Abstract
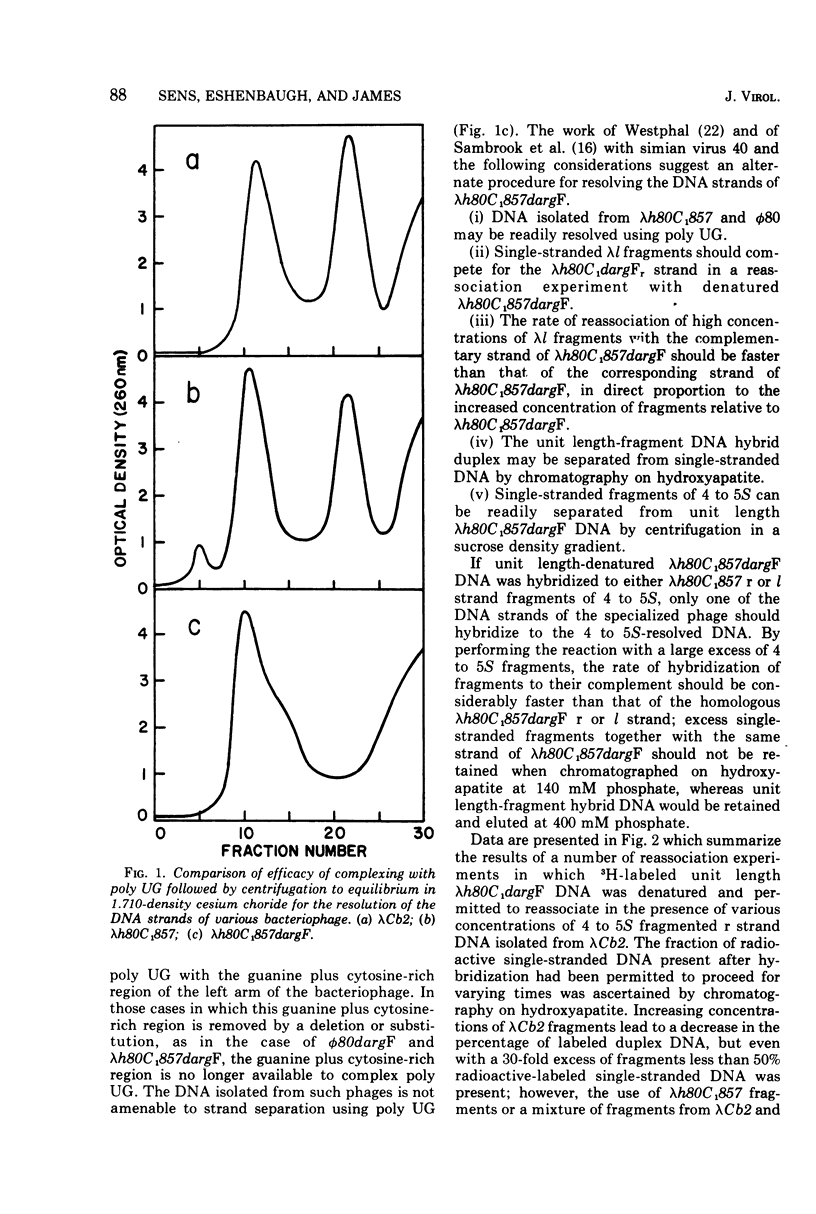
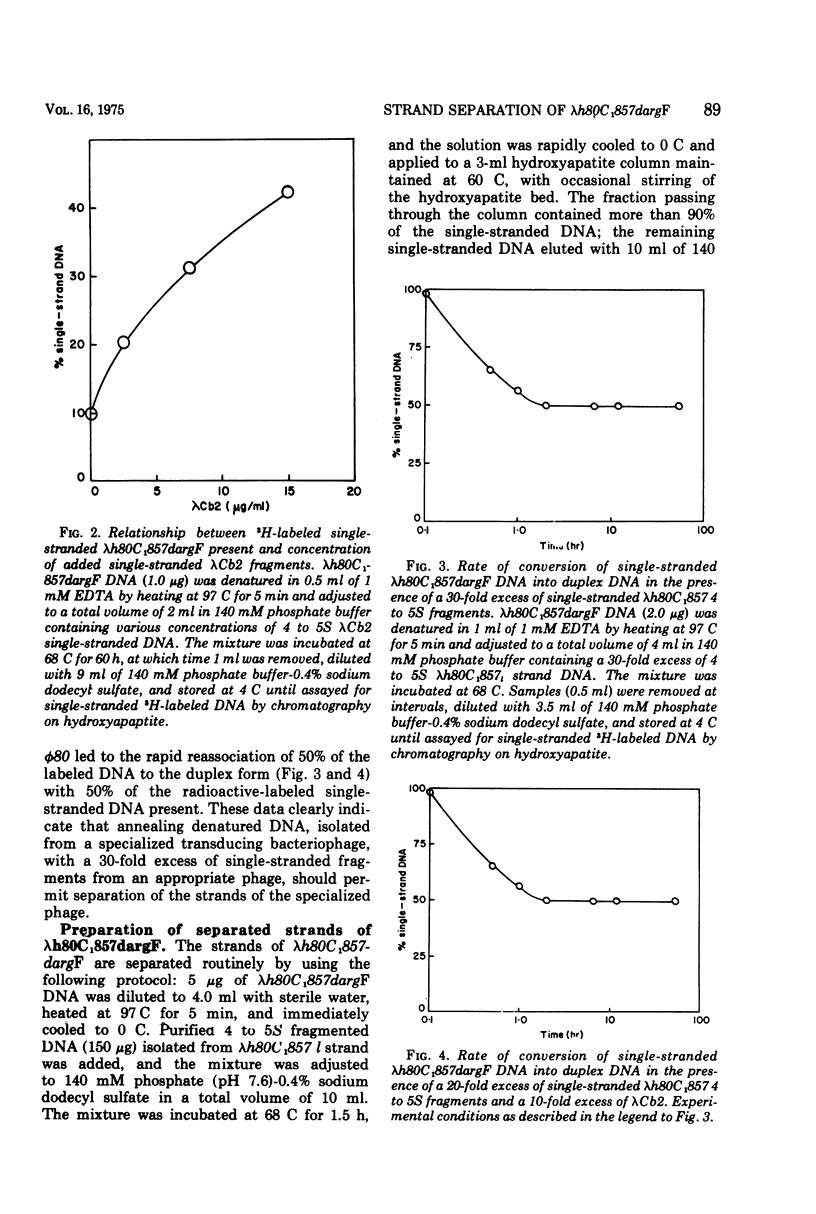
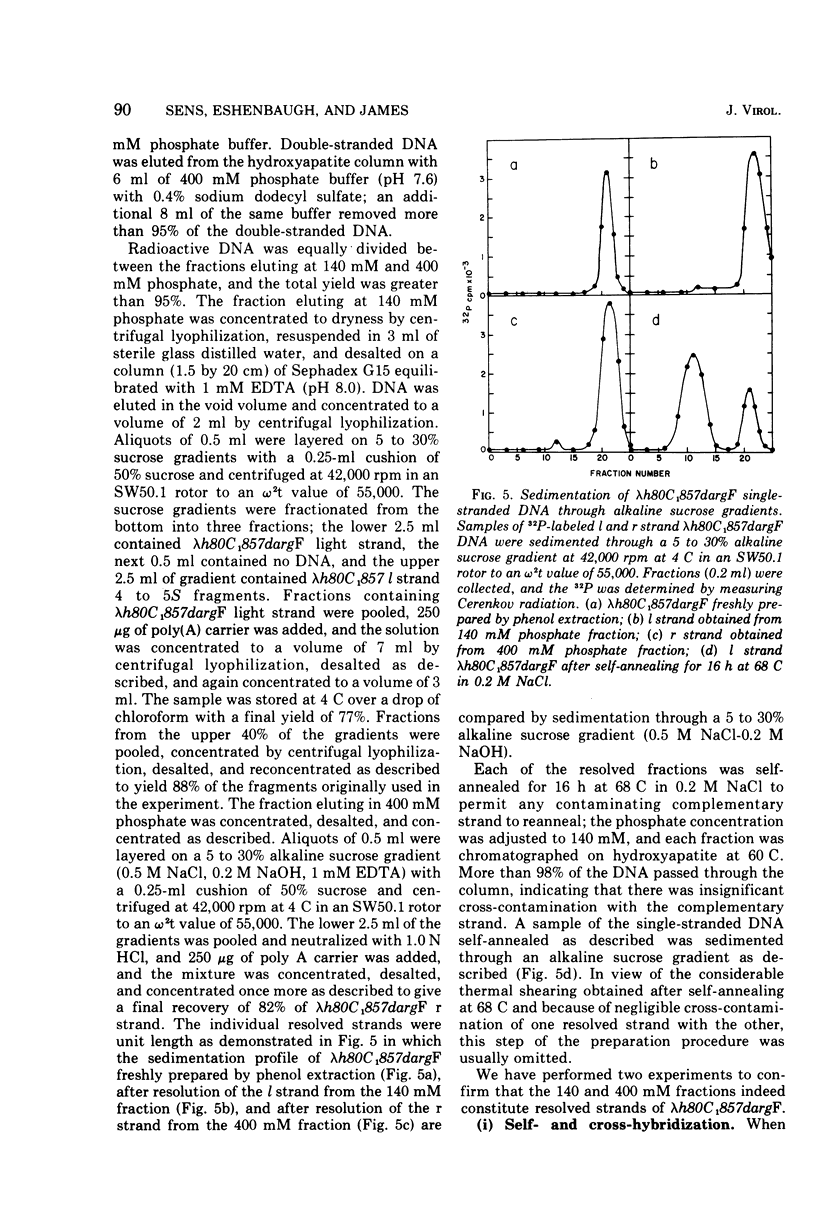
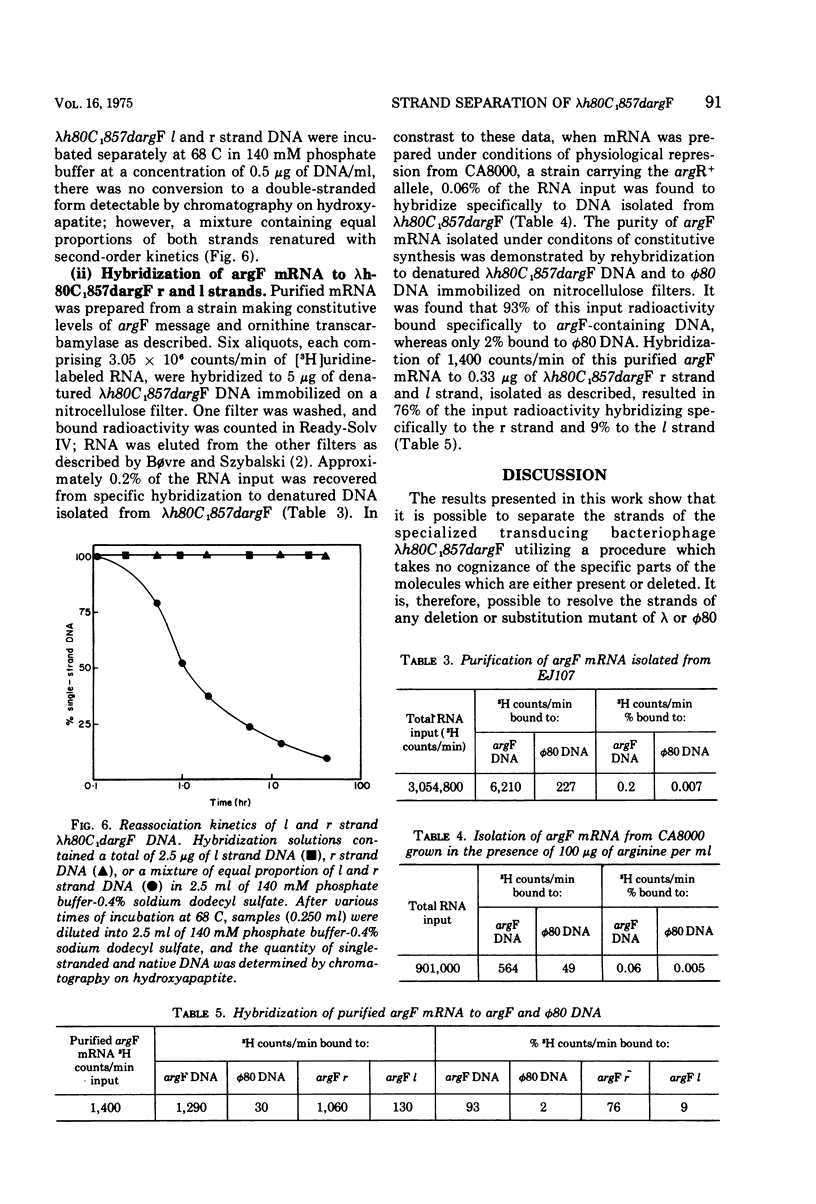
The DNA strands of lambdoid phages with deletions or substitutions of the guanine plus cytosine-rich region in the left arm are not resolvable by complexing with poly UG followed by centrifugation in CsCl. This work describes a completely general procedure for the strand resolution of these phages by hybridization with fragments of separated strands of the parent phage. In particular, resolution of the DNA strands of the specialized transducing phage lambda-h80C1-857dargF is described, and evidence is presented which indicates that argF is transcribed from the r strand.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bovre K., Szybalski W. Patterns of convergent and overlapping transcription within the b2 region of coliphage lambda. Virology. 1969 Aug;38(4):614–626. doi: 10.1016/0042-6822(69)90181-0. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. GENETIC RECOMBINATION BETWEEN LAMBDA PROPHAGE AND IRRADIATED LAMBDA DG PHAGE. Virology. 1964 Jun;23:234–251. doi: 10.1016/0042-6822(64)90287-9. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Segregants from lysogenic heterogenotes carrying recombinant lambda prophages. Virology. 1963 Jun;20:344–356. doi: 10.1016/0042-6822(63)90124-7. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Hurwitz J. Genetic transcription in bacteriophage lambda: studies of lambda m RNA synthesis in vivo. J Mol Biol. 1968 Nov 14;37(3):387–406. doi: 10.1016/0022-2836(68)90110-1. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elseviers D., Cunin R., Glansdorff N. Control regions within the argECBH gene cluster of Escherichia coli K12. Mol Gen Genet. 1972;117(4):349–366. doi: 10.1007/BF00333028. [DOI] [PubMed] [Google Scholar]
- Eshenbaugh D. L., Sens D., James E. A solid phase radioimmune assay for ornithine transcarbamylase. Adv Exp Med Biol. 1974;42(0):61–73. doi: 10.1007/978-1-4684-6982-0_5. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Beckwith J. R. Directed transposition of the arabinose operon: a technique for the isolation of specialized transducing bacteriophages for any Escherichia coli gene. J Mol Biol. 1969 Aug 28;44(1):117–127. doi: 10.1016/0022-2836(69)90408-2. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A. Control of the argECBH cluster in Escherichia coli. Mol Gen Genet. 1972;117(4):337–348. doi: 10.1007/BF00333027. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Ippen K., Scaife J. G., Beckwith J. R. The promoter-operator region of the lac operon of Escherichia coli. J Mol Biol. 1968 Dec;38(3):413–420. doi: 10.1016/0022-2836(68)90395-1. [DOI] [PubMed] [Google Scholar]
- Pouwels P. H., Cunin R., Glansdorff N. Letter: Divergent transcription in the argECBH cluster of genes in Escherichia coli K12. J Mol Biol. 1974 Mar;83(3):421–424. doi: 10.1016/0022-2836(74)90288-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Sharp P. A., Keller W. Transcription of Simian virus 40. I. Separation of the strands of SV40 DNA and hybridization of the separated strands to RNA extracted from lytically infected and transformed cells. J Mol Biol. 1972 Sep 14;70(1):57–71. doi: 10.1016/0022-2836(72)90163-5. [DOI] [PubMed] [Google Scholar]
- Summers W. C., Szybalski W. Size, number, and distribution of poly G binding sites on the separated DNA strands of coliphage T7. Biochim Biophys Acta. 1968 Sep 24;166(2):371–378. doi: 10.1016/0005-2787(68)90224-4. [DOI] [PubMed] [Google Scholar]
- Taylor K., Hradecna Z., Szybalski W. Asymmetric distribution of the transcribing regions on the complementary strands of coliphage lambda DNA. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1618–1625. doi: 10.1073/pnas.57.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Westphal H. SV40 DNA strand selection by Escherichia coli RNA polymerase. J Mol Biol. 1970 Jun 14;50(2):407–420. doi: 10.1016/0022-2836(70)90201-9. [DOI] [PubMed] [Google Scholar]
- Wolf R. E., Jr, Fraenkel D. G. Isolation of specialized transducing bacteriophages for gluconate 6-phosphate dehydrogenase (gnd) of Escherichia coli. J Bacteriol. 1974 Feb;117(2):468–476. doi: 10.1128/jb.117.2.468-476.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]



