Abstract
The present study comprised sarcomeric genotyping of the three most commonly involved sarcomeric genes: MYBPC3, MYH7, and TNNT2 in 192 unrelated Egyptian hypertrophic cardiomyopathy (HCM) index patients. Mutations were detected in 40 % of cases. Presence of positive family history was significantly (p = 0.002) associated with a higher genetic positive yield (49/78, 62.8 %). The majority of the detected mutations in the three sarcomeric genes were novel (40/62, 65 %) and mostly private (47/62, 77 %). Single nucleotide substitution was the most frequently detected mutation type (51/62, 82 %). Over three quarters of these substitutions (21/27, 78 %) involved CpG dinucleotide sites and resulted from C > T or G > A transition in the three analyzed genes, highlighting the significance of CpG high mutability within the sarcomeric genes examined. This study could aid in global comparative studies in different ethnic populations and constitutes an important step in the evolution of the integrated clinical, translational, and basic science HCM program.
Keywords: Sarcomeric genotyping, HCM genetics, Egypt
Background
Evaluation of genetic basis of inherited heart muscle disease is a prerequisite for further population, phenotypic, and mechanistic studies. Hypertrophic cardiomyopathy (HCM) is a model of common genetic heart disease attracting the interest of cardiologists, geneticists, scientists, and the public. Public interest is usually temporarily raised following the diagnosis of HCM as the cause of sudden death of a young competitive athlete [1]. Different population studies reported the prevalence of HCM, based on left ventricular (LV) wall thickness greater than 15 mm, as 1:500 (0.2 %) [2, 3]. However, this frequency represents the tip of the iceberg and is probably an underestimate since it represents only the clinically manifest HCM and does not include the prehypertrophic stage represented by the genotype-positive/phenotype-negative cases [4]. Based on the reported prevalence in different populations, an estimate of at least 160,000 individuals is expected to be affected with HCM within the over 80 million population of Egypt [5].
Although the incidence of this disease appears to be constant in different parts of the world, its phenotypic and genetic features seem to be quite heterogeneous. Laboratory genetic analysis undertaken over the past two decades have detected several hundred mutations in genes encoding proteins within and associated with the sarcomere [6–8].
Why various clinical manifestations are found among those who share the same sarcomeric gene mutation remains unknown [9]. Factors that account for this variability are only beginning to unravel. Diversity of the causal genes and mutations are partly responsible, but modifier genes and environmental factors also play a role in the phenotypic expression of HCM [10, 11]. Extensive and continuous research in different populations with different genetic backgrounds is imperative to understand the heterogeneity of this disease.
Mutation in any of the sarcomeric genes (MYBPC3, MYH7, TNNT2, TPM1, TNNI3, MYL2, MYL3, ACTC, and TTN) is usually identified in approximately half of the genetically screened HCM index cases [12, 13]. However, the proportion of detection of a pathogenic mutation is population-dependent and the reported frequency ranged from >50 % in France [12] and Italy [14] to <30 % in Sweden [15]. The three most commonly involved sarcomeric genes in HCM as reported in several populations in Europe and USA are myosin binding protein C (MYBPC3), β-myosin heavy chain (MYH7), and cardiac troponin T (TNNT2) [6, 16]. It is, therefore, reasonable to consider screening of those three genes as a first line strategy for molecular characterization of HCM patients in Egypt.
HCM diagnosis is usually delayed till complications develop and is, therefore, considered among the neglected diseases particularly in the developing world. Establishment of tertiary centers, which provide state-of-the-art management, can highly benefit these patients and allow establishment of clinical registries for systematized research and better understanding of the pathophysiology of this common genetic cardiovascular disorder [1]. Establishing national programs is invaluable for sharing disease knowledge and creating networks for optimizing HCM care and encouraging research, hence, the initiative of establishing the BA HCM national program in Egypt aiming for clinical and molecular characterization of HCM among Egyptian patients through nationwide collaboration between clinical satellites extending from Alexandria in the north to Aswan in the south and all being linked to molecular genetics laboratory hosted in the Bibliotheca Alexandrina premises.
Subjects and Methods
HCM Patient Population
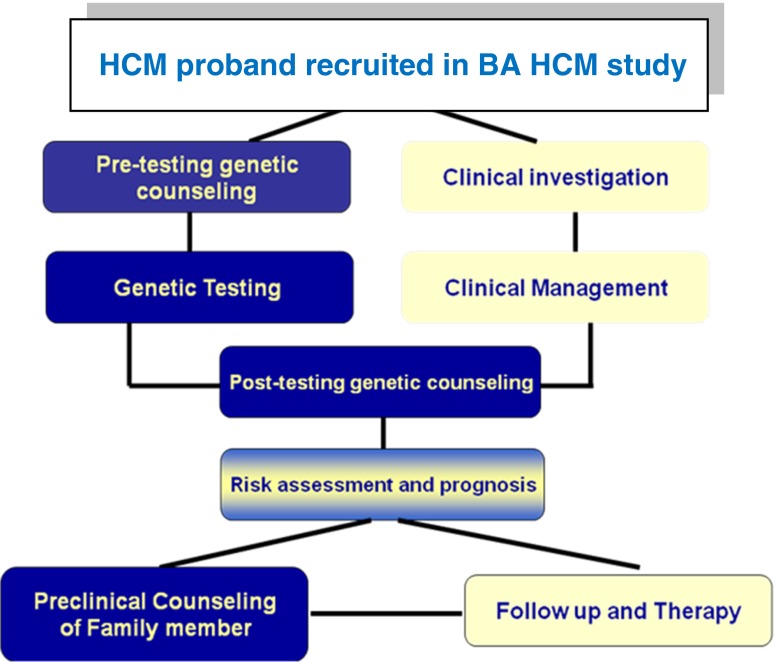
The present study comprised sarcomeric (MYBPC3, MYH7, and TNNT2) genotyping of 199 consecutive unrelated HCM index patients recruited from the different geographic regions of Egypt from July 2007 to June 2010 following standard clinical evaluation by 2D echocardiography at the satellite HCM clinics in Alexandria Faculty of Medicine, Cairo Faculty of Medicine, National Heart Institute, and Aswan Heart Center. The study was approved by the Alexandria Faculty of Medicine and Aswan Heart Center Research ethics committees and informed written consent was obtained from all patients following provision of pretesting genetic counseling. HCM patients' clinical assessment and genetic testing in the BA HCM study is undertaken through a multidisciplinary approach involving genetics and cardiology specialties as illustrated in Fig. 1.
Fig. 1.
Multidisciplinary approach involving genetics and cardiology in management of HCM patients and their families in BA HCM program
The diagnosis of HCM was based on the two-dimensional echocardiographic evidence of a hypertrophied, nondilated left ventricle (wall thickness ≥ 15 mm) in the absence of any other cardiac or systemic disease, including hypertension, capable of producing the magnitude of hypertrophy evident. The basic clinical and echocardiographic data was extracted from available patient clinical records and genotyping was undertaken blinded of patient clinical data. Detailed clinical characterization of the current patient cohort is beyond the scope of the current manuscript.
At least three generations of family pedigrees were constructed and cases were considered familial if there was a positive family history of clinically diagnosed HCM or considered suggestive to be familial through presence of family history of sudden cardiac death particularly if occurring at a young age <40 years. Also, history of consanguinity was always reported whenever present.
Molecular Genetic Analysis
Genomic DNA was extracted from 4 to 6 ml of blood samples donated from index HCM patients following written informed consent and was amplified for the coding exons and flanking intronic sequences of the three candidate sarcomeric genes: MYH7 (NM_000257.2, 38 coding exons were analyzed: third to 40th exons), MYBPC3 (NM_000256.3, 34 exons, first to 34th), and TNNT2 (NM_001001430.1, 16 exons, second to 17th). Primers sequences are available upon request.
Initial screening for mutations was undertaken using heteroduplex analysis by denaturing high-performance liquid chromatography (DHPLC) using WAVE™ DNA Fragment Analysis System. The conditions for DHPLC were developed on the basis of amplicon-specific melting profiles predicted by the Navigator software (Transgenomics, San Jose, CA, USA). Two melting temperatures were analyzed for most of the exons (54/88 = 61 %) and a single temperature for 24 (26 %) and three temperatures for 12 (13 %) amplicons. DHPLC sensitivity is reported as 95 % as compared to each amplicon-automatic sequencing [14].
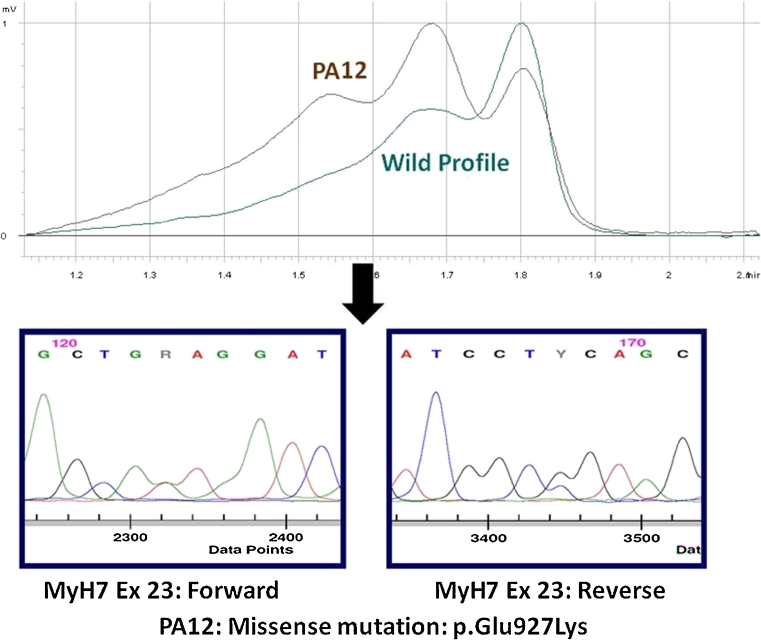
Samples showing a variant profile (different from the wild pattern) for any of the amplicons were subjected to bidirectional sequencing using automated dye terminator cycle–capillary electrophoresis (ABI 3500 Applied Biosystems, Foster City, CA, USA) to determine the nature of the sequence change (Fig. 2).
Fig. 2.
Strategy of genetic analysis in BA HCM study through initial screening with DHPLC (WAVE™ Transgenomics) followed by bidirectional automated sequencing of variant profile
Pathogenicity of variants detected in the present study was dependent on being previously reported to relate to HCM. For novel variants, pathogenicity was evaluated on several criteria including: absence in at least 200 unrelated chromosomes from age- and sex-matched healthy controls of the same ethnicity, cosegregation of the mutation with the disease phenotype in the proband's family, and the conservation of the mutated residue in different species in addition to pathogenicity prediction using in silico biosoftware analysis.
In Silico Analysis of Variants Using Alamut-2.0.2 Biosoftware
Predicting Damaging Effect of Amino Acid Substitution
Prediction of novel missense mutation pathogenicity was undertaken through analyzing the physicochemical properties of the substituted amino acid relative to the wild one through licensed online predictive Alamut-2 Biosoftware (http://www.interactive-biosoftware.com).
Splice Site Error Predictions
Substitutions within introns flanking the coding exons of the three sarcomeric genes (MYBPC3, MYH7, and TNNT2) were firstly excluded as being reported as single nucleotide polymorphisms (SNPs) and were also tested in at least 200 alleles of healthy subjects (Appendix). The variants that were not detected in controls were subjected to in silico analysis for prediction of possible splice site alteration using online programs such as splicesitefinder (SSF) and Human Splicing Finder splice site analysis (HSF). The variants with a predicted possible splice site effect were considered as “variants of uncertain significance” and were checked whenever possible for cosegregation within the family with HCM phenotype to determine their potential for pathogenicity.
Results
Study Population
One hundred and ninety-nine patients were consecutively enrolled in the present study. The patient cohort comprised: 192 clinically diagnosed HCM patients, a single patient had LV noncompaction cardiomyopathy without hypertrophy (P76), and six patients were clinically suspected as HCM phenocopies: five patients were suspected to be infiltrative due to storage disease based on symmetrical hypertrophy and highly reflective myocardium (data not shown) and/or presence of history of parental consanguinity and presence of horizontal transmission pattern and one patient was suspected to have mitochondrial cardiomyopathy due to associated bilateral optic neuritis. Another patient was clinically suspected of LEOPARD syndrome due to associated pigmented cutaneous lesions. The fact that no further biochemical analysis was undertaken at the time of sarcomeric genetic testing, it was decided to include all enrolled patients to the three sarcomeric gene analyses. All suspected HCM phenocopies were negative for mutations in the three sarcomeric genes analyzed in the present study and were, therefore, excluded in further frequency estimation and statistical analysis.
Demographics of the 192 Egyptian HCM patients are shown in Table 1. Males were twice as common as females (68 % vs. 32 %), patients' age of presentation ranged from 2 to 70 years, and 53 % of the cases presented at or below 40 years. Cases were considered familial in the presence of positive family history of a diagnosis of HCM detected in 60 cases (60/192, 31 %) or presence of familial incidence of sudden cardiac death (18/192, 9 %). Sarcomeric mutations were detected in 62.8 % (49/78) of familial HCM cases. Summary of the basic parameters measured by 2D echocardiography and wave Doppler are shown in Table 1.
Table 1.
Demographic data of the Egyptian hypertrophic cardiomyopathy patients
| Feature | (%) Nc or mean ± SD |
|---|---|
| Gender | |
| Male | 68 % (130/192) |
| Female | 32 % (62/192) |
| Age at diagnosis (in years) | 36.8 ± 16.80 |
| Median (range) | 38 (2–70) |
| ≤40 years | 53.1 % (102/192) |
| >40 years | 45.8 % (88/192) |
| Family history | 40.6 % (78/192) |
| Of diagnosed HCM | 31.2 % (60/192) |
| Of sudden death | 9.4 % (18/192) |
| +vea for sarcomeric mutation/+ve family history | 62.8 % (49/78) |
| Clinical presentation | |
| Shortness of breath (NYHA) | |
| I | 22.6 % (51/192) |
| II | 44.3 % (85/192) |
| III | 22.9 % (44/192) |
| IV | 6.2 % (12/192) |
| Chest pain | 65.6 % (126/192) |
| Palpitation | 66.1 % (127/192) |
| Syncope/presyncope | 52.6 % (101/192) |
| Baseline echocardiographic parameters | |
| Left atrium (mm) | 44.7 ± 9.3 |
| LV outflow gradient ≥ 30 mmHg | 56.7 % (103/185) |
| Max. LV thickness (mm) | 25.3 ± 19.7 |
| <30 mm | 80.9 % (149/184) |
| ≥30 mm | 19.0 % (35/184) |
| Symmetrical hypertrophy | 1.1 % (2/185) |
| Asymmetrical hypertrophy | 98.9 % (183/185) |
| Site of maximum thickness: | |
| Isolated septal hypertrophy | 56.2 % (104/185) |
| Septal and posterior wall hypertrophy | 28.7 % (53/185) |
| Apical hypertrophy | 2.7 % (5/185) |
| LV end-diastolic dimension (mm) | 44.2 ± 8.3 |
| LV end-systolic dimension (mm) | 26.6 ± 6.5 |
| Mitral regurgitation (MR): | 68 % (125/185) |
| Trivial/mild MR | 47 % (87/185) |
| Moderate MR | 14 % (26/185) |
| Severe MR | 7 % (12/185) |
| SAM | 62 % (114/185) |
| Major cardiac eventsb | |
| Sudden cardiac death | 2 % (2/100) |
| Previous cardiac arrest | 2 % (2/100) |
| Heart failure | 3 % (3/100) |
| Stroke | 1 % (2/100) |
| Interventions for obstruction/symptoms | |
| Alcohol septal ablation | 1 % (2/192) |
| Surgical septal myectomy | 27 % (52/192) |
| ICD | 1 % (2/192) |
| Pacemaker | 3 % (5/192) |
ICD implantable cardioverter-defibrillator, LV left ventricular, NYHA New York Heart Association, SAM systolic anterior motion
a+ve: positive
bTotal number includes cases with available follow-up data
cTotal number varies according to available clinical data of parameters assessed
Molecular Analysis
Prevalence of Mutations in MYBPC3, MYH7, and TNNT2 Sarcomeric Genes
Seventy-seven index HCM patients harbored one or more mutation in any of the three analyzed sarcomeric genes (77/192, 40 %) (Tables 2 and 3). Among the genopositive patients, the highest frequency of mutations was detected in MYBPC3 (48/77, ~60 %), followed by MYH7 (24/77, ~30 %), and the least involved was TNNT2 (8/77, ~10 %). Details of mutations detected are shown in Table 2 and potential splice site mutations predicted by in silico analysis are shown in (Table 3).
Table 2.
Mutational spectrum in MYBPC3, MYH7, and TNNT2 in Egyptian HCM cohort
| Genea | Ex | Mutation name In protein level | Mutation name in coding DNA level (cDNA) | Mutation type | Domain | Mode of mutation in CpG site | No. of patient (IDs) | Novelty N = novel, R = reported [Ref] (allele freq −EVS)d |
|---|---|---|---|---|---|---|---|---|
| MYBPC3 | 2 | Val94Ala | c.281T > C | Missense | C0 (cardiac-specific region) | – | 1 (P23) | N |
| MYBPC3 | 3 | Pro102Leu | c.305C > T | Missense | Binding site to cardiac actin | – | 2 (P19, PA69) | N |
| MYBPC3 | 4 | Ser139X | c.416C > G | Nonsense | Binding site to cardiac actin | – | 3 (P46, PA15, PU3) | N |
| MYBPC3 | 5 | Arg177His | c.530G > A | Missense | C1 (Ig-like C2-type 1) | CG > A | 1 (P62) | (0.003) |
| MYBPC3 | 5 | Ala179GlnfsX59 | c.534_541del | Frameshift | C1 (Ig-like C2-type 1) | NA | 3 (PA25, PA52, PA46) | N |
| MYBPC3 | 5 | Trp196X | c.587G > A | Nonsense | C1 (Ig-like C2-type 1) | – | 1 (PA19) | N |
| MYBPC3 | 5 | Trp196X | c.588G > A | Nonsense | C1 (Ig-like C2-type 1) | – | 1 (PA18) | N |
| MYBPC3 | 5 | Ala216Thr | c.646G > A | Missense | C1 (Ig-like C2-type 1) | CG > A | 1 (PA64) | R [17] |
| MYBPC3 | 6 | Glu258Lys | c.772G > A | Missense | MYBP-C motif (phosphorylation site) | CG > A | 2 (PA59, PU10) | R [18] |
| MYBPC3 | 9 | Ser296ThrfsX4 | c.887del(G) | Frameshift | MYBP-C motif (phosphorylation site) | NA | 1 (PA90) | N |
| MYBPC3 | 12 | Glu319Ala | c.956A > C | Missense | MYBP-C motif (phosphorylation site) | – | 1 (PA33) | N |
| MYBPC3 | 12 | Tyr333X | c.999C > A | Nonsense | MYBP-C motif (phosphorylation site) | A < CGc | 1 (P69) | N |
| MYBPC3 | 15 | Cys436X | c.1308C > A | Nonsense | C2 (Ig-like C2-type 2) | A < CGc | 1 (PA60) | N |
| MYBPC3 | 15 | Glu441Lys | c.1321G > A | Missense | C2 (Ig-like C2-type 2) | CG > A | 4 (PA38, PA63, PU12, PA123) | R [19] |
| MYBPC3 | 15 | Thr445Met | c.1334C > T | Missense | C2 (Ig-like C2-type 2) | T < CG | 1 (PA53) | N |
| MYBPC3 | 16 | Arg470Pro | c.1409G > C | Missense | C3 (Ig-like C2-type 3) | CG > Cc | 1 (P55) | N |
| MYBPC3 | 16 | Phe473_Glu474del | c.1418_1423del | In-frame | C3 (Ig-like C2-type 3) | NA | 1 (PA80) | N |
| MYBPC3 | 17 | Asp506ThrfsX7 | c.1516del (G) | Frameshift | C3 (Ig-like C2-type 3) | NA | 3 (P70, P62, P63) | N |
| MYBPC3 | 17 | Gly507Arg | c.1519G > A | Missense | C3 (Ig-like C2-type 3) | CG > A | 1 (PA53) | R [20] |
| MYBPC3 | 17 | Asn515Asp | c.1543A > G | Missense | C3 (Ig-like C2-type 3) | – | 1 (PA48) | N |
| MYBPC3 | 18 | Ala558lysfsX9 | c.1672_1673del | Frameshift | C4 (Ig-like C2-type 4) | NA | 1 (P48) | N |
| MYBPC3 | 19 | Glu619Lys | c.1855G > A | Missense | C4 (Ig-like C2-type 4) | CG > A | 2 (P48 ,P76) | R [21] |
| MYBPC3 | 23 | Ile769ThrfsX53 | c.2306del(T) | Frameshift | C5 (Ig-like C2-type 5) | NA | 1 (PA32) | N |
| MYBPC3 | 24 | Val771Met | c.2311G > A | Missense | C6 (fibronectin type-III 1) | CG > A | 1 (PA26) | R [22] |
| MYBPC3 | 25 | Glu832Gly | c.2495A > G | Missense | C6 (fibronectin type-III 1) | – | 2 (P4, P92) | N |
| MYBPC3 | 25 | Arg845Pro | c.2534G > C | Missense | C6 (fibronectin type-III 1) | CG > Cc | 2 (P4, P92) | N |
| MYBPC3 | 27 | Arg943X | c.2827C > T | Nonsense | C7 (fibronectin type-III 2) | T < CG | 1 (PA87) | R [23] |
| MYBPC3 | 28 | Leu993Phe | c.2977C > T | Missense | C8 (Ig-like C2-type 6) | – | 1 (PA16) | N |
| MYBPC3 | 30 | Trp1098X | c.3293G > A | Nonsense | C9 (fibronectin type-III 3) | – | 4 (PA6, PA42, PA56,PA93) | R [24] |
| MYBPC3 | 31 | Arg1138Cys | c.3412C > T | Missense | C9 (fibronectin type-III 3) | T < CG | 1 (PU11) | (0.0002) |
| MYBPC3 | 32 | Glu1179Lys | c.3535G > A | Missense | Connection between C9 and C10 | CG > A | 1 (PA51) | R [25] |
| MYBPC3 | 33 | Glu1239del | c.3715_3717del | In-frame | C10 (Ig-like C2-type 7) | NA | 1 (P39) | N |
| MYBPC3 | 33 | Thr1256AsnfsX10 | c.3766dup.(A) | Frameshift | C10 (Ig-like C2-type 7) | NA | 1 (P36) | N |
| MYH7 | 6 | Glu170Lys | c.508G > A | Missense | Myosin head | – | 1 (P28) | N |
| MYH7 | 9 | Arg249Gln | c.746G > A | Missense | Myosin head | CG > A | 1 (PA55) | R [26] |
| MYH7 | 10 | Leu267Val | c.799C > G | Missense | Myosin head | – | 1 (PA26) | N |
| MYH7 | 11 | Asp309Asn | c.925G > A | Missense | Myosin head | CG > A | 1 (PA8) | N |
| MYH7 | 12 | Glu379Lys | c.1135G > A | Missense | Myosin head | – | 1 (P26) | N |
| MYH7 | 13 | Asp394Glu | c.1182C > A | Missense | Myosin head | – | 1 (PA54) | N |
| MYH7 | 14 | Arg453Cys | c.1357C > T | Missense | Myosin head | T < CG | 1 (P79) | R [27] |
| MYH7 | 15 | Asn471Ser | c.1412A > G | Missense | Myosin head | – | 1 (PA58) | N |
| MYH7 | 15 | Gln498Arg | c.1493A > G | Missense | Myosin head | – | 1 (PA13) | N |
| MYH7 | 18 | Arg663Cys | c.1987C > T | Missense | Myosin head | T < CG | 1 (P8) | R [28] |
| MYH7 | 19 | Gly716Ala | c.2147G > C | Missense | Myosin head | – | 1 (PA30) | N |
| MYH7 | 19 | Arg719Gln | c.2156G > A | Missense | Myosin head | CG > A | 2 (PA4, P67) | R [29] |
| MYH7 | 20 | Pro731Leu | c.2192C > T | Missense | Myosin head | – | 1 (P29) | R [30] |
| MYH7 | 21 | Ala797Thr | c.2389G > A | Missense | IQ domain | CG > A | 1 (P16) | R [31] |
| MYH7 | 22 | Arg819Gln | c.2456G > A | Missense | Connection between head and coiled coil | CG > A | 1 (P73) | N |
| MYH7 | 22 | Lys847del | c.2539_2541del | In-frame | Myosin coiled coil | NA | 1 (P53) | R [13] |
| MYH7 | 23 | Glu927Lys | c.2779G > A | Missense | Myosin coiled coil | – | 1 (PA12) | R [32] |
| MYH7 | 23 | Met932Lys | c.2795T > A | Missense | Myosin coiled coil | – | 1 (P56) | R [33] |
| MYH7 | 25 | Glu1056Asp | c.3168G > C | Missense | Myosin coiled coil | – | 1 (PA57) | N |
| MYH7 | 35 | Arg1662His | c.4985G > A | Missense | Myosin coiled coil | CG > A | 1 (PU1) | (0.00008) |
| MYH7 | 39 | Ser1924AlafsX9 | c.5769del (G) | Frameshift | Myosin coiled coil | NA | 3 (P37, PA24, P85) | N |
| TNNT2 | 10 | Arg92Trp | c.274C > T | Missense | Troponin T chain | T < CG | 1 (P45) | R [34] |
| TNNT2 | 10 | Arg92Gln | c.275G > A | Missense | Troponin T chain | CG > A | 2 (P34, P98) | R [35] |
| TNNT2 | 11 | Asn142Tyr | c.424A > T | Missense | Troponin T chain | – | 1 (P22) | N |
| TNNT2 | 16 | Asn262Asp | c.784A > G | Missense | Troponin T chain | – | 1 (PA89) | N |
| TNNT2 | 16 | Asn269Lys | c.807C > A | Missense | Troponin T chain | A < CGc | 2 (P47, PA66) | N |
| TNNT2 | 17 | Arg278Leu | c.833G > T | Missense | Troponin T chain | CG > Tc | 1 (PU7) | N |
Caps and italics are names of genes according to HUGO nomenclature of genes
aGenBank reference: MYBPC3:NM_000256.3; MYH7: NM_000257.2; TNNT2: NM_001001430.1
bUniprot reference: cardiac myosin-binding protein C: Q14896; myosin heavy chain 7: P12883; cardiac troponin T: P45379; IQ domain: calmodulin binding domain: http://www.uniprot.org/uniprot/Q14896,P12883,P45379
cCpG substitution by a mutagenic mutation other than deamination of methylated cytosine to thymine
dAllele frequency obtained from Exome Variant Server (EVS) of Exome Sequencing Project: http://evs.gs.washington.edu/EVS/) [36]. Mutations in bold were reported in EVS
Table 3.
Possible splice site error associated with novel intronic substitution
| Gene | Intron | Mutation name according to coding DNA level (cDNA) | No. of patients (IDs) | Cosegregation | Detection in Egyptian controls (200 alleles) or reported in EVSa (allele freq) 1 = present 0 = Absent |
|---|---|---|---|---|---|
| MYBPC3 | 3 | c.407 − 7C > A | 2 (P14, P16) | Absent (P14′a) | 0 |
| MYBPC3 | 7 | c.821 + 3G > T | 1 (PU1) | Yes (PU1′A) | 0 |
| MYBPC3 | 16 | c.1458 − 7C > A | 1 (P89) | TBP* | 0 |
| MYBPC3 | 16 | c.1458 − 17C > G | 1 (P17) | TBP* | 0 |
| MYBPC3 | 22 | c.2149 − 8C > T | 1 (P100) | TBP* | 0 |
| MYBPC3 | 32 | c.3627 + 2del | 1 (PU2) | NT | 0 |
| TNNT2 | 14 | c.690 − 4G > T | 1 (P32) | TBP* | 1 (0.0002) a |
| TNNT2 | 15 | c.781 − 48_64del | 1 (PA78) | TBP* | 0 |
MYBPC3 NM_000256.3, MYH7 NM_000257.2, TNNT2 NM_001001430.1, NT not tested, TBP* To Be Pursued by family clinical screening to test cosegregation in affected family members
aAllele frequency obtained from Exome Variant Server (EVS) of Exome Sequencing Project (ESP): http://evs.gs.washington.edu/EVS/) [36]. Mutations in bold were reported in EVS
Missense mutation was the most commonly encountered mutation type in the three sarcomeric genes. There were 62 individual mutations detected in any of the three sarcomeric genes in the present cohort, the majority of which (47/62, 77 %) were detected in single index patient (i.e., private) (Tables 2 and 3). Most of the mutations detected in the studied cohort were novel (40/62, 65 %) and were not detected in 200 alleles of age-, sex-, and ethnically matched control series nor in the 1000 Exome Variant Server (EVS) [36]. Three missense mutations detected in our HCM patients (Ala177His, Arg1138 in MYBPC3, and Arg1662His in MYH7) were found in the EVS; however, they were reported at low allele frequencies (0.003, 0.0002, and 0.00008, respectively). Additionally, seven missense mutations previously reported as pathogenic in relevance to HCM and also found in the present cohort (Ala216Thr, Clu441Lys, Gly507Arg, Glu619Lys, Val771Met, Glu1179Lys in MYBPC3, and Ala797Thr in MYH7) were also reported in the EVS. This finding should be interpreted with caution. While on the one hand, these variants might be regarded as rare polymorphic nonpathogenic variants, on the other hand, it could still have a pathogenic and/or modifier effect in a disease with the heterogeneous nature of HCM having later age of onset and reduced penetrance especially when considering controls not thoroughly evaluated by electrocardiogram (ECG) and echo. Further screening in a larger number of ethnic-matched controls with thorough cardiologic evaluation is warranted.
MYBPC3 Mutation Spectrum
MYBPC3 was the most commonly involved in the present cohort (47 patients were MYBPC3-genopositive, 47/192, 24 %). Thirty-five individual mutations were detected in MYBPC3, 18 mutations were missense, seven were nonsense, eight were deletion mutation of which six caused frameshift effect resulting in premature stop codon and two mutations caused in-frame deletion of a single or more amino acids (Table 2).
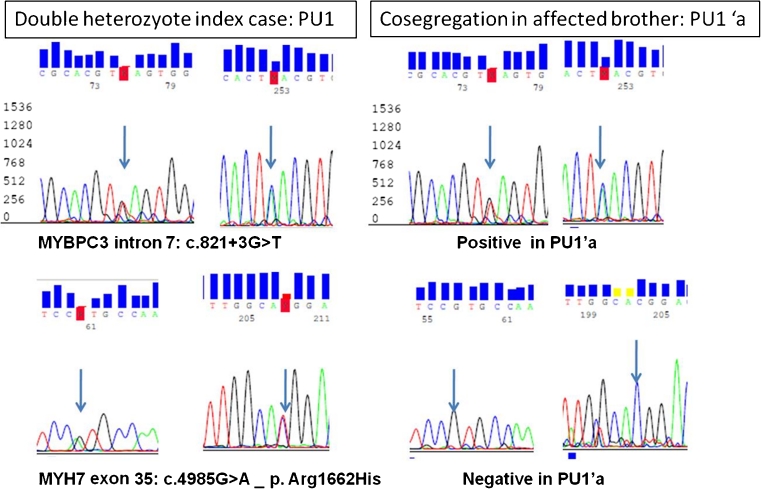
In addition, two mutations were detected in introns 32 and 7, namely, c.3627 + 2delT (PU2) and c.821 + 3G > T (PU1), respectively (Table 3). These variants are likely to result in splice site error since both were absent in 200 chromosomes of unrelated healthy subjects and the former is affecting the conserved 5′ donor splice site dinucleotide GT of intron 32 and the latter (c.821 + 3G > T) was predicted to result in splice error by in silico modeling and was found to cosegregate with the HCM phenotype in a HCM family member (PU1′a), thus, in favor of its potential pathogenicity. In further support of this speculation is the fact that the index patient PU1 was double heterozygote and also carried missense mutation in MYH7 exon39 (p.Arg1662His) that did not cosegregate in the affected family member (PU1′a) (Fig. 3).
Fig. 3.
Bidirectional automated sequencing of MYBPC3 exon7 and MYH7 exon35 and flanking intronic sequences in index patient PU1 and screening of an affected brother (PU1′a) showing cosegregation of MYBPC3 intron 7 substitution mutation: c.821 + 3G > T with potential splice effect as predicted by in silico tools and not MYH7 missense variant (Arg1662His) indicating likely pathogenicity of the MYBPC3 intronic variant
Twenty-five mutations (25/35, 71 %) of MYBPC3 were private (i.e., reported in a single index patient) and ten mutations were found in more than one patient: five mutations were detected in two unrelated index patients, three mutations were individually found in three unrelated patients, and two mutations were detected in four unrelated index patients suggesting a possible founder effect (Table 2).
A single patient had noncompaction LV cardiomyopathy without hypertrophy (P76) and carried missense mutation in MYBPC3 (p.Glu619Lys) that was also detected in another HCM patient in the present cohort and was previously reported in association with HCM [21].
MYH7 Mutation Spectrum
Twenty-one individual mutations of MYH7 were detected in 24 unrelated index patients (24/192, 12.5 %). Of these, 19 were missense, two were deletion mutations, one of which was in-frame deletion resulting in deletion of a single amino acid (Lys847del) and one resulted in frameshift mutation (p.Ser1924AlafsX9). All mutations were private except two that were detected in more than one index patient, Arg719Gln was detected in two and Ser1924AlafsX9 was detected in three unrelated index patients (Table 2).
TNNT2 Mutation Spectrum
TNNT2 was the least commonly involved gene in our cohort (8/192, 4 %). Six missense mutations were detected in TNNT2, most of which were novel (4/6, 67 %) (Table 2). In addition, two missense mutations involving Arg92 codon (Arg92Trp and Arg92Gln) [34, 35] were detected in one and two patients, respectively.
Occurrence of Double Hits
Seven patients in the present cohort (7/192, 4 %) carried double hit mutations in one or more of the analyzed sarcomeric genes. Five were compound heterozygotes within MYBPC3 (P62, PA53, P48, P4, and P92). Two patients were double heterozygotes (PA26 and PU1): PA26 carried missense mutation in MYBPC3 (p.Val771Met) and another in MYH7 (Leu267Val) and PU1 had substitution mutation within intron 7 of MYBPC3 (c.821 + 3G > T) with a predictive splice error effect and a missense mutation in MYH7 (Arg1662His), and only the intronic variant was cosegregating in an affected family member (PU1′).
Possible Founder Effect
Two novel MYBPC3 mutations showed a particular geographic distribution in Egypt indicating a possibility for a founder effect. The frameshift mutation p.Trp1098X in exon30 was detected in three unrelated patients from the northwest Mediterranean coast in Alexandria governorate inhabited by Bedouin tribes. Another frameshift mutation, p.Asp506ThrfsX7 in exon15, was detected in three unrelated index patients from the Nile River Delta region in Damietta governorate.
Interestingly, two unrelated index patients (P4 and P92) were compound heterozygotes for two mutations (p.Glu832Gly and p.Arg845Pro) within exons 25 and 26 of MYBPC3 gene. These two mutations are likely to be in cis as they were cosegregating in an affected family member of P4. This may also suggest a founder effect due to the fact that both mutations were cosegregating in unrelated index patients.
Bioinformatic Analysis of Substitution Mutations for Possible Splice Error Effect
All novel variants detected in intronic sequences flanking the coding exons of the three sarcomeric genes were initially checked through online genomic SNP database (http://snpper.chip.org/). Those which were not determined as a SNP were subjected to in silico analysis for prediction of splice error effect (Table 3).
Apart from the single variant that involved the conserved donor splice site of intron 32 of MYBPC3 (c.3627 + 2delT) in PU2 and was considered as pathogenic, nine intronic novel variants were predicted by in silico analysis to cause a splice site error. Three were determined as not likely to be pathogenic since two of which were detected in the Egyptian control series [c.2149-5C > T(P87) in intron 22 of MYBPC3 (detected in 1 % of the controls) and c.3337-3dup (PA2) in intron 27 of MYH7 (was found in 16 % of the controls series)], and one variant detected in MYBPC3 intron 3 (c.407-7C>A) was not cosegregating with HCM phenotype in an affected relative of index P14.
Cosegregation provides a direct method for excluding that a variant is the sole cause of disease within a family. It was, therefore, always sought whenever possible. In the present study, c.821 + 3G > T in intron 7 of MYBPC3 was cosegregating with HCM phenotype in a family member of index PU1, thus, providing evidence for potential pathogenicity, especially that the other mutation found in PU1 in MYH7 (p.Arg1662His) was not cosegregating in the affected relative (Fig. 3). The missense variant (p.Arg1662His in MYH7) was reported in the EVS with a very low allele frequency: 0.00008 (Table 2).
Three intronic substitutions in MYBPC3 (c.1458-7C > A, c.1458-17C > G, and c.2149-8C > T) and two in TNNT2 (c.690-4G > T and c.781-48_64del) were classified as variants of undetermined clinical significance since they were predicted by in silico analysis to result in splice error (Table 3) and were absent in 200 chromosomes of healthy subjects. It is worth mentioning that variant c. 690-4G > T was reported in EVS at an allele frequency of 0.0002 and that cosegregation was not tested till writing of the current manuscript and should be pursued prior to considering the potential pathogenicity of this variant. It is worth mentioning that these in silico predictions of splice errors should be validated by ribonucleic acid (RNA) analysis.
In Silico Prediction of Novel Missense Mutation Pathogenicity
The novel missense substitutions are initially excluded in at least 200 chromosomes of matched healthy volunteers and are also subjected to in silico bioinformatic analysis for prediction of pathogenicity, which included several scoring systems (Table 4). The Grantham Difference, which depends on the composition, polarity, and molecular volume of the wild and substituted amino acid, shows that the higher the score, the less tolerated the amino acid substitution. Align GVGD combines Grantham difference of amino acids and protein multiple sequence alignments to predict whether the missense substitution falls in a spectrum from enriched deleterious to enriched neutral (http://agvgd.iarc.fr/). Sorting Intolerant from Tolerant (SIFT) depends on the degree of conservation of amino acid residues in sequence alignments derived from closely related sequences in different species (http://sift.jcvi.org). Polymorphism Phenotyping (Polyphen-2) predicts possible impact of an amino acid substitution on the structure and function of a human protein using physical and comparative considerations (http://genetics.bwh.harvarad.edu/pph2/).
Table 4.
Bioinformatic analysis of novel missense mutations associated with hypertrophic cardiomyopathy among Egyptian patients
| Gene/mutation in protein level | Grantham difference (0–225) | SIFTb D = deleterious (score < 0.05) T = tolerated (score > 0.05) | Align GVGD classc | Polyphen-2 (score)d | Likelihood of pathogenicitye |
|---|---|---|---|---|---|
| MYBPC3/Val94Ala | 64 | D (0.01) | C25 | Probably damaging (0.999) | Likely |
| MYBPC3/Pro102Leua | 98 | D (0.04) | C0 | Benign (0.008) | Pathogenic |
| MYBPC3/Arg177His | 29 | D (0.01) | C0 | Possibly damaging (0.889) | Likely |
| MYBPC3/Glu319Ala | 107 | T (0.53) | C0 | Benign (0.014) | Uncertain |
| MYBPC3/Thr445Met | 81 | D (0.00) | C65 | Probably damaging (1.000) | Likely |
| MYBPC3/Arg470Pro | 103 | D (0.00) | C65 | Probably damaging (0.999) | Likely |
| MYBPC3/Asn515Asp | 23 | D (0.00) | C15 | Benign (0.040) | Uncertain |
| MYBPC3/Glu832Glya | 98 | D (0.00) | C65 | Probably damaging (1.000) | Pathogenic |
| MYBPC3/Arg845Proa | 103 | D (0.00) | C65 | Probably damaging (1.000) | Pathogenic |
| MYBPC3/Leu993Phe | 22 | D (0.03) | C0 | Probably damaging (0.983) | Likely |
| MYBPC3/Arg1138Cys | 180 | D (0.00) | C65 | Probably damaging (1.000) | Likely |
| MYH7/Glu170Lys | 56 | D (0.00) | C0 | Probably damaging (0.996) | Likely |
| MYH7/Leu267Val | 32 | D (0.00) | C0 | Benign (0.011) | Uncertain |
| MYH7/Asp309Asn | 23 | D (0.00) | C0 | Probably damaging (0.963) | Uncertain |
| MYH7/Glu379Lys | 56 | D (0.00) | C0 | Benign (0.315) | Uncertain |
| MYH7/Asp394Glu | 45 | D (0.00) | C0 | Probably damaging (1.000) | Likely |
| MYH7/Asn471Ser | 46 | D (0.00) | C45 | Benign (0.019) | Likely |
| MYH7/Gln498Arg | 43 | D (0.00) | C35 | Benign (0.370) | Likely |
| MYH7/Gly716Ala | 60 | D (0.00) | C55 | Possibly damaging (0.948) | Likely |
| MYH7/Arg819Gln | 43 | D (0.00) | C35 | Probably damaging (1.000) | Likely |
| MYH7/Glu1056Asp | 45 | D (0.00) | C0 | Probably damaging (0.997) | Likely |
| MYH7/Arg1662His | 29 | D (0.00) | C0 | Benign (0.001) | Uncertain |
| TNNT2/Asn152Tyr | 143 | T (0.06) | C0 | Possibly damaging (0.951) | Likely |
| TNNT2/Asn272Asp | 23 | T (0.32) | C0 | Possibly damaging (0.820) | Uncertain |
| TNNT2/Asn269Lysa | 94 | T (1.00) | C0 | Benign (0.028) | Pathogenic |
| TNNT2/Arg288Leu | 102 | T (1.00) | C0 | Probably damaging (0.972) | Uncertain |
aDe novo concurrent with HCM in more than a single family
bSorting intolerant from tolerant prediction
cScores include GV = 0 (invariable), 0 < GV < 62 = variable conservative, GV > 62 = variable nonconservative
dPolymorphism phenotyping
eLikelihood of pathogenicity based on in silico analysis and de novo concurrence of variant in unrelated HCM cases
The likelihood of pathogenicity of a novel missense variant is analyzed according to output of the available bioinformatic tools. At least two programs agreeing in predictive output is used to determine pathogenicity. The concurrent occurrence of novel variants of uncertain clinical significance in several unrelated families such as MYBPC3/Pro102Leu (in P19 and PA69), MYBPC3/Glu832Gly and MYBPC3/Arg845Pro (in P4 and P92), and TNNT2/Asn269Lys (in P47 and PA66) in addition to their absence in our control series and in the 1000 exome project (EVS) [36] favored their pathogenic potential.
Uncertain and likely pathogenic variants were always checked for cosegregation whenever possible. Interestingly, the MYH7/Arg1662His detected in a single index patient (PU1) detected as a second hit was determined by in silico analysis as a benign/tolerated variant and was not cosegregating with HCM phenotype in an affected family member, and its being reported in EVS (with an allele frequency of 0.00008) are in favor of its less likelihood of a direct role in pathogenicity of HCM and may have a modifier effect. The de novo concurrence of the double in cis variants of MYBPC3 exon25 (Glu832Gly) and exon26 (Arg845Pro) in two families (P4&P92) were found to cosegregate in the affected family member of P4 favoring the pathogenic effect of one or either of them.
Mutations Involving CpG Dincleotides
All substitution mutations were analyzed in context of the reference GenBank sequence of the three sarcomeric genes. It was observed that over half (27/51, 53 %) of all substitution mutations resulting in missense and nonsense effect occurred in CpG dinucleotides [60 % in MYBPC3 (15/25), 40 % in MYH7 (8/20), and 66.6 % in TNNT2 (4/6)] (Table 2). In relation to mode of nucleotide substitution in the CpG site, it was observed that the most commonly encountered types were the G > A and C > T transitions, 55.6 % (15/27) and 22.2 % (6/27), respectively. On the other hand, the C > A, G > C, and G > T transversion types were detected less frequently, [11.1 % (3/27), 7.4 % (2/27), and 3.7 % (1/27), respectively] (Table 5). The fact that over three quarters of these single nucleotide substitutions involving the CpG sites (21/27, 78 %) resulted from C > T or G > A transition in the three genes [73.3 %(11/15) in MYBPC3, 100 % (8/8) in MYH7, and 50 % (2/4) in TNNT2] highlights the significance of CpG high mutability resulting from deamination of 5-methylcytosine to thymine within the CpG sites in sarcomeric genes causing HCM among Egyptian patients.
Table 5.
Frequency of different substitution modes within CpG in the present HCM cohort
| Mode of substitution in CpG | Frequency in present cohort |
|---|---|
| G > A | 55.6 % (15/27) |
| C > T | 22.2 % (6/27) |
| C > A | 11.1 % (3/27) |
| C > G | 0 % (0/27) |
| G > C | 7.4 % (2/27) |
| G > T | 3.7 % (1/27 ) |
Influence of Genetic Mutation on Clinical Phenotype
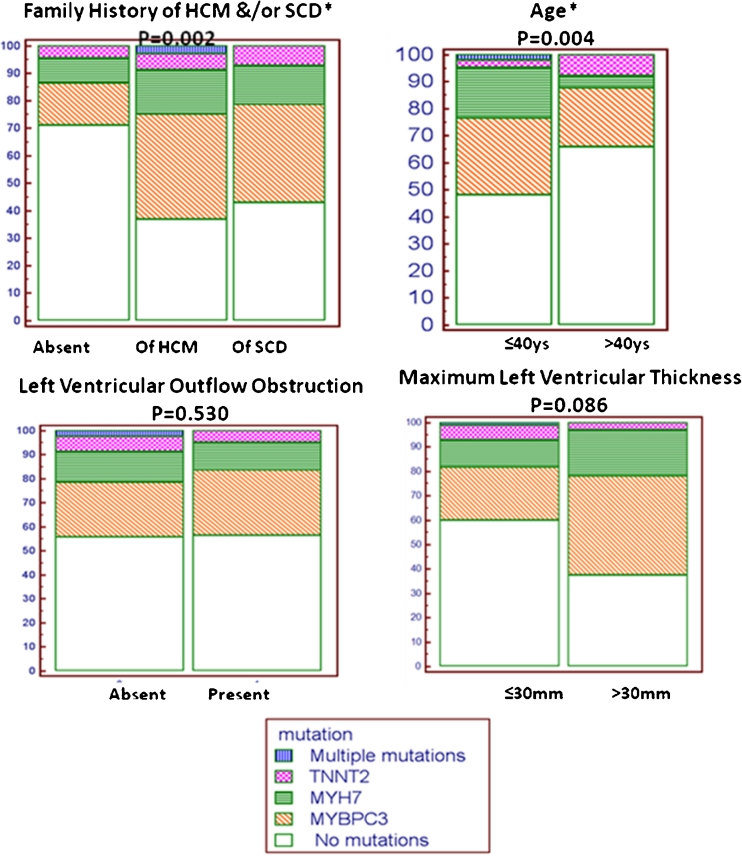
The clinical parameters comprised of presence of familial history of HCM and/or occurrence of sudden cardiac death (SCD) in the family, age of presentation at a cutoff ≤40 years, and presence/absence of LV outflow obstruction and maximum LV wall thickness being categorized into two groups (≤30 mm and >30 mm) were assessed in relevance to detection of positive mutation. It was observed that the positive genetic yield in any of the three sarcomeric genes was significantly associated with presence of family history of HCM and/or occurrence of SCD (p = 0.002) and with young age of presentation at ≤40 years (p =0.004) (Fig. 4). There was an observed trend of presence of mutation in association with increased LV maximum thickness (>30 mm), but this did not reach significance (p = 0.08). There was no statistically significant difference in the LV thickness among the genotype-positive cases of the three sarcomeric genes (using analysis of variance (ANOVA) test, p = 0.36, Table 6).
Fig. 4.
Univariate analysis of clinical parameters as predictors of positive sarcomeric genetic yield in Egyptian HCM patients using chi-square test (MedCalc-version 12.2.1) *Statistically significant association at p value <0.05
Table 6.
Comparison of LV maximum thickness among the genotype positive cases of the three sarcomeric genes
| MYBPC3 | MYH7 | TNNT2 | Double_hits | |
|---|---|---|---|---|
| Mean (SD) | 2.59 (0.61) | 2.57 (0.70) | 2.17 (0.42) | 2.39 (0.88) |
| ANOVA | 1.08 | |||
| P value | 0.36 | |||
There was also no statistically significant difference observed between the genopositive and genonegative patients with respect to initial clinical presentation with regards to severe degree of dyspnea (NYHA class III/IV), chest pain, and syncope (24 vs. 29 p = 0.39, 51 vs. 75 p = 0.95, and 19 vs. 22 p = 0.37, respectively). The overall prevalence of atrial fibrillation was not significantly different between both groups (p = 0.80). It was observed that prior cardiac arrests in this cohort (Table 1) occurred only in genopositive patients. As for incidence of other events as cardiac death and stroke, they were similar between the two groups. It is difficult at present to assess a correlation between adverse major cardiac events and genetic status due to the relatively short follow-up duration (median 38.8 months, range 30–49 months). Assessment of genotype–phenotype correlation is still under investigation.
Discussion
This study represents the first report of comprehensive screening of MYBPC3, MYH7, and TNNT2 in 192 Egyptian HCM patients, of whom 78 were familial (40.6 %). Initial screening of commonly involved sarcomeric genes is a justifiable first line approach of HCM genotype analysis [12, 14]. The frequency of positive sarcomeric genotyping detected among the Egyptian HCM index patients (40 %) is comparable to the frequency reported by Van Driest et al. in an earlier study [6], in which eight sarcomeric genes were analyzed in 389 HCM patients (38 % were genopositive). Also, recently, Curila et al. reported a similar frequency (40 %) analyzing four sarcomeric genes (MYBPC3, MYH7, TNNT2, and TNNI3) in 100 HCM patient [37]. However, the proportion of genetic yield varied among different population cohorts and ranged from >50 % in France and Italy [12, 14] to <30 % in Sweden [15]. This variation in frequency of positive genetic yield may probably relate to the proportion of familial cases included in the different cohorts. The positive sarcomeric genetic yield detected in the present cohort was significantly associated with presence of family history of HCM and/or occurrence of sudden cardiac death (p = 0.002) and with young age of presentation (p = 0.004).
MYBPC3 was the most commonly involved sarcomeric gene (24 %) and was twice as common as MYH7 (12 %), and TNNT2 was the least commonly involved in the present cohort. MYBPC3 and MYH7 accounted for 87 % of all genotype-positive patients. A similar genotype pattern was also observed in several European HCM cohorts [12, 14, 25, 37].
HCM is considered a highly heterogeneous disease not just at the phenotype but also showing marked genetic heterogeneity. This is reflected as a high frequency of novel mutations reported in different population studies [6, 12, 14] limiting the possibility of screening for specific genes or particular mutations in research and clinical practice. However, different ethnic cohorts including the present study revealed repetitive particular genotype association that may relate to a founder effect [14, 38–40], highlighting the importance of genetic screening in different populations.
A high proportion of the mutations detected in the present cohort were novel (65 %) and assessment of their potential pathogenicity was undertaken based on the practice guidelines for interpretation of unclassified variants in clinical molecular genetics, and the overall conclusion was based on more than a single line of evidence [41]. Accordingly, novel variants in the present study were classified as: uncertain, likely pathogenic, and pathogenic. This molecular stratification of variants was always considered in genetic counseling. An example is of a family pedigree with a novel variant (MYH7: Asp309Asn) in index patient (PA8). This novel variant, according to in silico bioinformatic analysis, was predicted as probably damaging by PolyPhen-2 (score 0.963) and was considered as unlikely to be damaging by other in silico tools (Table 4). It has not been detected in 200 chromosomes of matched healthy controls and was tested for cosegregation within the family. The mutation was detected in two sibs who were phenonegative by standard echocardiographic screening. Thus, confirmation of pathogenicity is not possible at the current stage; however, follow-up of the phenonegative sibs is warranted since the mutation pathogenicity is uncertain and reduced penetrance is a possibility.
Single nucleotide substitution was the most commonly encountered mutation type (51/62, 82 %). These substitutions were analyzed in context of the reference GenBank sequence of the three sarcomeric genes to determine whether deamination of five methyl cytosine and transition to thymine represented a common cause of mutagenesis in sense or antisense DNA strands of sarcomeric genes in our HCM cohort [42–45]. It was found that C > T and G > A transitions constituted 78 % (21/27) of the substitutions occurring within the CpG dinucleotides in the present cohort (as shown in Table 5). This is in agreement with the frequency of C > T and G > A transition reported by Meures and Mealey [46] in substitution mutations found in HCM in the literature (Table 7).
Table 7.
Frequency of different substitution modes within CpG in the present cohort in comparison to previously reported mutations in the 3 analyzed sarcomeric genes
| Mode of substitution in CpG | Present study | Previously reported CpG substitutionsa |
|---|---|---|
| G > A | 15/27 = 55.6 % | 113/251 = 45 % |
| C > T | 6/27 = 22.2 % | 56/251 = 22.3 % |
| C > A | 3/27 = 11.1 % | 13/251 = 5.2 % |
| C > G | 0/27 = 0 % | 18/251 = 7.2 % |
| G > C | 2/27 = 7.4 % | 30/251 = 12.0 % |
| G > T | 1/27 = 3.7 % | 21/251 = 8.4 % |
a[46]
This observation probably explains the theory behind some mutation recurrence in several HCM cohorts of different ethnicity: such as the functionally characterized [47] Arg92codon mutation of TNNT2 reported in several studies [34, 35, 48], the E258K mutation of MYBPC3 reported with a founder effect among the Italian population [14], and the Ala797Thr reported with a possible founder effect in South Africa [49]. All such missense mutations involve CpG sites and are, therefore, likely to be hypermutable spots and hence, were also detected in other studies including the present. The hypothesis of considering codon Arg92 as a putative hot spot with high mutation rate rather than recurrence among different populations due to a common founder effect was earlier proposed by Forissier et al. [48]. It is worth mentioning that other reported founder mutation resulting from deletion or insertion such as the Dutch 2373insG (Q791fs) in MYBPC3 [50] was not detected in the current study. The high mutability at the CpG sites within the sarcomeric genes probably explains also the high frequency of novel mutations detected in different studies [12, 14, 25, 37] including that observed in the present study (65 %).
Conclusion and Future Directions
This study is the first to provide genetic data of HCM among Egyptians. Positive genotype was detected in 40 % of our cohort. Expanding the genetic screening analysis through use of next-generation sequencing technology to enable simultaneous analysis of a broad panel of inherited cardiovascular disease-related genes is expected to increase the genetic-positive yield. Molecular characterization of HCM in different ethnic populations with different genetic backgrounds will aid in global comparative studies helping to understand the pathophysiology and heterogeneity of HCM. The true value of these studies lies in their translation to the clinical setting and their utilization towards improved HCM diagnosis, prognosis, and treatment [51, 52]. It is anticipated that the molecular screening of HCM patients and their family members is likely to improve the management of HCM in Egypt.
Acknowledgments
This study is sponsored by Magdi Yacoub Heart Foundation (MYF) and is under the auspice of Bibliotheca Alexandrina (BA). We acknowledge the Sawiris Foundation for support given during the initial stages of the program. We are thankful to patients and their families. We also acknowledge the support of the BA HCM Consortium that involves nationwide referring cardiology consultants and BA and MYF administrative staff. We thank Dr. Francesca Girolami, (Genetics Department Careggi Hospital, Florence) for consultation on mutation interpretation and Dr. Sherif Algendy (Alexandria Faculty of Medicine) for recruiting healthy ECG screened control subjects.
Abbreviations
- BA
Bibliotheca Alexandrina
- ECG
Electrocardiogram
- EVS
Exome Variant Server
- HCM
Hypertrophic cardiomyopathy
- MYF
Magdi Yacoub Foundation
- SCD
Sudden Cardiac Death
Appendix 1
Variants detected in Egyptian controls in comparison with European Americans and African Americans from Exome Variant Server database
| Gene | Ex./In. | Mutation name (c.DNA) | Amino acid change | MAF/minor allele count in EVS | dbSNP | ||
|---|---|---|---|---|---|---|---|
| Egyptian | European American | African American | |||||
| MYBPC3 | Ex.4 | c.472G > A | V158M | T = 0.06/12 | T = 0.0864/718 | T = 0.0171/69 | rs3729986 |
| MYBPC3 | In.5 | c.506-12delC | NA | Del. = 0.23/46 | – | – | rs11570050 |
| MYBPC3 | In.11 | c.927-46G > C | NA | G = 0.005/1 | G = 0.0001/1 | G = 0.016/62 | rs73451796 |
| MYBPC3 | In.13 | c.1223 + 29G > A | NA | T = 0.01/2 | T = 0.1174/958 | T = 0.0366/ 141 | rs11570078 |
| MYBPC3 | In.14 | c.1227-19C > A | NA | T = 0.02/4 | – | – | – |
| MYBPC3 | In.14 | c.1227-32T > A | NA | T = 0.005/1 | – | – | – |
| MYBPC3 | In.16 | c.1458-26C > G | NA | C = 0.005/1 | C = 0.0/0 | C = 0.0112/47 | rs11570081 |
| MYBPC3 | In.22 | c.2149-5C > T | NA | A = 0.005/1 | – | – | rs36211722 |
| MYBPC3 | In.22 | 2148 + 39T > C | NA | G = 0.005/1 | – | – | |
| MYBPC3 | In.23 | c.2308 + 18C > T | NA | A = 0.015/3 | – | – | – |
| MYBPC3 | In.26 | c.2737 + 12C > T | NA | A = 0.025/5 | A = 0.0257/206 | A = 0.0513/192 | rs3729936 |
| MYH7 | In.11 | c.999 + 44T > C | NA | G = 0.005/1 | G = 0.0886/762 | G = 0.4017/1770 | rs3729810 |
| MYH7 | In.16 | c.1888 + 23G > A | NA | T = 0.02/4 | – | – | – |
| MYH7 | In.20 | c.2287-43C > T | NA | A = 0.005/1 | A = 0.0001/1 | A = 0.0/0 | – |
| MYH7 | In.23 | c.2923-18G > A | NA | T = 0.02/4 | T = 0.0009/8 | T = 0.1512/666 | rs7157087 |
| MYH7 | In.27 | c.3337-3dup | NA | Dup. = 0.08/16 | – | – | rs45504498 |
| MYH7 | In.28 | c.3853 + 27T > A | NA | T = 0.01/2 | T = 0.3628/3120 | T = 0.6239/4406 | rs2277475 |
| MYH7 | In.32 | c.4520-3C > T | NA | A = 0.005/1 | – | – | – |
| MYH7 | Ex.32 | c.4472C > G | S1491C | C = 0.005/1 | C = 0.0115/99 | C = 0.003/13 | rs3729823 |
| TNNT2 | In.2 | c.41 + 75C > G | NA | C = 0.005/1 | C = 0.0/0 | C = 0.0275/38 | rs10920185 |
| TNNT2 | In.2 | c.41 + 86C > A | NA | T = 0.005/1 | – | – | – |
| TNNT2 | In.2 | c.42-20G > A | NA | T = 0.025/5 | T = 0.0001/1 | T = 0.035/154 | rs45561443 |
| TNNT2 | In.3 | c.53-7_11del | NA | Del. = 0.41/82 | – | – | rs45533739 |
| TNNT2 | In.6 | c.133 + 12G > A | NA | T = 0.025/5 | T = 0.0003/3 | T = 0.0502/221 | rs45580032 |
| TNNT2 | EX.6 | c.83C > T | A28V | A = 0.005/1 | – | – | – |
| TNNT2 | In.8 | c.203 + 7G > A | NA | T = 0.005/1 | – | – | – |
| TNNT2 | In.8 | c.203 + 67G > A | NA | T = 0.01/200 | T = 0.9991/3179 | T = 0.9234/1384 | rs1573230 |
| TNNT2 | In.9 | c.264 + 7G > A | NA | T = 0.005/1 | T = 0.0003/3 | T = 0.0311/137 | rs45490292 |
| TNNT2 | Ex.15 | c.740A > G | K247R | C = 0.03/6 | – | – | – |
| TNNT2 | In.15 | c.781-61A > G | NA | C = 0.02/4 | – | – | |
| TNNT2 | In.15 | c.781-33C > T | NA | A = 0.025/5 | A = 0.102/877 | A = 0.4639/2044 | rs2275863 |
| TNNT2 | In.16 | c.821 + 22C > T | NA | A = 0.005/1 | A = 0.0001/1 | A = 0.0402/177 | rs45495692 |
| TNNT2 | In.16 | c.821 + 43C > T | NA | A = 0.01/2 | A = 0.0003/3 | A = 0.032/141 | rs45570832 |
| TNNT2 | In.16 | c.821 + 46C > T | NA | A = 0.015/3 | A = 0.0283/243 | A = 0.0057/25 | rs45576635 |
Alleles frequency of European and African Americans were obtained from the Exome Variant Server (EVS) [36]
MYBPC3 NM_000256.3, MYH7 NM_000257.2, TNNT2 NM_001001430.1, Ex exon, In intron
Footnotes
BA HCM National Program is under the auspice of Bibliotheca Alexandrina and Magdi Yacoub Foundation.
References
- 1.Cecchi F, Yacoub MH, Olivotto I. Hypertrophic cardiomyopathy in the community: why we should care. Nature Clinical Practice. Cardiovascular Medicine. 2005;2(7):324–325. doi: 10.1038/ncpcardio0248. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–789. doi: 10.1161/01.CIR.92.4.785. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ. Hypertrophic cardiomyopathy: an important global disease. American Journal of Medicine. 2004;116(1):63–65. doi: 10.1016/j.amjmed.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Maron BJ, Ho CY. Hypertrophic cardiomyopathy without hypertrophy: an emerging pre-clinical subgroup composed of genetically affected family members. JACC. Cardiovascular Imaging. 2009;2(1):65–68. doi: 10.1016/j.jcmg.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 5.http://www.indexmundi.com/egypt/population.html (accessed July17, 2012).
- 6.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clinic Proceedings. 2005;80(6):739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 7.Konno T, Chang S, Seidman JG, Seidman CE. Genetics of hypertrophic cardiomyopathy. Current Opinion in Cardiology. 2010;25(3):205–209. doi: 10.1097/HCO.0b013e3283375698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathy. NEJM. 2011;364:1643–1656. doi: 10.1056/NEJMra0902923. [DOI] [PubMed] [Google Scholar]
- 9.Funada A, Masuta E, Fujino N, Hayashi K, Ino H, Kita Y, Ikeda H, Fujii T, Nakanuma Y, Yamagishi M. Heterogeneity of clinical manifestation of hypertrophic cardiomyopathy caused by deletion of lysine 183 in cardiac troponin I gene. International Heart Journal. 2010;51(3):214–217. doi: 10.1536/ihj.51.214. [DOI] [PubMed] [Google Scholar]
- 10.Daw EW, Chen SN, Czernuszewicz G, Lombardi R, Lu Y, Ma J, Roberts R, Shete S, Marian AJ. Genome-wide mapping of modifier chromosomal loci for human hypertrophic cardiomyopathy. Human Molecular Genetics. 2007;16:2463–2471. doi: 10.1093/hmg/ddm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalozoumi, G., Tzimas, C., Sanoudou, D. (2012). The expanding role of epigenetics. Global Cardiology Science and Practice, 7: http://dx.doi.org/10.5339/gcsp.2012.7 [DOI] [PMC free article] [PubMed]
- 12.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 13.Van Driest SL, Jaeger MA, Ommen SR, Will ML, Gersh BJ, Tajik AJ, et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. Journal of the American College of Cardiology. 2004;44(3):602–610. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 14.Girolami F, Olivotto I, Passerini I, Zachara E, Nistri S, Re F, et al. A molecular screening strategy based on beta-myosin heavy chain, cardiac myosin binding protein C and troponin T genes in Italian patients with hypertrophic cardiomyopathy. Journal of Cardiovascular Medicine. 2006;7(8):601–607. doi: 10.2459/01.JCM.0000237908.26377.d6. [DOI] [PubMed] [Google Scholar]
- 15.Morner S, Richard P, Kazzam E, Hellman U, Hainque B, Schwartz K, et al. Identification of the genotypes causing hypertrophic cardiomyopathy in northern Sweden. Journal of Molecular and Cellular Cardiology. 2003;35(7):841–849. doi: 10.1016/S0022-2828(03)00146-9. [DOI] [PubMed] [Google Scholar]
- 16.Marian AJ, Salek L, Lutucuta S. Molecular genetics and pathogenesis of hypertrophic cardiomyopathy. Minerva Medica. 2001;92(6):435–451. [PMC free article] [PubMed] [Google Scholar]
- 17.Fokstuen S, Lyle R, Munoz A, Gehrig C, Lerch R, Perrot A, et al. A DNA resequencing array for pathogenic mutation detection in hypertrophic cardiomyopathy. Human Mutation. 2008;29(6):879–885. doi: 10.1002/humu.20749. [DOI] [PubMed] [Google Scholar]
- 18.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. The New England Journal of Medicine. 1998;338(18):1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 19.Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clinic Proceedings. 2008;83(6):630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 20.Erdmann J, Daehmlow S, Wischke S, Senyuva M, Werner U, Raible J, et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clinical Genetics. 2003;64(4):339–349. doi: 10.1034/j.1399-0004.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 21.Frisso G, Limongelli G, Pacileo G, Del Giudice A, Forgione L, Calabro P, et al. A child cohort study from southern Italy enlarges the genetic spectrum of hypertrophic cardiomyopathy. Clinical Genetics. 2009;76(1):91–101. doi: 10.1111/j.1399-0004.2009.01190.x. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Castro M, Reguero JR, Alvarez V, Batalla A, Soto MI, Albaladejo V, et al. Hypertrophic cardiomyopathy linked to homozygosity for a new mutation in the myosin-binding protein C gene (A627V) suggests a dosage effect. International Journal of Cardiology. 2005;102(3):501–507. doi: 10.1016/j.ijcard.2004.05.060. [DOI] [PubMed] [Google Scholar]
- 23.Alders M, Jongbloed R, Deelen W, van den Wijngaard A, Doevendans P, Ten Cate F, et al. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. European Heart Journal. 2003;24(20):1848–1853. doi: 10.1016/S0195-668X(03)00466-4. [DOI] [PubMed] [Google Scholar]
- 24.Millat G, Bouvagnet P, Chevalier P, Dauphin C, Jouk PS, Da Costa A, et al. Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. European Journal of Medical Genetics. 2010;53(5):261–267. doi: 10.1016/j.ejmg.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Garcia MI, Monserrat L, Ortiz M, Fernandez X, Cazon L, Nunez L, et al. Screening mutations in myosin binding protein C3 gene in a cohort of patients with hypertrophic cardiomyopathy. BMC Medical Genetics. 2010;11:67. doi: 10.1186/1471-2350-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenzweig A, Watkins H, Hwang DS, Miri M, McKenna W, Traill TA, et al. Preclinical diagnosis of familial hypertrophic cardiomyopathy by genetic analysis of blood lymphocytes. The New England Journal of Medicine. 1991;325(25):1753–1760. doi: 10.1056/NEJM199112193252501. [DOI] [PubMed] [Google Scholar]
- 27.Watkins H, Rosenzweig A, Hwang DS, Levi T, McKenna W, Seidman CE, et al. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. The New England Journal of Medicine. 1992;326(17):1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 28.Song L, Zou Y, Wang J, Wang Z, Zhen Y, Lou K, et al. Mutations profile in Chinese patients with hypertrophic cardiomyopathy. Clinica Chimica Acta. 2005;351(1–2):209–216. doi: 10.1016/j.cccn.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Consevage MW, Salada GC, Baylen BG, Ladda RL, Rogan PK. A new missense mutation, Arg719Gln, in the beta-cardiac heavy chain myosin gene of patients with familial hypertrophic cardiomyopathy. Human Molecular Genetics. 1994;3(6):1025–1026. doi: 10.1093/hmg/3.6.1025. [DOI] [PubMed] [Google Scholar]
- 30.Kato M, Takazawa K, Kimura A, Ruegg JC, Amano K, Wang Y, et al. Altered actin binding with myosin mutation in hypertrophic cardiomyopathy and sudden death. Lancet. 1995;345(8959):1247. doi: 10.1016/S0140-6736(95)92034-X. [DOI] [PubMed] [Google Scholar]
- 31.Moolman-Smook JC, Brink PA, Corfield VA. Identification of a novel Ala797Thr mutation in exon 21 of the beta-myosin heavy chain gene in hypertrophic cardiomyopathy. Human Mutation. 1995;6(2):197–198. doi: 10.1002/humu.1380060219. [DOI] [PubMed] [Google Scholar]
- 32.Yu B, Sawyer NA, Caramins M, Yuan ZG, Saunderson RB, Pamphlett R, et al. Denaturing high performance liquid chromatography: high throughput mutation screening in familial hypertrophic cardiomyopathy and SNP genotyping in motor neurone disease. Journal of Clinical Pathology. 2005;58(5):479–485. doi: 10.1136/jcp.2004.021642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roncarati R, Latronico MV, Musumeci B, Aurino S, Torella A, Bang ML, et al. Unexpectedly low mutation rates in beta-myosin heavy chain and cardiac myosin binding protein genes in Italian patients with hypertrophic cardiomyopathy. Journal of Cellular Physiology. 2011;226(11):2894–2900. doi: 10.1002/jcp.22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moolman JC, Corfield VA, Posen B, Ngumbela K, Seidman C, Brink PA, Watkins H. Sudden death due to troponin T mutations. Journal of the American College of Cardiology. 1997;29(3):549–555. doi: 10.1016/S0735-1097(96)00530-X. [DOI] [PubMed] [Google Scholar]
- 35.Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O'Donoghue A, Spirito P, Matsumori A, Moravec CS, Seidman JG, et al. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. The New England Journal of Medicine. 1995;332(16):1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 36.Exome variant Server v.0.0.13, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/).
- 37.Curila K, Benesova L, Penicka M, Minarik M, Zemanek D, Veselka J, Widimsky P, Gregor P. Spectrum and clinical manifestations of mutations in genes responsible for hypertrophic cardiomyopathy. Acta Cardiologica. 2012;67(1):23–29. doi: 10.1080/ac.67.1.2146562. [DOI] [PubMed] [Google Scholar]
- 38.Moolman-Smook JC, De Lange WJ, Bruwer ECD, Brink PA, Corfield VA. The origins of hypertrophic cardiomyopathy-causing mutations in two South African subpopulations: a unique profile of both independent and founder events. American Journal of Human Genetics. 1999;65:1308–1320. doi: 10.1086/302623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Zheng D, Dong N, Yang X, Song J, Jiang T, Cheng X, Li H, Zhou B, Zhao C, Jiang W. Mutation of Arg723Gly in β-myosin heavy chain gene in five Chinese families with hypertrophic cardiomyopathy. Chinese Medical Journal. 2006;119:1785–1789. [PubMed] [Google Scholar]
- 40.Christiaans I, Nannenberg EA, Dooijes D, Jongbloed RJE, Michels M, Postema PG, Majoor-Krakauer D, Van den Wijngaard A, Mannens MMAM, Van Tintelen JP, Van Langen IM, Wilde AAM. Founder mutations in hypertrophic cardiomyopathy in the Netherlands. Netherlands Heart Journal. 2010;18:248–254. doi: 10.1007/BF03091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell, J., Bodmer, D., Sistermans, E., Ramsden, S. C. (2007). Practice guidelines for the interpretation and reporting of unclassified variants (UVs) in clinical molecular genetics. CMGS/VKGL.
- 42.Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. American Journal of Human Genetics. 1998;63(2):474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper, D. N., Krawczak, M. (1999). Single base pair substitutions. In Human gene mutations. Oxford: BIOS Scientific. pp 109–162.
- 44.Walsh CP, Xu GL. Cytosine methylation and DNA repair. Current Topics in Microbiology and Immunology. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- 45.Antonarakis SE, Krawczak M, Cooper DN. The nature and mechanisms of human gene mutation. In: Scriver CR, Sly WS, editors. The metabolic and molecular basis of inherited disease. New York: McGraw-Hill; 2001. pp. 343–377. [Google Scholar]
- 46.Meurs KM, Mealey KL. Evaluation of the flanking nucleotide sequences of sarcomeric hypertrophic cardiomyopathy substitution mutations. Mutation Research. 2008;642(1–2):86–89. doi: 10.1016/j.mrfmmm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Ertz-Berger BR, He H, Dowell C, Factor SM, Haim TE, Nunez S, Schwartz SD, Ingwall JS, Tardiff JC. Changes in the chemical and dynamic properties of cardiac troponin t cause discrete cardiomyopathies in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18219–18224. doi: 10.1073/pnas.0509181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forissier JF, Carrier L, Farza H, Bonne G, Bercovici J, Richard P, Hainque B, Townsend PJ, Yacoub MH, Faure S, et al. Codon 102 of the cardiac troponin T gene is a putative hot spot for mutations in familial hypertrophic cardiomyopathy. Circulation. 1996;94:3069–3073. doi: 10.1161/01.CIR.94.12.3069. [DOI] [PubMed] [Google Scholar]
- 49.Moolman-Smook JC, De Lange WJ, Bruwer ECD, Brink PA, Corfield VA. The origins of hypertrophic cardiomyopathy-causing mutations in two South African subpopulations: a unique profile of both independent and founder events. American Journal of Human Genetics. 1999;65:1308–1320. doi: 10.1086/302623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alders M, Jongbloed R, Deelen W, van den Wijngaard A, Doevendans P, Ten Cate F, Regitz-Zagrosek V, Vosberg HP, van Langen I, Wilde A, Dooijes D, Mannens M. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. European Heart Journal. 2003;24(20):1848–1853. doi: 10.1016/S0195-668X(03)00466-4. [DOI] [PubMed] [Google Scholar]
- 51.Kassem, H. S., Girolami, F., Sanoudou, D. (2012). Molecular genetics made simple. Global Cardiology Science & Practice, 2, http://dx.doi.org/10.5339/gcsp.2012.6 [DOI] [PMC free article] [PubMed]
- 52.Olivotto, I., Kassem H Sh, Girolami F. (2011). Genetic testing for hyptertrophic cardiomyopathy. From exploration to exploitation. Cardiogenetics [eISSN 2035–8148], Vol 1, e3.