcinoma in 2 case-controlled studies (Chin et al., 2008), as well as, increased risk for ovarian cancer (E. Ratner et al.), triple negative breast cancer (Paranjape et al.), melanoma (Chan et al.), and hereditary breast and ovarian cancer (Pilarski et al.). The T>G variant is coincident with a let-7 target site in the 3′ UTR of KRAS and attenuates let-7-mediated suppression (Chin, et al., 2008), resulting in KRAS overexpression.
Furthermore, the rs61764370 SNP was recently shown to associate with resistance to platinum-based therapy in ovarian cancer and increased cancer specific death in these patients (E. S. Ratner et al., 2011). Additionally, evidence from Graziano et al., indicated that metastatic colorectal cancer patients with the rs61764370 SNP undergoing salvage cetuximab-irinotecan therapy displayed chemotherapy resistance, with poor overall survival and progression-free survival (Graziano et al., 2010). These findings suggest that this germline, non-coding sequence variant in the KRAS 3′ UTR phenocopies somatic, activating (gain-of-function) KRAS mutations found in the open-reading-frame in treatment response. This data highlights the potential utility for the rs61764370 SNP as a companion diagnostic in the clinic.
While the rs61764370 SNP was identified by direct sequencing of the KRAS 3′ UTR, other groups have utilized in silico analysis to identify candidate SNPs in microRNA target sites for genotype-phenotype correlations (Nicoloso et al., 2010; Sethupathy, Giang, Plotkin, & Hannenhalli, 2008). Analysis of microRNA target site SNPs in genes associated with the DNA damage repair pathway demonstrated that a heterozygous T>C SNP (rs8679) in the PARP1 3′ UTR associated with increased risk for developing bladder cancer, but not breast cancer (Teo et al., 2012). This SNP is coincident with several predicted microRNA target sites in the PARP1 3′ UTR, in particular miR-145, which is frequently down-regulated in bladder cancer (Ichimi et al., 2009). It is possible that the rs8679 SNP in combination with reduced miR-145 expression contribute to increased bladder cancer risk.
In this same report a heterozygous A>G SNP (rs7180135) in the RAD51 3′ UTR associated with a favorable response (improved 5-year cancer specific survival) to radiation therapy in muscle-invasive bladder cancer (Teo, et al., 2012). This SNP is predicted to be coincident with a miR-197 target site and disrupt microRNA targeting. Interestingly, miR-197 is downregulated in cells following exposure to ionizing radiation (Weidhaas, et al., 2007). It is possible that the rs7180135 SNP and downregulation of miR-197 following IR therapy work synergistically to enhance the cellular DNA damage response, resulting in increased survival. These reports indicate that microRNA target site SNPs can function similarly to protein-coding mutations that associate with not only disease risk, but treatment response and outcome as well.
4. Looking into the future: utilizing SNPs as companion diagnostics
There is sufficient proof-of-principle evidence that microRNA SNPs can play a critical role in predicting cancer risk, treatment response and outcome. Understanding the factors that contribute to cancer risk can be a powerful future tool for clinicians and genetic counselors, as well as in advancing our understanding of cancer biology. If a risk allele is identified clinicians could advise patients to begin earlier, more frequent and intensive screening or even stronger preventative measures, in hopes of preventing disease or catching it at an earlier and more treatable stage. More interestingly, as microRNAs are stimulated by external stimuli, it may also be possible to manage patients with such SNPs by modifying lifestyle factors to maintain homeostasis of their inherited differences. This is an avenue of active research that may prove most promising.
While assessing an individual’s risk can be a useful tool to catch cancer at an earlier time, the question regarding what is the best treatment for individual cancer patients still remains. There is mounting evidence that microRNA SNPs can predict treatment response and outcome. For example, the miR-34 family protects cells against cytotoxic therapy (Kato, et al., 2009). Therefore, the miR-34b/c promoter SNP (rs4938723) that inhibits miR-34 expression (Y. Xu, et al., 2011), could be hypothetically utilized as a companion diagnostic with treatment. The KRAS 3′ UTR SNP (rs61764370) associates with poor response to platinum-based therapy in ovarian cancer (E. S. Ratner, et al., 2011) and cetuximab-irinotecan treatment in colorectal cancer patients (Graziano, et al., 2010). Consistent with these findings patients harboring the SNP displayed poor outcome and poor overall survival. These results indicate the potential utilization of microRNA-associated SNPs as companion diagnostics. Application of these SNPs into treatment decisions will require further confirmation in prospective randomized trials, yet the evidence for their potential as a new class of inherited markers that could bring clinicians one-step closer to providing tailored/personalized care for the treatment of cancer is already very promising.
Acknowledgments
This work was supported by research grants from the NIH (1 R01 CA157749-01A1) and the Kalimeris Fund to Yale Therapeutic Radiology Clinical Investigators (to JBW).
Abbreviations
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- miRNA
microRNA
- SNP
single nucleotide polymorphism
- 5′ UTR
5′ untranslated region
- 3′ UTR
3′ untranslated region
- C. elegans
Caenorhabditis elegans
- DGCR8
DiGeorge syndrome critical region 8 (gene)
- Ago
Argonuate
- SLITRK1
SLIT and NTRK-Like family member 1 (gene)
- KRAS
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (gene)
- HMGA2
high-mobility group AT-hook 2 (gene)
- PARP1
Poly [ADP-ribose] polymerase 1 (gene)
- HCC
hepatocellular carcinoma
- CLL
chronic lymphocytic leukemia
- BCL2
B-cell CLL/lymphoma 2 (gene)
- BRCA1
breast cancer 1, early onset (gene)
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells (gene)
- THP1
Human acute monocytic leukemia cell line
Footnotes
5. Conflict of Interest Statement
Dr. Weidhaas has patented IP surrounding SNP rs61764370 through Yale University, and has co-founded a company that has licensed this IP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310(5746):317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower PE, Verducci JS, Lin S, Zhou J, Chung JH, Dai Z, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6(5):1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Buonocore F, Hill MJ, Campbell CD, Oladimeji PB, Jeffries AR, Troakes C, et al. Effects of cis-regulatory variation differ across regions of the adult human brain. Hum Mol Genet. 2010;19(22):4490–4496. doi: 10.1093/hmg/ddq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16(21):2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Chan E, Patel R, Nallur S, Ratner E, Bacchiocchi A, Hoyt K, et al. MicroRNA signatures differentiate melanoma subtypes. Cell Cycle. 2011;10(11) doi: 10.4161/cc.10.11.15777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38(12):1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68(20):8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4(7):e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38(7):813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36(3):445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325(5945):1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1(1):45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135(7):1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Felice KM, Salzman DW, Shubert-Coleman J, Jensen KP, Furneaux HM. The 5′ terminal uracil of let-7a is critical for the recruitment of mRNA to Argonaute2. Biochem J. 2009;422(2):329–341. doi: 10.1042/BJ20090534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano F, Canestrari E, Loupakis F, Ruzzo A, Galluccio N, Santini D, et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacogenomics J. 2010;10(5):458–464. doi: 10.1038/tpj.2010.9. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Wang K, Xiong G, Hu H, Wang D, Xu X, et al. A functional varient in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han. Fam Cancer. 2010;9(4):599–603. doi: 10.1007/s10689-010-9370-5. [DOI] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16(1):4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12(5):414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29(9):462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69(14):5970–5977. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118(7):2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69(4):1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125(2):345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28(25):2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen Regulation of MicroRNA Expression. Curr Genomics. 2009;10(3):169–183. doi: 10.2174/138920209788185289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. Rna. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li Y, Vandenboom TG, 2nd, Wang Z, Kong D, Ali S, Philip PA, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70(4):1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. Rna. 2008;14(3):417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27(6):2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36(16):5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311(5758):195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18(9):975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70(7):2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36(3):431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10(21):2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12(4):377–386. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Ge B, Hudson TJ. Influence of human genome polymorphism on gene expression. Hum Mol Genet. 2006;15(Spec No 1):R9–16. doi: 10.1093/hmg/ddl044. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306(5696):647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Peng S, Kuang Z, Sheng C, Zhang Y, Xu H, Cheng Q. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55(8):2288–2293. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26(5):611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Pilarski R, Patel DA, Weitzel J, McVeigh T, Dorairaj JJ, Heneghan HM, et al. The KRAS-variant is associated with risk of developing double primary breast and ovarian cancer. PLoS One. 2012;7(5):e37891. doi: 10.1371/journal.pone.0037891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309(5740):1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70(16):6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner ES, Keane FK, Lindner R, Tassi RA, Paranjape T, Glasgow M, et al. A KRAS variant is a biomarker of poor outcome, platinum chemotherapy resistance and a potential target for therapy in ovarian cancer. Oncogene. 2011 doi: 10.1038/onc.2011.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109(12):5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Hong SJ, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2011;43(6):993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21(9):327–335. [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409(6822):928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. 2007;104(45):17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman DW, Shubert-Coleman J, Furneaux H. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem. 2007;282(45):32773–32779. doi: 10.1074/jbc.M705054200. [DOI] [PubMed] [Google Scholar]
- Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104(9):3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeier S, Schaefer U, MacPherson CR, Bajic VB. dPORE-miRNA: polymorphic regulation of microRNA genes. PLoS One. 2011;6(2):e16657. doi: 10.1371/journal.pone.0016657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106(29):12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Giang H, Plotkin JB, Hannenhalli S. Genome-wide analysis of natural selection on human cis-elements. PLoS One. 2008;3(9):e3137. doi: 10.1371/journal.pone.0003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38(6):789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Teo MT, Landi D, Taylor CF, Elliott F, Vaslin L, Cox DG, et al. The role of microRNA-binding site polymorphisms in DNA repair genes as risk factors for bladder cancer and breast cancer and their impact on radiotherapy outcomes. Carcinogenesis. 2012;33(3):581–586. doi: 10.1093/carcin/bgr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18(10):1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18(2):132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Chu H, Li P, Yuan L, Fu G, Ma L, et al. Genetic variants in microRNAs predict bladder cancer risk and recurrence. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-0688. [DOI] [PubMed] [Google Scholar]
- Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67(23):11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Xu B, Feng NH, Li PC, Tao J, Wu D, Zhang ZD, et al. A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate. 2010;70(5):467–472. doi: 10.1002/pros.21080. [DOI] [PubMed] [Google Scholar]
- Xu B, Wang N, Wang X, Tong N, Shao N, Tao J, et al. MiR-146a suppresses tumor growth and progression by targeting EGFR pathway and in a pERK-dependent manner in castration-resistant prostate cancer. Prostate. 2011;72(11):1171–1178. doi: 10.1002/pros.22466. [DOI] [PubMed] [Google Scholar]
- Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29(11):2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu L, Liu J, Zhang Y, Zhu J, Chen J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer. 2011;128(2):412–417. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1(6):460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32(16):4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhou Y, Yin B, Hao W, Zhao L, Ju W, et al. 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol Rep. 2010;23(1):121–128. [PubMed] [Google Scholar]